SUMMARY
Maturation of iron-sulfur proteins is achieved by the SUF machinery in a wide number of Eubacteria and Archaea as well as eukaryotic chloroplasts. This machinery is encoded in E. coli by the sufABCDSE operon, where three Suf components, SufB, SufC, and SufD, form a complex and appear to provide an intermediary site for the iron-sulfur cluster assembly. Here we report the quaternary structure of SufC2-SufD2 complex in which SufC is bound to the C-terminal domain of SufD. Comparison with the monomeric structure of SufC revealed conformational change of the active site residues: SufC becomes competent for ATP-binding and hydrolysis upon association with SufD. The two SufC subunits were spatially separated in the SufC2-SufD2 complex, whereas cross-linking experiments in solution have indicated that two SufC molecules associate with each other in the presence of Mg2+ and ATP. Such dimer formation of SufC may lead to a gross structural change of the SufC2-SufD2 complex. Furthermore, genetic analysis of SufD revealed an essential histidine residue buried inside the dimer interface, suggesting that conformational change may expose this crucial residue. These findings together with biochemical characterization of the SufB-SufC-SufD complex have led us to propose a model for the iron-sulfur cluster biosynthesis in the complex.
Keywords: ATP-binding cassette ATPase, crystallography, iron-sulfur protein maturation, scaffold, SUF machinery
INTRODUCTION
Iron-sulfur (Fe-S) proteins that contain an Fe-S cluster as prosthetic group are widely utilized in organisms for a variety of cellular processes including respiratory and photosynthetic electron transport and in the regulation of gene expression.1,2 The most common Fe-S clusters have the forms of [4Fe-4S], [3Fe-4S] and [2Fe-2S], which are ligated to the polypeptides via the thiolate side chains of cysteine, and occasionally, imidazole nitrogen of histidine or carboxyl oxygen of aspartic acid. Fe-S clusters are not spontaneously formed in the cells. Genetic and biochemical studies have so far revealed three distinct systems responsible for Fe-S cluster biosynthesis, termed NIF, ISC and SUF, which are encoded by nif (nifSU), isc (iscSUA-hscBA-fdx), and suf (sufABCDSE) operons, respectively, in bacteria.3–5 The NIF machinery was initially considered to be specialized for the maturation of nitrogenase but is also distributed in some anaerobic organisms lacking nitrogenase.6 From lower to higher eukaryotes, the components of the ISC machinery are found in mitochondria,7 whereas the SUF machinery is conserved in the plastids of algae and higher plants.8 The homologs of the Suf components are also found in diverse organisms including Archaea, suggesting that the SUF machinery is an ancient system responsible for Fe-S cluster biosynthesis.
The enterobacterium Escherichia coli and related species possess both the ISC and the SUF systems. In E. coli, ISC functions as a general pathway for the assembly of a variety of Fe-S proteins, whereas SUF plays a role in Fe-S cluster biosynthesis under adverse conditions such as oxidative stress and iron starvation.6,9–11 Both systems contain a cysteine desulfurase (the paralogous components, IscS and SufS) that eliminates a sulfur atom from the substrate cysteine and provides it for the construction of an Fe-S cluster.12 Both systems contain an A-type protein, IscA and SufA, that can bind iron or an Fe-S cluster, although their exact role is still elusive.13–15 The remaining proteins involved in the two systems share no apparent similarity. In particular, the E. coli SUF system does not possess a homologue of IscU, which functions as a scaffold protein in the ISC system on which a nascent Fe-S cluster is formed during the course of the Fe-S biosynthesis.3,5,16 While SufE is structurally related to IscU, SufE acts as a sulfur shuttle protein in the SUF system and does not function as a scaffold.17–19 The three additional components of the SUF system, SufB, SufC, and SufD have attracted much attention because they are essential for in vivo Fe-S biosynthesis.6,10,20 SufC is a soluble ATPase that exhibits striking structural similarity to the ATPase subunits of ABC-transporters (ABC-ATPase).21,22 SufB and SufD share limited sequence similarity with each other and they both interact with SufC to form a SufB-SufC-SufD ternary complex (SufBCD).9,17 It was also demonstrated that the basal ATPase activity of SufC is enhanced in the presence of SufB or SufD.23 Based on these studies, it is anticipated that ATP binding and hydrolysis by SufC can drive a power stroke to induce a conformational change within the SufBCD complex. However, the functional purpose of such a conformational change is unknown. The SufBCD complex also interacts with SufS-SufE to facilitate sulfur liberation from cysteine, in which the sulfur atom is transferred from SufS to SufE and then to SufB.19 In addition, in vitro reconstitution experiments have suggested that SufB can form an Fe-S cluster, although it is not clear if SufB can function as a scaffold protein.19 Despite progress in elucidating biochemical properties of the Suf components, our understanding of the molecular mechanism underlying the Fe-S cluster biosynthesis is fragmentary and is confined to a very few events.
We previously determined the crystal structure of a monomeric form of SufC. Curiously, the local conformation of SufC, in particular the ATP binding segments, is unique and distinct from that of most other ABC-ATPase family members. E171, an invariant catalytic residue in the Walker B motif of SufC, is rotated away from the ATP binding pocket and forms a salt bridge with K152 in a neighboring domain.21,22 This conformation of the active site within the monomeric SufC is unfavorable for ATPase activity and seems to represent an inactive, resting form of SufC that prevents wasteful ATP hydrolysis. The Structural GenomiX project team has also determined the crystal structure of the SufD homodimer, and demonstrated that SufD has a novel fold in which 20 β-strands are assembled into a right-handed parallel β-helix.24 Structural study of the SufB protein has been hampered by the insoluble, aggregated nature of this molecule. Thus, further structural information is needed to answer several important questions such how the SufB, SufC, and SufD proteins interact with each other and how the SufBCD complex undergoes ATPase-induced conformational changes during the course of Fe-S cluster biosynthesis.
In the present study, we have identified a novel complex consisting of SufC2-SufD2, and determined its quaternary structure by X-ray crystallography and electron microscopy. Furthermore, cross-linking experiments in solution have indicated that two SufC subunits, which are spatially separated in the SufC2-SufD2 crystals, can associate with each other in the presence of Mg2+ and ATP. We also identified, by mutational analysis, a functionally essential residue of SufD that is buried inside at the dimer interface. These findings, together with the sequence similarity between SufD and SufB, have led us to propose a model whereby the SufBCD complex works in Fe-S cluster biosynthesis at the expense of ATP.
RESULTS
Co-expression and co-purification of the SufCD complex
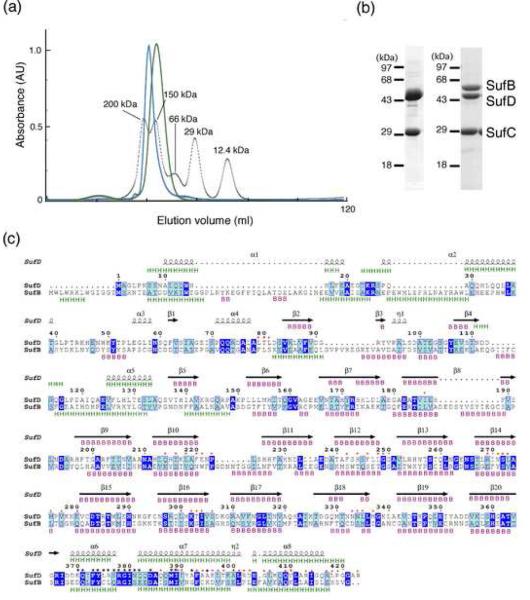
The SufBCD complex has previously been purified from E. coli cells overexpressing the entire sufABCDSE operon.17 We co-expressed pairwise combinations of the genes using a tandem expression plasmid to characterize protein-protein interactions between SufB, SufC and SufD. Using this approach we found that SufC-SufD formed a soluble binary complex. Co-expression of SufB with either SufC or SufD resulted in inclusion body formation, probably due to the insoluble, aggregated nature of SufB (data not shown). The SufCD complex was stable during the purification steps including Ni affinity chromatography and gel filtration. An approximate 1:1 stoichiometry of SufC and SufD was estimated for the complex on a Coomassie-stained SDS-PAGE gel. The molecular size of the whole complex was estimated to be 150 kDa by gel filtration chromatography (see Fig. 5a) and DLS analysis (not shown). Since the monomer sizes of SufC and SufD are 30 kDa and 46 kDa, respectively, the SufCD complex most likely forms a SufC2-SufD2 heterotetramer. The purified complex was then subjected to X-ray crystallographic and electron microscopic analyses.
Fig. 5. Characterization of the SufBCD complex.
(a) Comparison of molecular size between the SufC2-SufD2 complex (green) and the SufBCD complex (blue). The elution profiles from the gel-filtration column (Sephacryl S-200) are shown with those of the size marker proteins including cytochrome c, carbonic anhydrase, albumin, alcohol dehydrogenase and β-amylase (dotted line) monitored by absorbance at 280 nm. (b) The SDS-PAGE of the SufC2-SufD2 complex (left) and the SufBCD complex (right). The gel was stained with Coomassie Blue. The SufC band in the left panel was slightly larger than that in the right panel because of the His10-tag sequence. (c) Sequence comparison between E. coli SufD and SufB. Identical and similar residues are highlighted in blue and cyan, respectively. The secondary structures of SufD in the crystal structure are shown above the sequence with spirals (α-helices) and arrows (β-strands). The secondary structures of SufD and SufB were predicted by JPRED server 43, and shown above and below the sequence alignment, respectively, with `H' for α-helices and `B' for β-strands. The SufD residues involved in binding with SufC are shown by black asterisks. Residues at the interface between the β-helical core domain and the C-terminal helical domain of SufD are marked with red asterisks.
Overall structure of the SufCD complex
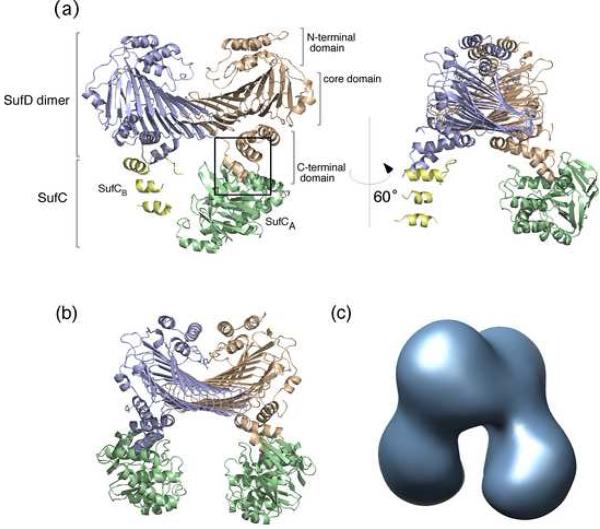
We have determined the crystal structure of the SufCD complex from E. coli at 2.2 Å resolution (Fig. 1a). An asymmetric unit contains one complex comprising two SufC molecules (termed SufCA and SufCB) and one SufD homodimer with a subunit stoichiometry of 2:2 that agrees well with the biochemical experiments described above. Though each SufC subunit is bound to each subunit of SufD homodimer, one SufC subunit (SufCB) was mostly disordered. Electron densities for the three SufC helices (α3, α6 and α7) were clearly visible in the region occupied by SufCB, whereas the remaining segments appeared as fragmentary densities. One possible explanation for why SufCB was less clearly seen than SufCA is that a large portion of SufCB is relatively mobile due to the crystal packing: SufCB is much more loosely packed in the crystal lattice compared to SufCA. Another possible reason for the less clear electron densities of SufCB was the partial dissociation of SufC from the complex during the course of crystallization or in the crystal lattice, which resulted in an occasional packing of the complex lacking one SufC subunit.
Fig. 1. Overall Structure of the SufC2-SufD2 Complex from E. coli.
(a) Ribbon representation of the crystal structure of the SufC2-SufD2 complex. The subunits are depicted by different colors. The view in the right panel is rotated by 60° about the vertical axis relative to the left panel. The interaction between SufC and SufD is indicated in the square box, and the close up view corresponding to this region is represented in Fig. 2 to provide details of the interaction. (b) The entire SufC2-SufD2 complex including the computer modeled SufCB subunit. Details of the model construction are shown in Fig. S1. (c) The 3D-reconstitution image of the SufC2-SufD2 complex obtained by electron microscopy.
Since the SufC2-SufD2 complex exhibited an apparent 2-fold symmetry, we modeled the invisible segments of the SufCB subunit by rotating the determined SufCA structure 180 degrees along the non-crystallographic 2-fold axis in the SufD homodimer (Fig. 1b). Indeed, the three visible helices of SufCB (α3, α6 and α7) superimposed well on the corresponding helices of SufCA with the r.m.s. deviation of 0.86 Å between the main-chain atoms when the 2-fold symmetry operation was applied to them (Fig. S1). This model structure of SufC2-SufD2 complex was further examined by comparing it with the 3D-reconstitution image of the complex derived from negative-stain electron microscopy (Fig. 1c). Excellent agreement was found between the structures, which confirmed the quaternary structure of the SufC2-SufD2 complex. The dissociation of the SufC subunit was negligible in the electron microscopy observations for as-isolated SufCD complex.
The SufD subunit of the SufC2-SufD2 complex has three domains; the N-terminal helical domain, the core-domain comprising a right-handed parallel β-helix, and the C-terminal helical domain (Fig. 1). The structure of the SufD homodimer in the SufC2-SufD2 complex was almost identical to that reported for SufD homodimer crystallized alone. The two structures are superimposable with r.m.s. deviation of 0.61 Å between Cα atoms. The novel core-domain of SufD is composed of a flattened β-helix consisting of nine β-helical turns with two strands per turn. This folding has been classified into a new superfamily of “superhelix turns that are made of two very long strands each” in the SCOP, a database for the structural classification of proteins.25 The two subunits of the SufD homodimer were related by non-crystallographic 2-fold symmetry and the dimer interface was held primarily by 23 hydrogen bonds that form the two anti-parallel β-sheets. To our knowledge, this is the first example of an interaction strategy between two β-helices.
The SufC subunit has two domains as observed in the members of the ABC-ATPase family; a RecA-like catalytic α/β domain that has the nucleotide-binding Walker A and Walker B motifs, and a helical domain specific to ABC-ATPases containing an ABC signature motif (Fig. S2). The two domains are connected by a Q-loop that contains a strictly conserved glutamine residue. Although the overall structure of SufC in the SufC2-SufD2 complex was similar to that of the monomeric form previously reported 21, notable structural changes occurred in the ATP-binding segments upon complex formation as described below.
Interaction between SufC and SufD in the SufC2-SufD2 complex
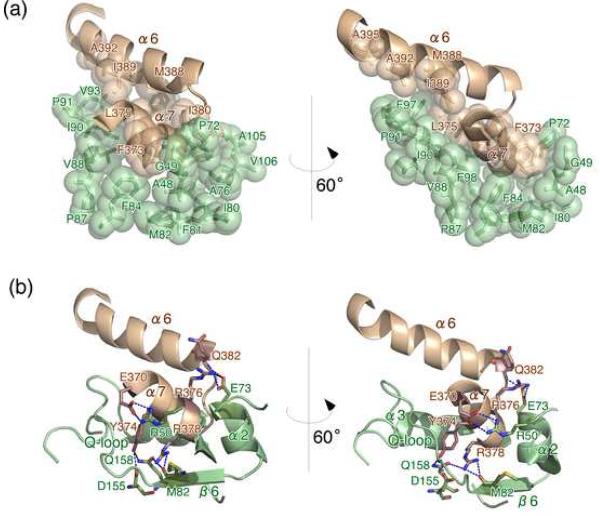
The electron densities for SufCA and its binding partner SufD were clear enough to determine their interactions in molecular detail. The SufCA was bound to the C-terminal helical domain of SufD by extensive hydrophobic interactions as well as by eight hydrogen bonds and one salt-bridge (Fig. 2), with the buried interface area of 962 Å2. The SufD residues involved in the hydrophobic interactions with SufC are Phe373, Leu375, Ile380, Met388, Ile389, Ala392 and Ala395 in the helices α6 and α7. Notably, most of these residues are conserved not only among the SufD orthologs but also in the SufB sequences (discussed later). The helices in the C-terminal helical domain of SufD interacted with the β6 strand, the α2 and α3 helices, and the Q-loop of SufC, which are located between the α/β and helical domains of SufC (Fig. 2). Interestingly, the Q-loop residues involved in the interaction with SufD are highly conserved among the SufC orthologs whereas they are not conserved in the sequences of more divergent ABC-ATPase (Fig. S2), suggesting that the Q-loop may contribute to the recognition of the partner protein.
Fig. 2. The close-up view of the SufC-SufD interface.
Residues involved in the interaction between SufC and SufD are depicted with space-filling model (a) and with stick model (b). SufC and SufD are shown in green and brown, respectively.
Although the overall structure of SufD is quite different from that of transmembrane subunits of the ABC transporters, the local structure at the site of interaction between SufC and SufD shows significant similarity to the interface between the transmembrane subunit and the ABC-ATPase subunit of other structurally characterized ABC transporters.26 When the Walker A motifs of SufC and ABC-ATPases are superimposed, the helices α6 and α7 of SufD also superimpose on the corresponding helices in the transmembrane subunits that are interacting with ABC-ATPases (Fig. S3). These helices in the ABC transporter form the “transmission interface”, which plays a role in transmitting the dynamic motion of the ABC-ATPase to the transmembrane subunit.27 The conserved structure of the interaction interface suggests that the SufC2-SufD2 complex may undergo a gross conformational change in which SufC drives a power stroke coupled with ATP binding and hydrolysis.
Conformational changes of SufC upon complex formation with SufD
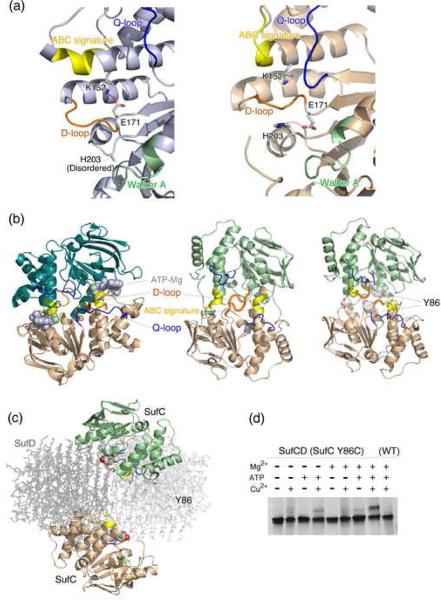
Upon complex formation between SufC and SufD, several significant structural changes occurred in the SufC subunit (Fig. 3a). The Q-loop is moved ≅5 Å toward the ABC signature motif due to the extensive interaction with the C-terminal helical domain of SufD. The protruding D-loop on the molecular surface (that follows the catalytic E171 residue) is moved toward the SufC interior. The α5 helix containing the ABC-signature motif became shorter in the complex than in the SufC monomer.
Fig. 3. Conformational changes and possible dimer formation of SufC.
(a) Comparison of the active site structures of SufC between the SufC monomer (left panel) (PDB id: 2D3W) and the SufCA subunit in the SufC2-SufD2 complex (right panel). The Walker A motif, ABC-signature, D-loop and Q-loop are shown in green, yellow, orange and blue, respectively. The K152 and E171 resides involved in the unique salt-bridge in the SufC monomer are shown in stick models. (b) (Left) Dimeric structure of ABC-ATPase HlyB (H662A variant) (PDB:1XEF). Two subunits are shown in different colors. The bound ATP and Mg are shown with van del Waals surfaces. Color coding for the conserved motifs is the same as in (a). (Middle and Right panel) Comparison of the putative dimer models of SufC. The docking models were constructed by superimposing two SufC molecules onto the ATP-bound HlyB (H662A) dimer using the structures of monomeric SufC (middle), or the structures of the SufCA subunit in the SufC2-SufD2 complex (right). The Y86 residues in the right panel are depicted with their van del Waals surfaces. (c) The spatial location of the two SufC subunits in the entire model of the SufC2-SufD2 complex. Structures of the SufD homodimer are depicted in pale gray with stick representation. (d) The disulfide bond formation between the two subunits of SufC (Y86C variant) in the SufC2-SufD2 complex. Electrophoresis was carried out under non-denaturing conditions and the gel was stained with Coomassie Blue.
Importantly, the unique salt bridge observed in the monomeric SufC between E171 in the Walker motif and K152 is cleaved, allowing the rotation of the E171 side-chain toward the ATP-binding pocket. Furthermore, H203, another key residue for the activity of ABC-ATPases,28 is shifted ≅5 Å toward E171. These structural changes remodel the catalytic pocket of SufC to be suitable for ATP binding and hydrolysis and result in a SufC local structure that more closely resembles that of active ABC-ATPases. Thus, the monomeric SufC is the “latent form” with weak ATPase activity, whereas SufC in the SufCD complex appears to represent the “competent form”. The findings are also consistent with the recent kinetic experiments reporting that ATPase activity of SufC is enhanced by complex formation with SufD.23 Thus,SufC is a novel ATPase whose activity is carefully regulated through conformational changes that occur upon binding to its cognate partner protein, SufD.
Dimer formation of SufC revealed by cross-linking experiments
According to current models of ABC-ATPase function, two ABC-ATPase subunits form a transient dimer during the catalytic step of ATP binding and hydrolysis. This dimerization drives gross structural changes of the transmembrane subunits of the ABC-transporters.26 In the dimeric state, the two ABC-ATPase subunits are aligned to form a “head-to-tail dimer” where the two nucleotides are sandwiched at the dimer interface between the Walker motifs of one subunit and the ABC signature motif of the other subunit (Fig. 3b, left). A docking model for a SufC dimer was generated by superimposing the structure of SufC monomer or SufC from the SufC2-SufD2 complex onto the dimeric form of the H662A variant of the HlyB ATP-bound ABC-ATPase.28 In the dimer model derived from the monomeric structure of SufC, steric hindrances blocked the potential SufC dimer interface due to the protruding D-loop on the molecular surface and the long α5 helix preceding the ABC-signature motif (Fig. 3b, middle). In contrast, in the SufC2-SufD2 complex these steric hindrances are eliminated (see above). Thus, the conformational changes in SufC upon complex formation with SufD would presumably allow association of SufC subunits to form a putative “head-to-tail dimer” (Fig. 3b, right). Despite the modeling results and the favorable conformational changes in SufC, the SufCA and the SufCB subunits are spatially separated in the SufC2-SufD2 complex with their ATP-binding motifs facing each other (Fig. 3c). The two SufC monomers would have to move about ≅20 Å towards the 2-fold axis of the complex for the formation of the head-to-tail dimer.
To determine if this discrepancy is a result of crystallization artifacts and/or the failure to capture a transient dimer, we examined the dimer formation of SufC (as part of SufC2-SufD2) under solution conditions. To detect such a transient dimer, if any, disulfide cross-linking experiments were conducted. First, we searched for a residue of SufC that would be located at the interface of a putative SufC dimer based on our docking model (Fig. 3b). The model predicts that the distance between the Cβ atoms of Y86 in each SufC monomer would be less than 4.8 Å in a SufC dimer. Y86 was substituted with a cysteine to allow for covalent trapping of transient SufC dimer via disulfide bond formation. The tandem expression vector carrying the wild-type SufD and the SufC-Y86C mutant was used to purify the mutant SufCD complex from E. coli cells lacking the chromosomal sufABCDSE operon. The purified mutant SufCD complex showed properties indistinguishable from the native complex. After the complex was incubated in the presence of Mg2+, ATP, and an oxidant (CuSO4) to stimulate disulfide bond formation, it was subjected to native-PAGE analysis. By native-PAGE we observed an additional band on the gel which migrated more slowly than the as-isolated SufCD complex (Fig. 3d). This novel band was not observed when reducing agent (DTT) was included in the sample (not shown), indicating that disulfide-bond formation is involved in the mobility shift. The intensity of the novel band was also diminished if Mg2+, ATP, or CuSO4 was omitted from the reaction cocktails (Fig. 3d). In another control experiment using the wild-type SufCD complex, the novel band was virtually undetected. The novel band was excised and the proteins extracted from the gel were further analyzed by denaturing SDS-PAGE, which showed both SufC and SufD present at equimolar stoichiometry (data not shown), confirming that disulfide formation trapped a conformationally distinct form of the complex. These findings strongly support the notion that SufC forms a transient dimer in the SufC2-SufD2 complex in the presence of ATP and Mg2+. The mobility shift on the native-PAGE suggests a conformational change of the SufC2-SufD2 complex upon SufC dimerization, which was captured in this case by the disulfide-bond formation at the dimer interface of SufC.
Residues of SufD essential forin vivofunction
To better understand the unique structure of SufD in the SufC2-SufD2 complex, a series of SufD mutations were generated and the mutant phenotypes were characterized. Mutations were generated in a plasmid pBBR-SufD and introduced into strain UT109. UT109 contains deletions of the chromosomal isc (ΔiscUA-hscBA) and suf (ΔsufABCDSE) operons. Normally deletion of both pathways in E. coli is lethal20 but UT109 also contains plasmids pKO3-NIF and pRK-sufABC-SE (ΔsufD). The plasmid pKO3-NIF, carrying the nifSU genes from Helicobacter pylori, allows UT109 to grow but is temperature-sensitive for replication.6 Upon shift to the non-permissive temperature (43°C) pKO3-NIF is lost and the cells are unable to grow without introduction of a functional sufD gene (in this case from pBBR-SufD) to complete the partial SUF system provided on pRK-sufABC-SE (ΔsufD).
A mutant containing a truncation of the N-terminal helical domain (Δ2–81 residues, Fig. 4a) of SufD in pBBR-SufD was still able to complement UT109 at the non-permissive temperature (data not shown), indicating this domain is dispensable for in vivo SufD function. In contrast, the C-terminal helical domain of SufD was indispensable for complementation. Stepwise truncation experiments revealed that a SufD variant lacking only ten C-terminal residues (Δ414–423) failed to support growth of UT109 at 43°C (data not shown). The truncation resulted in marked destabilization of SufD as determined by immunoblot analysis. In crystal structures, several residues at the C-terminus contact the β-helical core domain of SufD (denoted by red asterisks in Fig. 5c) via extensive hydrophobic interactions, suggesting that this intramolecular domain-domain interaction contributes to fixing the SufD structure.
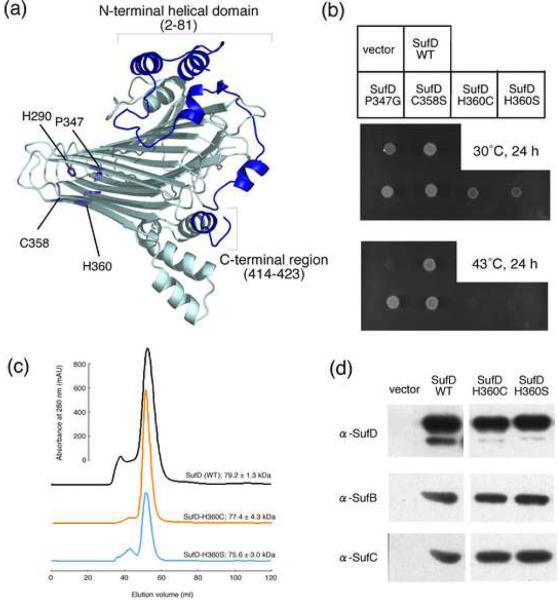
Fig. 4. Mutational Analysis of the SufD protein.
(a) The representation of a SufD subunit with its dimer interface in the front side. The amino acid residues changed by site-directed mutagenesis are depicted in the structure by stick representations. (b) Phenotypic characterization of the SufD mutations. Middle panel shows growth of the mutant (UT109) cells at the permissive temperature (30°C) of the temperature-sensitive plasmid, pKO3-NIF. Complementation for the loss of sufD is shown in the lower panel by the growth at 43°C. (c) Gel-filtration analysis of the mutant SufD proteins. The SufD proteins carrying N-terminally fused His-tag and the H360C or H360S mutation were expressed in the mutant E. coli cells (YT2512) in which the chromosomal sufABCDSE operon was deleted. The proteins were purified with Ni resin and then subjected to the gel-filtration (Sephacryl S-200) chromatography. The molecular sizes were estimated from the three experiments, and the values are the mean ± SD. (d) Pull-down assays to examine the interactions of the SufD mutant proteins with SufB and SufC. The SufD proteins carrying N-terminally fused His-tag and the H360C or H360S mutation were co-expressed in the YT2512 cells with intact SufB and SufC proteins using the plasmid pRK-sufABC-SE (ΔsufD). The proteins were purified with Ni resin and subjected to Western blot analysis using specific antibodies. Detection was with an ECL Plus kit (GE Healthcare).
We also focused on the highly conserved residues in the β-helical core domain (P347 and H360) that are located at the dimer interface of SufD homodimer (Fig. 4a), as well as the nearby C358 residue and also a moderately conserved H290 residue. Site-directed mutants P347G, C358S and H290A were still functional based on UT109 complementation. In contrast, when H360 was replaced with any other 19 amino-acid residues, the resulting mutant SufD proteins were completely inactivated (Fig. 4b for H360C and H360S and not shown for other mutations). Since the wild-type and variant molecules of SufD were present at equal levels in the cells as confirmed by immunoblot analysis (not shown), protein destabilization does not explain the loss of function. Likewise, overexpression of N-terminal His-tag SufD with H360C or H360S mutations resulted in purification of a stable homodimer as in the case for the wild-type SufD, judging from the gel-filtration chromatography of the purified proteins (Fig. 4c). Furthermore, co-expression and co-purification experiments indicated that the SufD H360 mutant proteins could still interact with SufB and SufC (Fig. 4d). Collectively, the mutations of H360 abolished in vivo function despite the fact that the mutation did not impair the tertiary nor quaternary structure of SufD as well as its interaction with partner proteins. It is worth mentioning that the side chain of H360 is buried inside the β-helix at the dimer interface of SufD homodimer. This would imply that the SufC2-SufD2 (or SufBCD) complex undergo significant structural changes so as to expose this functional residue during their operation (discussed later).
Biochemical characterization of the SufBCD complex
Despite the fact that several lines of biochemical evidence have suggested that the SufBCD complex provides the assembly site for the Fe-S cluster,19 structural analysis of the whole complex has been hampered by its instability. The three-dimensional structure of the SufB component is also unavailable, primarily due to the insoluble, aggregated nature of this molecule. Thus, understanding the architecture of the SufBCD complex may provide critical insight into the molecular mechanism underlying Fe-S cluster biosynthesis. SDS-PAGE analysis of the SufBCD complex suggested that the subunit stoichiometry was approximately SufB:SufC:SufD=1:2:1 (Fig. 5b).19 The molecular size of the SufBCD complex was estimated to be 160 kDa on a gel-filtration chromatography (Fig. 5a),17 slightly larger than that for the SufC2-SufD2 complex (ca. 150 kDa). This difference in molecular size may arise from the fact that SufB (56.3 kDa) is larger than SufD (46.8 kDa) by ≅10 kDa. Since SufB and SufD share limited sequence similarity with each other (17% identity and 37% similarity), the secondary structure was predicted for SufB and compared with that of SufD. This prediction suggests that the structures of SufB and SufD also share striking similarity, in particular in the regions corresponding to the β-helix core-domain and the C-terminal helical domain (Fig. 5c) involved in the subunit-subunit interactions in the SufC2-SufD2 complex. Indeed, the amino acid residues of SufD involved in the interaction with SufC are conserved in the corresponding segments of SufB (denoted by black asterisks in Fig. 5c), suggesting that SufC is bound to SufB in a similar manner that was observed in the SufC2-SufD2 complex.
DISCUSSION
In the present study, we have demonstrated the quaternary structure of the SufC2-SufD2 complex. Although one SufC subunit was not well defined by the electron density of the crystals, the two-fold symmetry of the complex in combination with the electron microscopy imaging allowed us to elucidate the entire structure of the complex. The structure revealed intriguing architecture of the SufC2-SufD2 tetramer, where each SufC subunit is bound to the C-terminal helical domain of the SufD homodimer. Comparison with the monomeric structure of SufC uncovered local conformational changes around the nucleotide binding pocket as well as at the putative SufC-SufC dimer interface, suggesting that SufC becomes competent for active dimer formation upon association with SufD. Although two SufC subunits were spatially separated from each other in the crystals, transient formation of the head-to-tail dimer of SufC was supported by cross-linking experiments, in which the SufC2-SufD2 complex carrying the Y86C mutation of SufC formed a disulfide bond in the presence of ATP and Mg2+. Transient dimer formation of SufC during the course of ATP binding and hydrolysis may elicit a significant conformational change of the entire SufC2-SufD2 complex.
In addition to the structural conservation between SufC and other ABC-ATPases, the present study also revealed a similarity of local structures in the intersubunit contact sites (Fig. S3). Despite the lack of structural correlation between SufD and the transmembrane subunits, structural motifs involved in interaction between SufC and SufD bear striking similarity to the corresponding motifs in the ABC-transporter complexes. The intersubunit contact regions in the ABC-transporter (the so-called “transmission interface”) transmits the dynamic motion of the ABC-ATPase to the transmembrane subunits during ATP binding and hydrolysis.26,27 The conserved binding motifs at the interface of SufC and SufD as well as the tight binding of the helices α6 and α7 of SufD to the cleft of SufC between the α/β domain and the helical domain suggest that SufC ATPase activity drives a power stroke leading to a structural change of the SufD homodimer. This view is consistent with the actual formation of the head-to-tail dimer of SufC described above, and also with the findings that a functionally essential residue of SufD (H360) is buried inside at the homodimer interface, where splitting of the SufD homodimer is mechanistically plausible. It should also be noted that the C-terminal helical domain of SufD forms a rigid three-helix bundle structure and is tightly anchored to the core domain, suggesting that the dynamic motions of SufC subunits are directly transmitted to the β-helix core domains of SufD.
What then is the role of the SufC2-SufD2 complex in the biogenesis of Fe-S clusters? We are currently skeptical about its physiological role per se, since the complex was detected under non-physiological conditions where only the two genes for sufC and sufD were overexpressed. Previous studies have shown that controlled expression from the intact sufABCDSE operon results in preferential formation of the SufBCD ternary complex, where no SufC2-SufD2 complex has been detected.17 Interestingly, SufBCD forms a complex with a stoichiometry of approximately 1:2:1. Since SufB and SufD share similarity in both primary and secondary structures, in particular, at the regions corresponding to the β-helix core domain and the C-terminal helical domain that are involved in intersubunit interactions, the quaternary structure of the SufC2-SufD2 complex likely mimics the structure of SufBCD. This model is further supported by the fact that the basal ATPase activity of SufC is enhanced by either SufB or SufD,23 suggesting that association of either SufB or SufD elicits similar conformational changes to SufC. Thus, the SufB1-SufC2-SufD1 complex most likely shares a common architecture with the SufC2-SufD2 complex where one SufD subunit is replaced by the SufB subunit and SufB interacts with both SufD and SufC. It is also likely that gross structural changes, described in this study for the SufC2-SufD2 complex, may also occur in the SufB1-SufC2-SufD1 complex at the possible interface between the β-helix core domains of SufB and SufD. Such structural changes should be relevant to the functional significance of the H360 residue of SufD that otherwise is buried inside the complex. In addition, invariant residues of SufB including C418 and E447 are predicted to be near the putative interface with SufD, suggesting that these residues might work in concert, serving as the binding site for an iron ion or an Fe-S cluster in the SufB1-SufC2-SufD1 complex. Further biochemical and mutational studies as well as the structural characterization are required to establish a clear picture of Fe-S cluster assembly mediated by the structural changes of the SufB1-SufC2-SufD1 complex.
EXPERIMENTAL PROCEDURES
Construction of the expression plasmids
The individual coding regions of sufB, sufC and sufD were amplified by PCR using the primers listed in Table SI, and cloned into the pCR2.1-TOPO vector (Invitrogen) by the TA cloning method. The NdeI-BamHI fragments carrying the respective coding regions were excised and cloned into the two plasmids, pET-21a(+) and pET-19b (Novagene). Then, the XbaI-XhoI fragments containing the ribosome binding sequence and the respective coding regions were excised from the pET-21a(+) derivatives and cloned into the HindIII (modified to NheI site by linker ligation)-SalI sites of a low-copy plasmid vector, pMW219 (Nippon Gene Co., Ltd.). The XbaI-XhoI fragments excised from the pET-19b derivatives contained the ribosome binding sequence and the coding region for (His)10 sequence fused to the respective Suf proteins, and were cloned into the XbaI-SmaI (modified to XhoI site by linker ligation) sites. The tandemly arranged two genes were expressed under the control of the lac promoter. Both of the two constructs, pMW219-sufD-His10-SufC and pMW219-sufC-His10-SufD, were equally functional in expressing the SufCD complex.
Another expression plasmid pBBR-SufD was constructed by transferring the XbaI-SacI fragment carrying the ribosome binding sequence and the SufD coding region from the pET-21a(+) derivative to the pBBR1MCS-4 plasmid,29 in which the expression was driven from the lac promoter.
Overproduction and purification of SufC-SufD complex
The SufD protein was co-expressed with the N-terminally His10-tagged SufC protein from the pMW219-sufD-His10-SufC plasmid in E. coli DH5α cells and were purified using HIS-Select Nickel Affinity resin (SIGMA) by batch method according to the manufacturer's protocol. The SufD protein was co-eluted from the resin with His10-SufC and the complex was further purified by gel filtration using a HiPrep 16/60 Sephacryl S-200 HR column equipped with the ÄKTA explorer 10S system (GE Healthcare) and developed at a flow rate of 0.5 ml/min with 50 mM Tris-HCl (pH7.8) buffer containing 150 mM NaCl. The SufB-SufC-SufD complex was expressed and purified as described elsewhere.17
Crystallization and data collection of SufC-SufD complex
Crystallization conditions of His10-SufC-SufD complex were surveyed by hanging-drop vapor diffusion method using commercial screening kits. Small needle-shaped crystals were produced at 20°C when Grid Screen PEG 6000 A3 (Hampton Research) was used as the reservoir solution. Improvement of crystal quality was achieved by optimizing conditions and the application of micro-seeding technique. The crystals suitable for X-ray diffraction experiments were produced in a 2 μl drop containing 1 μl protein solution [20 mM Tris-HCl (pH 7.8), 50 mM NaCl and 5 mM AMPPNP] and 1 μl reservoir solution [5%(w/v) PEG 6000, 0.1 M MES (pH 8.0 – 8.5)] that was equilibrated against a 0.2 ml reservoir solution. Crystals grew to a typical size of ~ 0.2 × 0.3 × 0.1 mm3 in a week.
Crystals were soaked in the cryo-protectant solution containing 20% (v/v) glycerol and then flash-cooled with a nitrogen-gas stream at 100 K. Diffraction data were collected with oscillation method (Δϕ=1.0°) using the ADSC Quantum 315 detector and synchrotron radiation (λ=1.000 Å) at beamline BL41XU, SPring-8 (Hyogo, Japan). The data were processed and scaled using the HKL2000 suite. Results of the data collection are summarized in Table I.
Table I.
Crystallographic Data and Refinement Statisticsa
| Crystallographic data | |
| Space group | P212121 |
| Cell parameters (Å) | a = 96.1, b= 106.2, c= 171.7 |
| Resolution range (Å) | 50.0 − 2.20 (2.28 − 2.20) |
| Unique reflections | 87226 |
| Redundancy | 12.2 (12.3) |
| Completeness (%) | 100 (100) |
| Rsym (%)b | 7.2 (29.1) |
| Refinement statistics | |
| Rcryst (%)c | 23.4 |
| Rfree (%)d | 25.9 |
| Number of molecules | |
| Water | 289 |
| 2-(N-morpholino)ethane-sulfonic acid (MES)e | 1 |
| Disordered regionsf | |
| SufCA | 0 |
| 1–85, | |
| SufCB | 107–154, 165–182, 194–247 |
| SufDA | 1–7 |
| SufDB | 1–8, 423 |
| RMSD from ideal values | |
| Bond length (Å) | 0.006 |
| Bond angle (°) | 1.3 |
| Average B-factor (Å2) | 34.5 |
| Ramachandran plot | |
| Most favored (%) | 88.9 |
| Additionally allowed (%) | 10.6 |
| Generously allowed (%) | 0.5 |
Values in parentheses are for the outermost shell.
Rsym = ΣhklΣi|li(hkl) − <l(hkl)>|/ΣhklΣi li (hkl), where <l(hkl)> is the average intensity over equivalent reflections.
R-factor = Σ∥Fobs(hkl)| − |Fcalc(hkl)∥/Σ|Fobs(hkl)|.
Rfree is the R-factor computed for the test set of reflections that were omitted from the refinement process.
MES molecule used in the crystallization buffer may bind to the active site of SufCA.
Numerals shown are invisible residue numbers. SufC and SufD are composed of 247 and 423 residues, respectively.
Structure determination of SufC-SufD complex
Structure determination was initiated by the molecular replacement method using MOLREP,30 in which the structure of the SufD homodimer was used as a search model. The electron density map phased by SufD model was ambiguous to locate the SufC moiety, although the SufD moiety was clearly located in the unit cell. Thus, the structure of SufD homodimer was improved by alternating rounds of CNS refinement31 and model revision using Xtalview/Xfit.32 The phase angles derived from the SufD homodimer were further improved by solvent flattening method using RESOLVE.33 The resulting electron density map revealed the entire polypeptide chain for one SufC molecule and only three helices for another SufC. The model of SufC including the side-chains was constructed using ARP/warp34 and LAFIRE.35 The crystal structure of SufCD complex was refined by iterative rounds of CNS refinement and model building with picking up ordered water molecules. Invisible electron density for one SufC subunit was not significantly improved during the progress of the structural refinement. The results of structural refinement are summarized in Table 1.
Secondary structures were assigned using PROMOTIF,36 and the geometry of the final model was analyzed using PROCHECK.37 Superposition and r.m.s. deviations of the structures were calculated using LSQMAN.37 All structural figures were prepared with PyMOL (http://pymol.sourceforge.net/).
Electron microscopy of SufC-SufD complex
We acquired negatively stained images of the SufC-SufD complex on a GATAN 4k × 4k charge coupled device camera using an EF-2000 transmission electron microscope equipped with a γ-type energy filter38 (Hitachi High-Tech, Tokyo, Japan) operated at 200 kV. Microscope magnification was × 60,000, resulting in a resolution of 0.21 nm/pixel. 3-D reconstruction of the particles was performed using software package SPIDER,39 following the reference-based method.40 In the 3D projection matching scheme, initial reference was produced from the atomic model of SufC2-SufD2 calculated from the crystal structure by 2-fold symmetry operation. Through the image-processing, 2-fold symmetry was considered.
Cross-linking between SufC-Y86C residues in SufC-SufD complex
Y86C site-directed mutagenesis was performed by the QuickChange mutagenesis method (Stratagene) using the plasmid pMW219-sufD-His10-SufC as a template. The genes were expressed in the mutant cells (YT2512) in which the entire sufABCDSE operon was deleted from the chromosome,20 and the SufC(Y86C)-SufD complex was purified as described for the wild-type complex. The complex showed identical properties with the wild-type complex during the chromatography on the gel-filtration column and the migration on a native PAGE. For the cross-linking experiments, the purified complex (1 mg/ml) was incubated at 37°C for 30 minutes in the presence of 5 mM ATP, 5 mM MgCl2 and 1 mM CuSO4, and the resultant products were analyzed by native-PAGE (10% gel) and SDS-PAGE (12.5% gel) according to the methods of Davis41 and Laemmili42, respectively.
In vivo complementation assay with mutated SufD
The complementation tests were carried out essentially as described previously6using the E. coli mutant strain UT109 (Δ iscU-hscBA∷Kmr; Δ sufABCDSE∷Gmr) that harbored the temperature-sensitive plasmid pKO3-NIF that carries the nif operon (nifSU) cloned from H. pylori. Mutagenesis of SufD was performed using the pBBR-SufD plasmid as a template and the primers listed in Table S1. The plasmid pRK-sufABC-SE (ΔsufD) was constructed from pRKSUF07820 by PCR-mediated in-frame deletion of the coding sequence for SufD (Δ9–417) using the primers EcSufC-R and S-FSc2 (Table S1).
Protein Data Bank accession code
Structure has been deposited in the RCSB Protein Data Bank under the ID code 2ZU0.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. N. Shimizu, M. Kawamoto, H. Sakai and K. Hasegawa (Japan Synchrotron Radiation Research Institute) for their aid with data collection using the synchrotron at SPring-8 (Hyogo, Japan). The synchrotron-radiation experiments were performed with the approval of the JASRI (Proposal No. 2007A1610 and 2006B2671). We also thank Drs T. Kato and K. Namba for the use of the EF-2000 electron microscope (Graduate School of Frontier Biosciences, Osaka University). This work was supported by a grant from the National Project on Protein Structural and Functional Analyses (to K.F.), Grants-in-Aid for Scientific Research 18054018 (to Y.T.) and 18054016 (to K.F.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of the Japanese Government, and a Grant-in-Aid for Young Scientists 18770084 (to K.W.) from MEXT, by a Grants-in-Aid from Protein Research Foundation (to K.W.), and by National Institutes of Health grant GM81706 (to F.W.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kiley PJ, Beinert H. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 2003;6:181–185. doi: 10.1016/s1369-5274(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 2.Rees DC, Howard JB. The interface between the biological and inorganic worlds: iron-sulfur metalloclusters. Science. 2003;300:929–931. doi: 10.1126/science.1083075. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 4.Ayala-Castro C, Saini A, Outten FW. Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 2008;72:110–125. doi: 10.1128/MMBR.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontecave M, Ollagnier-de-Choudens S. Iron-sulfur cluster biosynthesis in bacteria: Mechanisms of cluster assembly and transfer. Arch. Biochem. Biophys. 2008;474:226–237. doi: 10.1016/j.abb.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Tokumoto U, Kitamura S, Fukuyama K, Takahashi Y. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J. Biochem. (Tokyo) 2004;136:199–209. doi: 10.1093/jb/mvh104. [DOI] [PubMed] [Google Scholar]
- 7.Lill R, Mühlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 8.Balk J, Lobréaux S. Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 2005;10:324–331. doi: 10.1016/j.tplants.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Nachin L, Loiseau L, Expert D, Barras F. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 2003;22:427–437. doi: 10.1093/emboj/cdg061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 11.Yeo WS, Lee JH, Lee KC, Roe JH. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol. Microbiol. 2006;61:206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 12.Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Appl. Microbiol. Biotechnol. 2002;60:12–23. doi: 10.1007/s00253-002-1107-4. [DOI] [PubMed] [Google Scholar]
- 13.Ollagnier-de-Choudens S, Sanakis Y, Fontecave M. SufA/IscA: reactivity studies of a class of scaffold proteins involved in [Fe-S] cluster assembly. J. Biol. Inorg. Chem. 2004;9:828–838. doi: 10.1007/s00775-004-0581-9. [DOI] [PubMed] [Google Scholar]
- 14.Wada K, Hasegawa Y, Gong Z, Minami Y, Fukuyama K, Takahashi Y. Crystal structure of Escherichia coli SufA involved in biosynthesis of iron-sulfur clusters: implications for a functional dimer. FEBS Lett. 2005;579:6543–6548. doi: 10.1016/j.febslet.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 15.Lu J, Yang J, Tan G, Ding H. Complementary roles of SufA and IscA in the biogenesis of iron-sulfur clusters in Escherichia coli. Biochem. J. 2008;409:535–543. doi: 10.1042/BJ20071166. [DOI] [PubMed] [Google Scholar]
- 16.Shimomura Y, Wada K, Fukuyama K, Takahashi Y. The asymmetric trimeric architecture of [2Fe-2S] IscU: Implications for its scaffolding during iron-sulfur cluster biosynthesis. J. Mol. Biol. 2008;383:133–143. doi: 10.1016/j.jmb.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Outten FW, Wood MJ, Munoz FM, Storz G. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J. Biol. Chem. 2003;278:45713–45719. doi: 10.1074/jbc.M308004200. [DOI] [PubMed] [Google Scholar]
- 18.Goldsmith-Fischman S, Kuzin A, Edstrom WC, Benach J, Shastry R, Xiao R, Acton TB, Honig B, Montelione GT, Hunt JF. The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes. J. Mol. Biol. 2004;344:549–565. doi: 10.1016/j.jmb.2004.08.074. [DOI] [PubMed] [Google Scholar]
- 19.Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de Choudens S, Lascoux D, Fontecave M, Outten FW. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J. Biol. Chem. 2007;282:13342–13350. doi: 10.1074/jbc.M608555200. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 21.Kitaoka S, Wada K, Hasegawa Y, Minami Y, Fukuyama K, Takahashi Y. Crystal structure of Escherichia coli SufC, an ABC-type ATPase component of the SUF iron-sulfur cluster assembly machinery. FEBS Lett. 2006;580:137–143. doi: 10.1016/j.febslet.2005.11.058. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe S, Kita A, Miki K. Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8. J. Mol. Biol. 2005;353:1043–1054. doi: 10.1016/j.jmb.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 23.Petrovic A, Davis CT, Rangachari K, Clough B, Wilson RJ, Eccleston JF. Hydrodynamic characterization of the SufBC and SufCD complexes and their interaction with fluorescent adenosine nucleotides. Protein Sci. 2008;17:1264–1274. doi: 10.1110/ps.034652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badger J, Sauder JM, Adams JM, Antonysamy S, Bain K, Bergseid MG, Buchanan SG, Buchanan MD, Batiyenko Y, Christopher JA, Emtage S, Eroshkina A, Feil I, Furlong EB, Gajiwala KS, Gao X, He D, Hendle J, Huber A, Hoda K, Kearins P, Kissinger C, Laubert B, Lewis HA, Lin J, Loomis K, Lorimer D, Louie G, Maletic M, Marsh CD, Miller I, Molinari J, Muller-Dieckmann HJ, Newman JM, Noland BW, Pagarigan B, Park F, Peat TS, Post KW, Radojicic S, Ramos A, Romero R, Rutter ME, Sanderson WE, Schwinn KD, Tresser J, Winhoven J, Wright TA, Wu L, Xu J, Harris TJ. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins. 2005;60:787–796. doi: 10.1002/prot.20541. [DOI] [PubMed] [Google Scholar]
- 25.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 26.Hollenstein K, Dawson RJ, Locher KP. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 28.Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 30.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 31.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta. Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 33.Terwilliger TC. Maximum-likelihood density modification. Acta. Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen SX, Morris RJ, Fernandez FJ, Ben Jelloul M, Kakaris M, Parthasarathy V, Lamzin VS, Kleywegt GJ, Perrakis A. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- 35.Yao M, Zhou Y, Tanaka I. LAFIRE: software for automating the refinement process of protein-structure analysis. Acta Crystallogr. D Biol. Crystallogr. 2006;62:189–196. doi: 10.1107/S0907444905038965. [DOI] [PubMed] [Google Scholar]
- 36.Hutchinson EG, Thornton JM. PROMOTIF--a program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleywegt GJ, Jones TA. A super position. ESF/CCP4 Newsletter. 1994;31:9–14. [Google Scholar]
- 38.Taya S, Taniguchi Y, Nakazawa E, Usukura J. Development of γ-type energy filtering TEM. J. Electron. Microsc. 1996;45:307–313. [Google Scholar]
- 39.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 40.Frank J. Three-dimensional Electron Microscopy of Macromolecular Assemblies. Oxford University Press; New YorK: 2006. [Google Scholar]
- 41.Davis BJ. Disc Electrophoresis. II. Method and Application to Human Serum Proteins. Ann. N.Y. Acad. Sci. 1964;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 42.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.