Abstract
The [PSI+] prion is a self-propagating amyloid of the Sup35 protein, normally a subunit of the translation termination factor, but impaired in this vital function when in the amyloid form. The Sup35 N, M and C domains are the amino-terminal prion domain, a connecting polar domain and the essential C-terminal domain resembling eukaryotic elongation factor 1alpha, respectively. Different [PSI+] isolates (prion variants) may have distinct biological properties, associated with different amyloid structures. Here we use solid state NMR to examine the structure of infectious Sup35NM amyloid fibrils of two prion variants. We find that both variants have an in-register parallel β - sheet structure, both in fully hydrated and in lyophilized form. Moreover, we confirm that some leucine residues in the M domain participate in the in-register parallel β-sheet structure. Transmission of the [PSI+] prion by amyloid fibrils of Sup35NM and of the [URE3] prion by amyloid fibrils of recombinant full length Ure2p are similar whether they have been lyophilized or not (wet or dry).
A prion is an infectious protein, able to transmit a disease or trait without any essential nucleic acid. This concept arose from studies of the mammalian transmissible spongiform encephalopathies (TSEs), but there are now six known distinct prions in yeast, [URE3], [PSI+], [PIN+], [β], [SWI+], [MCA], and [OCT+], based on self-propagating altered forms of Ure2p, Sup35p, Rnq1p, Prb1p, Swi1p, Mca1p, and Cyc8p, respectively (1–6). Extensive evidence, culminating in transfection by the corresponding amyloid of the recombinant protein, has shown that at least [PSI+], [URE3], and [PIN+] are based on self-propagating amyloids (7–10). Amyloid is a fibrillar protein aggregate characterized by partial protease resistance, birefringence on staining with Congo Red and a cross-β-sheet structure (11).
The Sup35 protein is a subunit of the translation termination factor that is inactivated by its aggregation as amyloid in cells infected with the [PSI +] prion. The diminished levels of Sup35p lead to inefficient translation termination and thus more frequent read-through of premature termination codons, for example, suppressing a nonsense mutation in ADE2 and allowing adenine biosynthesis. Sup35p includes an N-terminal 123 residue prion domain (N), whose normal function is in mRNA turnover (12), a middle 130 residue charged domain (M), and the C-terminal 432 residue translation termination domain (13–16). The N domain is both necessary and sufficient for prion propagation (14).
A single prion protein sequence can determine several biologically distinguishable infectious entities. In mice, over a dozen TSE variants are recognized, distinguished by incubation period, distribution of pathology in the brain, species barriers, protease sensitivity of PrPSc and glycoform ratios (reviewed in refs. (17, 18)). Variants of the yeast prions [PSI+], [URE3] and [PIN+] have also been found, distinguished by intensity of the prion phenotype, stability of prion propagation, response to excess or deficiency of some chaperones and ability to cross species barriers (9, 19–25). Prion variants are a consequence of different amyloid structures which are faithfully propagated (26, 27)
Solid state NMR has been used to study amyloid structure (reviewed in (28)) and different amyloids have been found with in-register parallel β-sheet(29–32), antiparallel β-sheet (33, 34) and parallel β-helix-like (35, 36) structures. Solid state NMR structural studies of infectious amyloid of Sup35NM, Ure21–89 and Rnq1153–405 (the prion domains) have indicated that each is an in-register parallel β-sheet, meaning that adjacent peptide chains line up in the same N to C orientation, and with corresponding residues opposite each other (Fig. 1) (37–39). The β-sheets are folded along the fibril axis as shown by the diameter of fibrils of the prion domain. Mass per unit length measurements for Ure2p and Sup35p are also consistent with this structure, with each molecule contributing one layer to the fibril (one monomer per 4.7 angstroms fibril length) (40, 41). The material used in the solid state NMR studies (37–39) was prepared in a manner known to produce a mixture of prion variants on transfection into yeast cells (8–10). Although this was interpreted to mean that the in-register parallel structure is shared by different variants, it is clearly important to verify that this is the case by direct experiments. Weissman’s group has found that cells infected with Sup35NM fibrils produced at 4°C are mostly of a strong variant while those made at 37°C are nearly all a weak variant (8). The 37°C fibrils are more resistant to heat denaturation and breakage and show slow H-D exchange extending further toward the C-terminal part of the prion domain than do fibrils made at 4°C (27). Here we prepare 37°C and 4°C fibrils, verify the expected phenotypes of their transfectants and show that both have in-register parallel β-sheet structure.
Fig. 1. Model of in-register parallel β-sheet structure.
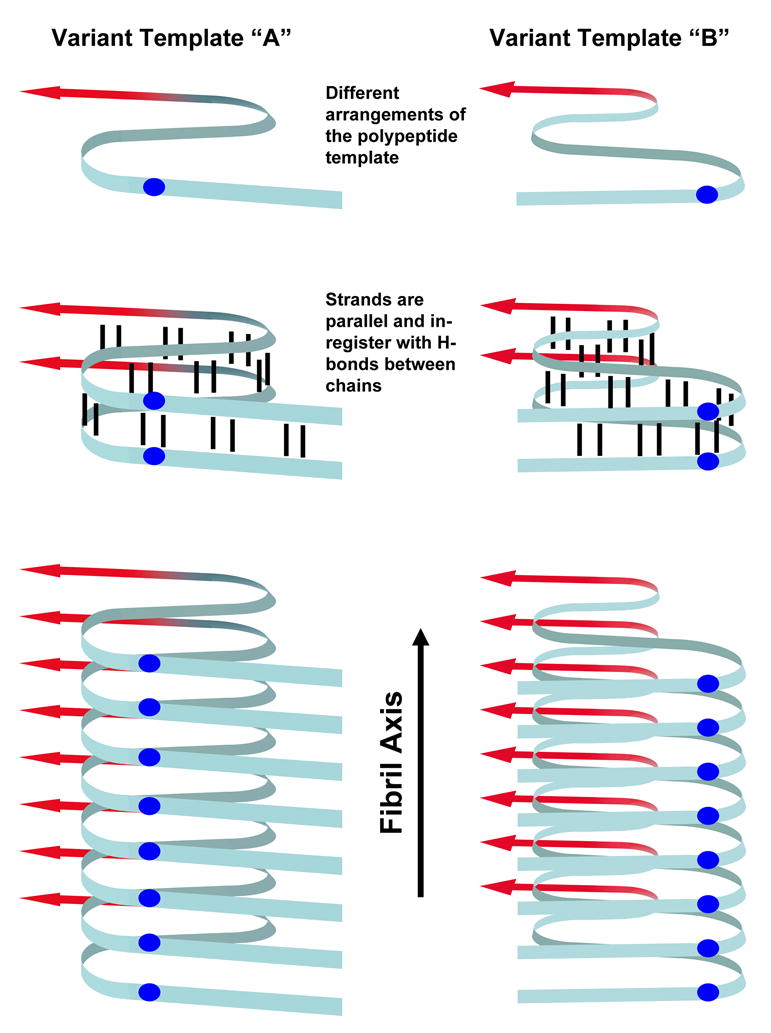
The yeast prions [PSI+], [URE3] and [PIN+] are structurally based on in-register parallel β-sheets. The ribbon represents the Prion domain (blue) along with some of the Middle domain (red) of Sup35. The prion conformation is propagated through in-register pairing of the polypeptides thus forming fibrils composed of long parallel β-sheets with the β-strands stacked perpendicularly to the fibril axis. Different prion variants are based on different arrangements of the polypeptide template which are propagated by incorporating free polypeptide into the in-register parallel β-sheets. Residue i of chain n is opposite residue i of the next polypeptide (blue balls) and hydrogen bonds (black lines) to residues i ± 1 of an adjacent chain are parallel to the fibril axis.
NMR experiments require relatively large amounts of material packed into a small volume. Lyophilized fibrils are routinely used for this purpose, and previous solid state NMR studies have shown that lyophilization does not perturb the molecular structures of β-amyloid (34, 42) and HETs(218–289) fibrils (38). Nonetheless, particularly in light of a previous report that hydrated and lyophilized Sup35NM fibrils may have different x-ray diffraction patterns (43), it is important to verify that drying does not perturb the Sup35NM fibril structure. We have therefore analyzed fibrils which have never been dried and find that never-dried fibrils are also in parallel β-sheet structure. Importantly, drying does not produce a difference in infectivity for yeast of either Sup35NM or Ure2p fibrils.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Sup35NM was expressed from pFPS167, which codes residues 1–253 with an additional carboxy-terminal histidine tag (see figure 2A). Freshly-transformed BL21-CodonPlus® (DE3) RIPL cells with pFPS167 were grown in Defined Amino-Acid Medium (DAM) for isotopic labeling using adaptations of established methods (37, 44).
Fig. 2. Sup35NM amyloid fibrils/prions formed at 4° and 37°C.
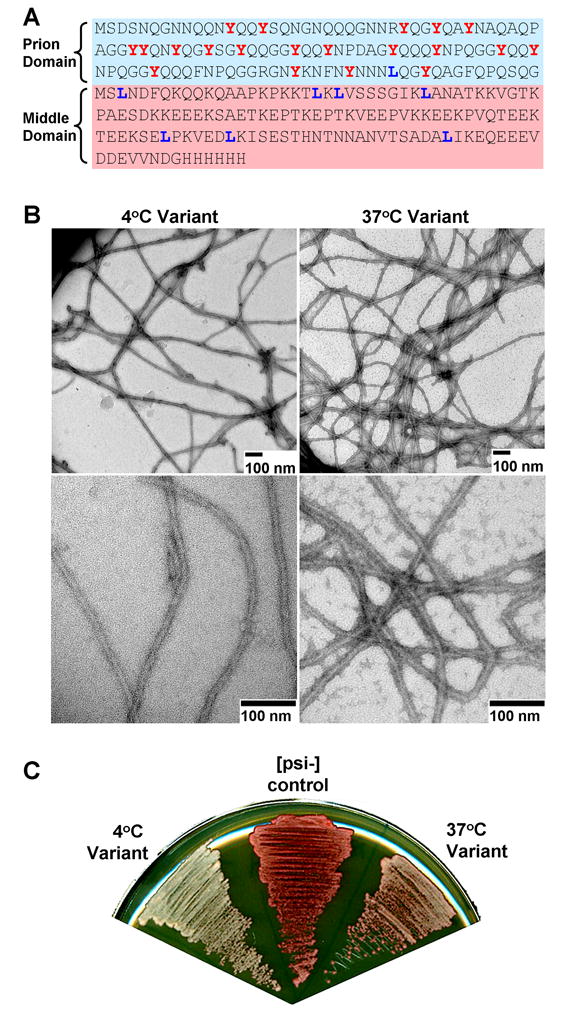
A. All experiments were performed using Sup35NM, which is composed of the prion (N) and middle (M) domain of Sup35, but lacks the C-terminal eRF3 domain. Tyrosine and leucine residues (highlighted in red and blue respectively) were selectively labeled for NMR experiments. B. Following purification in denaturant, Sup35NM was exchanged into phosphate buffer at either 4° or 37°C whereupon it formed long fibrils. C. Fibrils formed at both 4° or 37°C could infect yeast with the [PSI+] prion following transfection. Fibrils grown at 4°C conferred a stronger [PSI+] variant, as reported by whiter yeast colonies on low-adenine medium (see methods).
Cells were grown overnight in 50 ml LB medium with 50 μg/ml ampicillin, 34 μg/ml chloramphenicol and 100 μg/ml streptomycin, then harvested and re-suspended into 2 liters DAM with 50 μg/ml ampicillin. One liter DAM contains: 100 ml 10x salts (130g/L KH2PO4, 100g/L K2HPO4, 90g/L Na2HPO4, 20g/L NH4Cl), 10 ml 100x trace elements (per/100ml: 0.6g FeSO4, 0.6g CaCl2, 0.12g MnCl2, 0.08g CoCl2, 0.07g ZnSO4, 0.03g CuCl2, .002g H3BO3, .025g (NH4)6Mo7O24, 0.5g EDTA), 10ml 100x MgSO4 (1M), 12ml 40% (w/v) glucose, 100ml 10x TAU mix (0.3g/L thiamine, 2g/L adenine sulfate, 2g/L uracil), and 70ml AA mix (2g/L each amino acid). The cells were grown in DAM at 37°C with vigorous shaking until they reached A600 ≈ 1.0, harvested by centrifugation at ~8,000 × g for 7 minutes, and resuspended in 4 liters pre-warmed DAM with the same amino-acid composition, except the amino acid with the desired label had been substituted for the un-labeled counterpart at a concentration of 0.2 g/L. After 15 min shaking at 37°C, protein expression was induced by adding of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). After ~4 hours, the cells were harvested at ~8,000 × g and stored at −80°C or taken directly to the first purification step.
Sup35NM was purified as previously described (44). Following protein over-expression, cell pellets were re-suspended in 80ml of 8 M Guanidine, 150 mM NaCl, 100 mM K2HPO4, pH8 and incubated at room temperature for about 2 hours. The cell lysate was cleared by high-speed centrifugation (~150,000 × g) for 1 hour. The supernatant was mixed with 6ml nickel-nitrilotriacetic acid agarose (NiNTA from Qiagen) and incubated at 4°C for 1 hour. The sample and NiNTA were poured into a gravity column and washed twice with 10 ml of 8.5 M urea, 100 mM Na2HPO4 pH 8, 10 mM Tris pH 8, 150 mM NaCl, 20 mM imidazole and twice with 10 ml of 8.5 M urea, 10 mM Tris pH 7.5, 80 mM NaCl, 20 mM imidazole. The protein was eluted twice with 10 ml of 8.5M urea, 10 mM Tris pH7.5, 80 mM NaCl, 200 mM imidazole.
The eluate from NiNTA was passed through an equilibrated Q Sepharose ion exchange column. Following loading, the column was washed with one column volume of 8.5 M urea, 10 mM Tris pH 7.5, 80 mM NaCl (also used to equilibrate the column). Sup35NM was eluted with an 80 – 160 mM NaCl gradient in the same buffer. The Sup35NM-containing fractions were pooled and dialyzed several times against pre-warmed/cooled 5mM K-PO4, pH7.4, 150mM NaCl using Pierce dialysis cassettes (20,000 molecular weight cut off). Dialysis was always performed at 4°C or 37°C accordingly. Sup35NM samples were incubated at their respective temperatures for at least 1 week.
Fibrils were harvested at by high-speed centrifugation (~150,000 × g), which also served to separate fibrils from remaining soluble contaminants. Fibrils were washed several times with water. To achieve very dense samples for solid state NMR, the fibrils were spun at ~280,000 × g for 30 minutes.
Ure2p was expressed from pKT41-1 (40), purified by affinity chromatography, dialyzed against 50 mM sodium phosphate pH 8.0, 300 mM NaCl and fibers formed at room temperature with agitation.
Transfection
Sup35NM fibrils were tested for infectivity by transfection into Saccharomyces cerevisiae strain 74-D694 (MATa ade1–14 ura3 leu2 trp1 his3 [psi−](45) and Ure2p fibrils into S. cerevisiae strain DK174 (MATa kar1 PDAL5ADE2 ura3 trp1 leu2 his3) as previously described (9, 46).
Electron Microscopy
The formation of Sup35NM fibrils was confirmed by electron microscopy of negatively stained samples. Aqueous suspensions of fibrils were dispensed to carbon-coated copper grids and incubated for several minutes. The fibril suspensions were blotted away, the grids were briefly washed with water and then 2% uranyl acetate stain was applied to the grids for ~ 2 minutes. The stain was blotted away and the grids were left to air dry. Fibrils were visualized using an FEI Morgagni transmission electron microscope operating at 80kV.
Nuclear Magnetic Resonance
Solid state NMR experiments on selectively labeled Sup35NM fibrils were performed at room temperature at 9.39 T (100.4 MHz 13C NMR frequency) using an InfinityPlus spectrometer (Varian, Palo Alto, CA) and MAS spinning probes with 3.2 mm diameter rotors (Varian). 13C NMR spectra were recorded at an MAS frequency of 20 kHz with 1H-13C cross -polarization (47) and two-pulse phase-modulated decoupling (48). PITHIRDS-CT measurements were done at a MAS frequency of 18 or 20 kHz (49), using spin-lock detection for an improved signal-to-noise ratio (50). Each PITHIRDS-CT data point is the result of 1024 or more scans with a 4s recyle delay.
The raw PITHIRDS-CT data, Sraw(t), (with Sraw(0) = 100) were corrected for the 1.1% natural abundance of 13C based on the roughly linear decay of the signal to 70% of the initial value by 76.8 ms with dry unlabeled samples (37). The corrected signal due to the specific label, Scor, was calculated as Scor(t) = [Sraw(t) − fna(100-0.39t)]/(1-fna), where fna is the fraction of the 13C signal due to natural-abundance spins. For dry samples, we assume all natural abundance and specifically labeled residues contribute to the signal. For wet samples we estimate fna assuming that natural abundance carbonyl 13C from all of N and 1/3 of M contribute to the signal. This estimate is based on our finding that part of the M domain is in β-sheet structure and contributes to the signal, but part is not, and is presumed to be unstructured (see Results). If all of M were structured, the natural abundance carbonyl 13C correction would increase by one residue equivalent, implying that more leucines are in an in-register parallel beta sheet structure. Thus this is a conservative assumption. The raw PITHIRDS-CT data are shown in supporting information (Fig. S1). Even in the raw data for leucine-1-13C fibrils, it is clear that a significant fraction of the 13C NMR signal decays on the 35 ms time scale typical of in-register parallel beta-sheets with 5 A° intermolecular 13C-13C distances.
Estimation of Leu residues in in-register parallel β sheet structure
With N0.5 = number of Leu-1-13C residues ~0.5 nm from its nearest Leu-1-13C neighbor and therefore presumably having in-register parallel β sheet structure, Siso = signal from isolated residues (as in the natural abundance sample), S′ = observed signal from labeled amyloid, S0.5 = simulated signal from linear array of 13C atoms 4.7 angstroms apart, and Nres = total Leu residues (8),
That is, the location of the signal between that predicted for 0.5 nm spacing (S0.5) and that known for isolated labels (Siso) is assumed proportional to the fraction in these forms. For example, if all residues had the 0.5 nm spacing, then S′ would equal S0.5 and N0.5 would equal Nres. If all labeled residues were isolated, then S′ would equal Siso and N0.5 would be zero.
Using values at 40 ms:
The increased fraction of Leu-1-13C residues estimated to be within 0.5 nm of their nearest neighbor in wet or rehydrated filaments can be accounted for if ~2.6 residues are mobile in these preparations and therefore not contributing to the NMR signal. The remaining 1.8 residues are assumed to be structured, but not in in-register parallel β sheets.
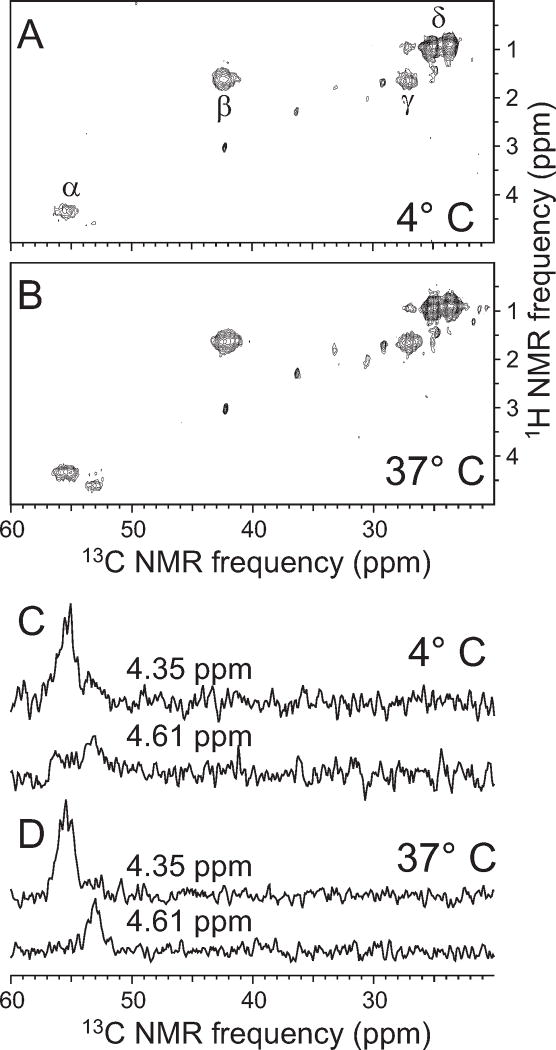
2D 1H-13C NMR spectra in Fig. 5 were obtained with MAS at 9.5 kHz. 1H-13C spin polarization transfer between the t1 and t2 dimensions was carried out with a refocused INEPT sequence (51), with a total transfer period of 3.4 ms. 1H-13C scalar couplings were refocused in the t1 dimension by a single 13C π pulse at t1/2, and removed in the t2 dimension by WALTZ decoupling, with a 7.8125 kHz 1H rf field.
Fig. 5. Two-dimensional1H-13C NMR spectra.
A,B. Spectra of Sup35NM fibrils formed at 4°C and 37°C, with uniform 15N- and 13C-labeling of all leucine residues. Spectra were obtained under conditions appropriate for solution NMR, so that only signals from highly mobile residues are observed. Crosspeaks arising from 1Hα/13Cα, 1Hβ/13Cβ, 1Hγ/13Cγ, and 1Hδ/13Cδ sites of leucines are indicated. C,D. 1D slices at 1H chemical shifts of 4.35 ppm and 4.61 ppm, corresponding to major and minor components of the 1Hα/13Cα signals. The minor component is relatively stronger in the spectrum of 37°C fibrils. Chemical shifts in these spectra are relative to DSS.
RESULTS
Sup35NM fibrils were prepared as described (8) by incubation at either 4°C or 37°C. Although not clearly distinguishable by electron microscopy (Fig. 2B), the variants produced on transfection into yeast cells were as described, with a strong [PSI+] variant produced by 4°C fibrils and a weak variant by 37°C fibrils (Fig. 2C).
37°C fibrils and 4°C fibrils both have in-register parallel β-sheet structure
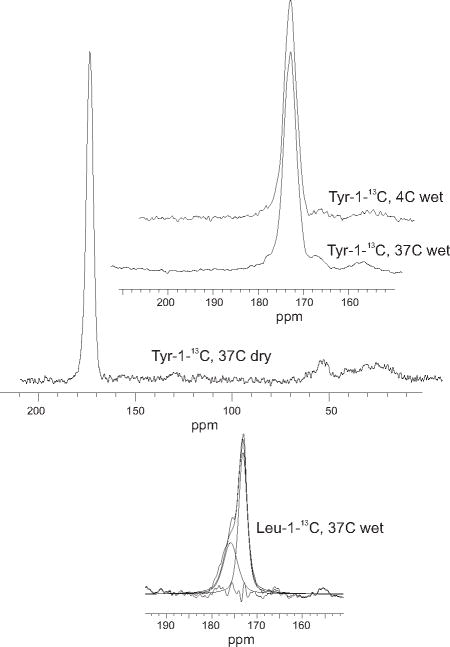
We prepared 4°C and 37°C amyloid fibrils of Sup35NM fully labeled with 13C at the carbonyl position of all tyrosine residues (Fig. 2A), and concentrated them by centrifugation but without drying. The one dimensional spectrum of each (Fig. 3) shows a single carbonyl peak at 172.84 ppm with a full width at half height of ~3ppm (Table 1). The tyrosine carbonyl 13C frequency expected for random coil residues is 174.2, and residues in β sheet conformation are shifted 1–3 ppm to lower values while α - helical residues show shifts to higher values (33, 52–54). The tyrosine residues of Sup35NM are distributed throughout the N domain but are absent from the M domain (Fig. 2A); this result indicates that all or nearly all are in β sheet conformation for amyloids of both prion variants.
Fig. 3. One dimensional solid state NMR spectra.
Spectra of Tyr- and Leu-labeled Sup35NM were recorded at 9.39 T using magic angle spinning at 9.0 kHz. The full spectrum of Tyr-1-13C labeled Sup35NM is shown along with the expanded carbonyl peaks of some of the samples showing the decomposition into major and minor components listed in Table 1.
Table 1.
One-dimensional solid state NMR experiments on Sup35NM. The carbonyl peak of each 1D spectrum was fit to a sum of two Gaussian curves. Chemical shifts are relative to tetramethylsilane and random coil shifts are from Wishart et al. (58)
| Major component | Minor components | ||||||
|---|---|---|---|---|---|---|---|
| label, hydration | Chemical shift ppm | Line width FWHM ppm | Fraction | Chemical shift ppm | Line width FWHM ppm | Fraction | Random coil ppm |
| 37°C Leu-1-13C dry | 173.24 | 4 | 0.88 | 176.45 | 2.9 | 0.12 | 175.9 |
| 37°C Leu-1-13C dry+H2O | 173.29 | 2.8 | 0.89 | 175.8 | 2.4 | 0.11 | 175.9 |
| 37°C Leu-1-13C wet | 172.61 | 2.2 | 0.59 | 174.93 177.14 |
2.4 3.1 |
0.25 0.16 |
175.9 175.9 |
| 4°C Leu-1-13C wet | 173.12 | 2.3 | .58 | 175.36 | 1.3 | .42 | 175.9 |
| 37°C Tyr-1-13C wet | 172.84 | 3.6 | 1.00 | 174.2 | |||
| 4°C Tyr-1-13C wet | 172.84 | 3.3 | 1.0 | 174.2 | |||
| 37°C Tyr-1-13C dry | 172.68 | 4.3 | 1.0 | 174.2 | |||
The rapid molecular motion of proteins in solution averages out dipole-dipole interactions and chemical shift anisotropies (CSA), but in solid state 13C NMR, magic angle spinning (MAS) of the sample is needed to average out dipole-dipole interactions and CSA. Pulse sequences that selectively restore 13C-13C dipole-dipole interactions are useful for measuring nearest neighbor distances because the magnitude of the interaction between 13C-labeled atoms is proportional to 1/r3. Such pulse sequences are called “dipolar recoupling” sequences. We used the PITHIRDS-CT sequence(49), which has been shown to be particularly effective in measurements of relatively large 13C-13C distances (> 4 Å), even among 13C-labeled sites with large CSA. In PITHIRDS-CT measurements, a radio-frequency (rf) pulse occupying one third of each MAS rotation period and rotating the spins of interest by π (thus “PITHIRDS”) recouples the dipole-dipole interactions. The effective time of recoupling is varied by shifting certain pulses within the rotation period(49), but with a constant total time before the signal is measured (hence CT = constant time) to minimize effects of T2 relaxation on the experimental data.
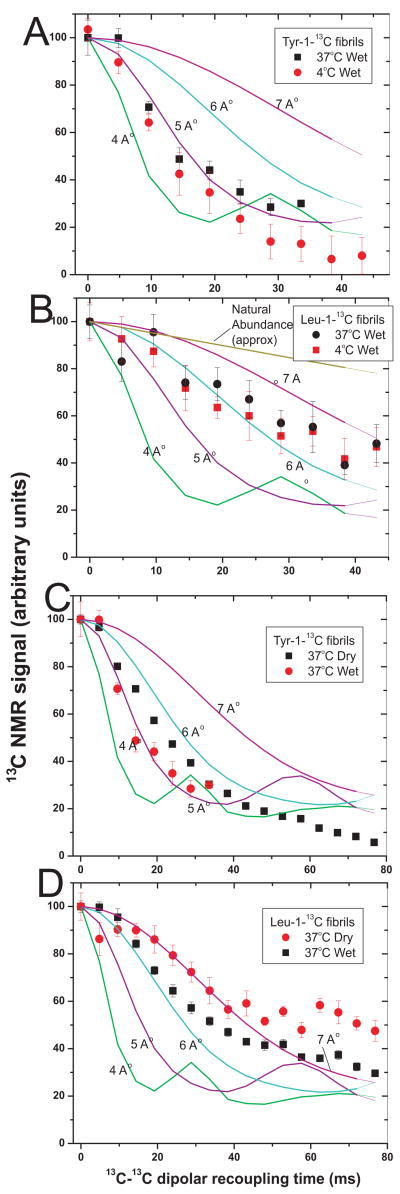
The rate of decay of 13C NMR signals under the PITHIRDS-CT sequence depends on distances among 13C nuclei, as shown by the simulations in Fig. 4. Wet (never dried) fibrils of Sup35NM labeled with Tyr-1-13C and formed at 4°C or at 37°C each show rates of signal decay indicating a 4.5 – 5.0 Å distance to the nearest neighbor (Fig. 4A), indicating an in-register parallel β-sheet structure as previously shown for fibrils formed at 20°C (37).
Fig. 4. Dipolar recoupling experiments.
Measurements of 13C-13C nuclear magnetic dipole-dipole couplings using the PITHIRDS-CT method at 9.39T. A. Comparison of Tyr-1-13C labeled Sup35NM fibrils formed at 4°C or 37°C using magic angle spinning (MAS) at 18 kHz. Fibrils were compacted by centrifugation, but were never dried. Simulated PITHIRDS-CT curves are shown for ideal linear chains of 13C nuclei with the indicated spacings. B. Comparison of Leu-1-13C labeled Sup35NM fibrils formed at 4°C or 37°C and never dried. C. Comparison of Tyr-1-13C labeled Sup35NM fibrils formed at 37°C and either never dried (wet) or dried by lyophilization (dry). D. Comparison of Leu-1-13C labeled Sup35NM fibrils formed at 37°C and dried by lyophilization (dry) or rehydrated by addition of water (wet).
The Sup35 M domain has partial β sheet structure in both 4°C and 37°C filaments
There are 8 leucine residues in Sup35NM, only one of which is in the N domain (Fig. 2A). Sup35NM leucine-1-13C filaments formed at 4°C or 37°C were examined, without drying, by solid state NMR 1D analysis (Fig. 3, Table 1). The carbonyl peak was best fit by two (or in one case three) Gaussian peaks. The major peak for both samples, accounting for 58% of the signal (~4.6 residues), has a chemical shift of ~173 ppm which is a frequency typical of β sheet structure.
PITHIRDS-CT experiments on leucine-1-13C filaments, carried out as for tyrosine-1-13C labeled filaments (above), showed less rapid decay, indicating that not all of the leucine residues had the nearest-neighbor 13C-13C distances of approximately 5 Å characteristic of in-register parallel β-sheets. Assuming that the signal is a sum of residues in in-register parallel β-sheets and residues outside the β-sheets, with >9 Å nearest-neighbor distances, the results can be explained if ~3.6 leucines are in the β-sheets. This result, taken with that for the 1D spectra above, suggests ~4 leucine residues are in in-register parallel β-sheets. The filaments formed at 4°C and at 37°C showed nearly the same signal decay rate (Fig. 4B), indicating similar numbers of leucine residues in theβ-sheets in the two samples.
Sup35NM filaments with uniform 15N- and 13C-labeling of leucine residues were also grown at 4°C and 37°C. Two-dimensional (2D) 1H-13C NMR spectra of these samples, obtained at room temperature with measurement conditions appropriate for solution NMR rather than solid state NMR (i.e., 1H-13C polarization transfers mediated by scalar couplings rather than dipole-dipole couplings, and low 1H decoupling powers during detection of 13C NMR signals (55–57)) showed strong crosspeaks at chemical shifts that were within 0.04 ppm and 0.3 ppm (for 1H and 13C, respectively) of random coil values(58) (Fig. 5A). The 2D 1H-13C spectra therefore indicate that a subset of the leucine residues (presumably those leucines that are not involved in β-sheet structure) exist in highly mobile segments of the M domain. 2D spectra of 4°C and 37°C filaments were quite similar, although 37°C filaments showed a more intense minority component in the 1Hα/13Cα crosspeak signal (Fig. 5B).
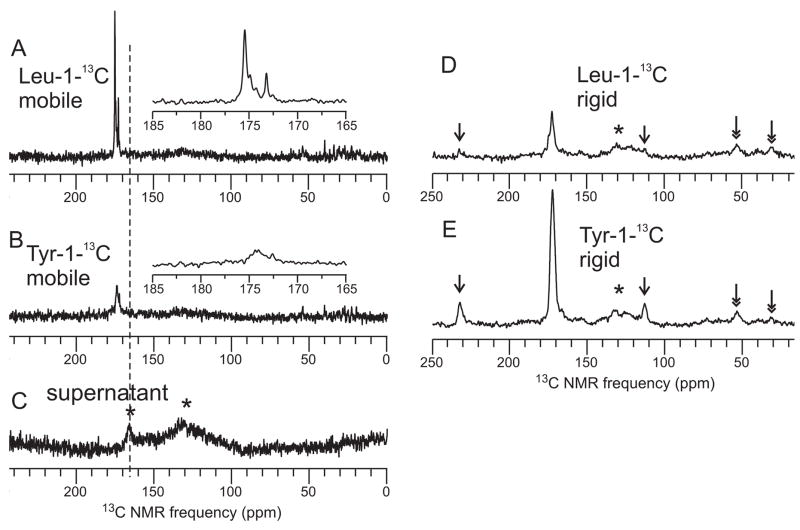
It was possible that the NMR signals observed under “solution NMR” conditions arose from Sup35NM molecules in solution, rather than from mobile segments of Sup35NM in fibrils. We thus directly compared 13C NMR spectra of Leu-1-13C-labeled Sup35NM fibrils and Tyr-1-13C-labeled Sup35NM fibrils, both as fully hydrated, never dried, centrifuged pellets. Spectra were obtained both with “solution NMR” conditions, i.e., direct pulsing of 13C spins and relatively weak proton decoupling (Fig. 6A and 6B), and with “solid state NMR” conditions, i.e., Hartmann-Hahn cross-polarization of 13C spins and relatively strong proton decoupling (Fig. 6D and 6E). The Leu-labeled sample shows strong, sharp carbonyl 13C signals under solution NMR conditions, while the Tyr-labeled sample shows only weaker, broad carbonyl signals under solution NMR conditions. Both samples show carbonyl 13C signals under solid state NMR conditions, but the signals from the Tyr-labeled sample are stronger. These results support our claim that the mobile Leu residues are from the fibrils themselves, not from free Sup35NM in solutionas the latter possibility would result in mobile Tyr residues as well. In addition, as shown in the figure 6C, we have looked directly for Leu carbonyl signals from free Sup35NM molecules by resuspending and repelleting the fibrils, and then recording a 13C NMR spectrum of the supernatant. We see no signals from free Sup35NM, despite extensive signal averaging. Therefore, we conclude that free Sup35NM does not make a measurable contribution to our NMR measurements.
Fig. 6. Comparisons of solution NMR and solid state NMR spectra.
A. 1D 13C NMR spectrum of Leu-1-13C labeled Sup35NM fibrils formed at 4° C, obtained with 512 scans under conditions appropriate for observation of highly mobile protein segments, i.e., direct pulsing of 13C, continuous-wave proton decoupling with a 11 kHz rf field, 6.00 kHz MAS frequency, 2.0 s recycle delay. The inset is an expansion of the carbonyl signal region. Sample is a wet pellet (never dried) in a thin-wall 3.2 mm MAS rotor with 36 μl volume, containing 4.6 mg of protein. B. Same as for A, but with 4.9 mg of Tyr-1-13C labeled Sup35NM fibrils formed at 4° C. All sample and measurement conditions are identical, and the vertical scale is the same as in A. C. Same as A, but for the supernatant obtained by suspending the Leu-1-13C labeled Sup35NM fibril pellet in 50 μl H2O and repelleting. The MAS rotor contains 36 μl of supernatant, 16384 scans were acquired, and the vertical scale is reduced by a factor of four relative to A. Asterisks indicate NMR probe background signals at 130 ppm and 165 ppm. D. Spectrum of Leu-1-13C labeled Sup35NM fibrils formed at 4° C (same sample as in A), obtained with 2048 scans under conditions appropriate for observation of rigid protein segments, i.e., 1H-13C cross-polarization for 1.5 ms, TPPM proton decoupling with an 85 kHz rf field, 6.00 kHz MAS frequency, 2.0 s recycle delay. E. Same as D, but for Tyr-1-13C labeled Sup35NM fibrils formed at 4° C (same sample as in B). Arrows indicate MAS sideband lines, expected from rigid (but not mobile) carbonyl 13C sites due to their chemical shift anisotropy. Double-headed arrows indicate signals from natural-abundance 13C in Sup35NM. Vertical scales in D and E are the same. Nearly equal natural-abundance signal intensities in D and E indicate nearly equal sample quantities.
Dried and never dried samples have similar infectivity for yeast
In order to address the possibility that drying of protein samples may cause irreversible damage to the structure of amyloid fibrils, we compared the infectivity rates for fibrils with and without drying. In this functional assay amyloid fibrils were mixed with plasmid DNA and used to transform non-prion yeast cells by a procedure similar to DNA transformation (8, 9). Together with selection for the DNA plasmid, limited adenine in the medium allowed detection of colonies with a prion induced by the fibrils. In these experiments, freshly formed amyloid fibrils were compared with the same fibrils that underwent drying using conditions that were normally used to prepare samples for solid state NMR experiments. Just before transformation, the dried fibrils were dissolved in water and these re-hydrated fibrils as well as wet (never dried) fibrils were each sonicated. For these experiments we used Sup35NM fibrils formed at 4°C as well as full length Ure2p fibrils formed at room temperature. The result shown in Table 2 clearly indicates that the infectivity rates of dry and wet fibrils are similar for both proteins, so we conclude that drying of amyloid fibrils before the NMR experiments does not cause an irreversible alteration of their biological effects, consistent with prior evidence that lyophilization does not alter the molecular structures of amyloid fibrils significantly (i.e., NMR chemical shifts of lyophilized and fully hydrated fibrils are the same(34, 38, 42)).
Table 2.
Comparison of prion induction by wet and dry amyloid fibrils. Data shown are the average of two experiments. Sup35NM fibrils formed at 4°and Ure2p fibrils formed at room temperature were either used without ever having been dried, or lyophilized to dryness and rehydrated by addition of water. Fibrils were mixed with pRS425 (LEU2) and introduced into spheroplasts selecting plasmid transformants on -Leu with limiting adenine. Over 300 colonies appeared for each fibril type and tests of Ade+ and curability by guanidine showed the indicated fraction of prion-containing transformants. None of the control plasmid transformants without fibrils were Ade+ and curable
| protein | Sup35NM | Ure2p | none (control) |
|---|---|---|---|
| final concentration of wet and dry fibrils, μM | 1.3 | 0.8 | 0 |
| conversion to prion, by wet fibrils (%) | 31 | 22 | 0 |
| conversion to prion, by dry fibrils (%) | 31 | 20 | 0 |
Solid state NMR of dried and never dried fibrils
We compared Sup35NM Tyr-1-13C fibrils made at 37°C that had been either dried by lyophilization, as in our previous report (37), or had never been dried (Fig. 4C). The wet fibrils showed, if anything, a more rapid decay of signal in the PITHIRDS-CT experiment, showing that the in-register parallel structure is not a consequence of drying. The one-dimensional solid state NMR spectrum of wet 37°C or 4°C fibrils each showed a single peak with the shift to lower frequencies indicative of β-sheet structure (Table 1). We previously showed that dry Sup35NM Tyr-1-13C 20C fibrils show a major peak indicatingβ-sheet (91%) and a minor non-β-sheet peak (9%).
Dry Sup35NM Leu-1-13C 37°C fibrils showed slower decay in PITHIRDS-CT experiments than did fibrils to which water had been added (Fig 4D), and the one dimensional spectra showed a higher proportion of β-sheet content. Both results can be explained by loss of some non-β-sheet signal in wet samples; unstructured residues should be mobile in the wet sample and then not contribute to the NMR signal. Note that never dried Sup35NM Leu-1-13C 37°C fibrils (Fig. 4B) show essentially the same kinetics as those which had been dried and rehydrated (Fig. 4D).
DISCUSSION
Prions of yeast or mammals are genes made of protein, in the sense that they carry inherited information and, like genes of DNA or RNA, can even have an array of alleles. The alleles are called “strains” in the mammalian literature and “variants” for yeast prions (to avoid confusion with yeast strains). The molecular basis of prion propagation and prion variants clearly involves self-propagating amyloids differing in structure between variants, but for no variant is the atomic structure yet known. Our earlier work has shown that infectious amyloids of Sup35NM, Ure2p1–89, and Rnq1p153–405 each have an in-register parallel β-sheet structure(37–39). Mutations in the N domain affect different prion variants in different ways suggesting different regions are involved in the β-sheet structure (59, 60). H-D exchange experiments show that fibrils formed at 4°C and 37°C had significantly different distributions of the fastest- and slowest- exchanging components (27), indicating significant differences in structure between variant amyloids. In the H-D exchange experiments, most amino acid residues showed a mixture of exchange faster than one minute and exchange slower than one week, indicating that although 4°C and 37°C fibrils each include infectious particles producing mostly a single prion variant, the amyloids in each case are significantly heterogeneous.
Our earlier experiments used dried Sup35NM and Ure21–89 fibrils which, in principle, might alter their structure or infectivity. Here, we show that wet and dry fibrils have the same infectivity, and that, as previously shown for the dry fibrils, the wet Sup35NM fibrils have an in-register parallel β-sheet structure. Our earlier experiments used infectious fibrils, but those fibrils produced a mixture of prion variants. Here, we prepared fibrils at 4°C and 37°C following Tanaka et al. (8), and similarly found that they produced different variants on transfection into yeast. We showed that in both cases the N (prion) domain had in-register parallel β-sheet structure based on data with Tyr-1-13C labeled fibrils.
We previously estimated that 6–7 of the 8 Leu residues of Sup35NM fibrils formed at 20°C were in β-sheets, and that all β-sheet Leu residues were in in-register parallel structure (37). This indicated that, unexpectedly, the in-register parallel structure extended into the highly charged M domain. Our present data on wet fibrils formed at 4°C or 37°C yields an estimate of 4 residues in in-register parallel β-sheet, consistent with results of Toyama et al. (27), both confirming that the in-register parallel β-sheet structure extends into the charged M domain which contains 7 of the 8 leucines. The different estimates may reflect the different temperatures of sample preparation, a factor shown to be critical in determining structure (27).
Of course, an important caveat of all studies of infectious amyloids is that although yeast prion amyloids are highly infectious, the particle/infectious unit ratio must be quite high, so that it is always possible that the physical properties of the bulk of the amyloid are not those of the minority infectious fibrils.
Given that different variants have in-register parallel β-sheet structure, what is the nature of the difference? The ~7–10 nm diameter of Sup35NM fibrils is a fraction of the length of the extended 123 residue N domain (~40 nm) implying that the sheet must be folded several times along the fibril axis. We have previously suggested that variants may differ by the location of the folds or the length of the loops. Moreover, the data of King and of Weissman (27, 60) suggest that the extent of β-sheet structure differs among variants. Differences in the staggering of β-strands in the multilayered structure are also possible (28).
The most important feature of our finding the in-register parallel β-sheet structure for yeast prions of Sup35p, Ure2p and Rnq1p is that this structure provides a clear explanation for how variant information is inherited (39, 61, 62). The end of the fibril provides a template guiding the conformation assumed by the new monomer joining the fibril to be the same as the last one. The ‘polar zipper’ bonds of Asn and Gln residues(63–65), possible similar interactions between Ser and Thr residues, and the hydrophobic interactions all favor the in-register parallel structure. Only the repulsion of like-charged residues work against forming this structure, and such residues are few in the prion domains of Sup35p, Ure2p and Rnq1p.
Supplementary Material
Acknowledgments
We thank Brian Kimble for help with protein purification and our colleagues for critical reading of the manuscript.
Abbreviations
- CSA
chemical shift anisotropy
- NMR
nuclear magnetic resonance
- MAS
magic angle spinning
Footnotes
This work was supported by the Intramural Program of the National Institute of Diabetes Digestive and Kidney Diseases.
Supporting Information Available: Supplementary Fig. 1 shows the PITHIRDS-CT data uncorrected for signal from natural abundance 13C. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in S. cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 2.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 3.Roberts BT, Wickner RB. A class of prions that propagate via covalent auto-activation. Genes Dev. 2003;17:2083–2087. doi: 10.1101/gad.1115803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nemecek J, Nakayashiki T, Wickner RB. A prion of yeast metacaspase homolog (Mca1p) detected by a genetic screen. Proc Natl Acad Sci USA in press. 2008 doi: 10.1073/pnas.0812470106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009 doi: 10.1038/ncb1843. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 9.Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: amyloid of Ure2p is infectious. Embo J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel BK, Liebman SW. “Prion proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+] J Mol Biol. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sipe JD. Amyloidosis. Annu Rev Biochem. 1992;61:947–975. doi: 10.1146/annurev.bi.61.070192.004503. [DOI] [PubMed] [Google Scholar]
- 12.Hosoda N, Kobayashii T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S, Katada T. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J Biol Chem. 2003;278:38287–38291. doi: 10.1074/jbc.C300300200. [DOI] [PubMed] [Google Scholar]
- 13.Wilson PG, Culbertson MR. SUF12 suppressor protein of yeast: a fusion protein related to the EF-1 family of elongation factors. J Mol Biol. 1988;199:559–573. doi: 10.1016/0022-2836(88)90301-4. [DOI] [PubMed] [Google Scholar]
- 14.TerAvanesyan A, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frolova L, LeGoff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni A-L, Celis JE, Philippe M, Justesen J, Kisselev L. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 16.Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruce ME. Scrapie strain variation and mutation. Br Med Bull. 1993;49:822–838. doi: 10.1093/oxfordjournals.bmb.a072649. [DOI] [PubMed] [Google Scholar]
- 18.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 19.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlumpberger M, Prusiner SB, Herskowitz I. Induction of distinct [URE3] yeast prion strains. Mol Cell Biol. 2001;21:7035–7046. doi: 10.1128/MCB.21.20.7035-7046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kryndushkin D, Wickner RB. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2149–2154. doi: 10.1091/mbc.E07-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kryndushkin D, Shewmaker F, Wickner RB. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 2008;27:2725–2735. doi: 10.1038/emboj.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kushnirov VV, Kryndushkin D, Boguta M, Smirnov VN, Ter-Avanesyan MD. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- 25.Edskes HK, McCann LM, Hebert AM, Wickner RB. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics. 2009 doi: 10.1534/genetics.108.099929. published ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 28.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Quart Revs Biophys. 2006;1:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 29.Benzinger TL, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC. Propagating structure of Alzheimer’s beta-amyloid(10–35) is parallel beta-sheet with residues in exact register. Proc Natl Acad Sci U S A. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antzutkin ON, Balbach JJ, Leapman RD, Rizzo NW, Reed J, Tycko R. Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of beta-sheets in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2000;97:13045–13050. doi: 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balbach JJ, Petkova AT, Oyler NA, Antzutkin ON, Gordon DJ, Meredith SC, Tycko R. Supramolecular structure in full-length Alzheimer’s beta-amyloid fibrils: Evidence for a parallel beta-sheet organization from solid-state nuclear magnetic resonance. Biophys J. 2002;83:1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci USA. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balbach JJ, Ishii Y, Antzutkin ON, Leapman RD, Rizzo NW, Dyda F, Reed J, Tycko R. Amyloid fibril formation by A beta 16–22, a seven-residue fragment of the Alzheimer’s beta-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 2000;39:13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 34.Petkova AT, Buntkowsky G, Dyda F, Leapman RD, Yau WM, Tycko R. Solid state NMR reveals a pH-dependent antiparallel beta-sheet registry in fibrils formed by a beta-amyloid peptide. Journal of Molecular Biology. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 35.Ritter C, Maddelein ML, Siemer AB, Luhrs T, Ernst M, Meier BH, Saupe SJ, Riek R. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–279) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 37.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baxa U, Wickner RB, Steven AC, Anderson D, Marekov L, Yau W-M, Tycko R. Characterization of β-sheet structure in Ure2p1–89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 39.Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc Natl Acad Sci U S A. 2008;105:2403–2408. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baxa U, Taylor KL, Wall JS, Simon MN, Cheng N, Wickner RB, Steven A. Architecture of Ure2p prion filaments: the N-terminal domain forms a central core fiber. J Biol Chem. 2003;278:43717–43727. doi: 10.1074/jbc.M306004200. [DOI] [PubMed] [Google Scholar]
- 41.Diaz-Avalos R, King CY, Wall JS, Simon M, Caspar DLD. Strain-specific morphologies of yeast prion amyloids. Proc Natl Acad Sci U S A. 2005;102:10165–10170. doi: 10.1073/pnas.0504599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paravastu AK, Petkova AT, Tycko R. Polymorphic fibril formation by residues 10–40 of the Alzheimer’s beta-amyloid polypeptide. Biophys J. 2006;90:4618–4629. doi: 10.1529/biophysj.105.076927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kishimoto A, Hasegawa K, Suzuki H, Taguchi H, Namba K, Yoshida M. beta-Helix is a likely core structure of yeast prion Sup35 amyloid fibers. Biochem Biophys Res Commun. 2004;315:739–745. doi: 10.1016/j.bbrc.2004.01.117. [DOI] [PubMed] [Google Scholar]
- 44.Shewmaker F, Ross ED, Tycko R, Wickner RB. Amyloids of shuffled prion domains that form prions have a parallel in-register β-sheet structure. Biochemistry. 2008;47:4000–4007. doi: 10.1021/bi7024589. [DOI] [PubMed] [Google Scholar]
- 45.Chernoff YO, Lindquist SL, Ono B-I, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 46.Brachmann A, Toombs JA, Ross ED. Reporter assay systems for [URE3] detection and analysis. Methods. 2006;39:35–42. doi: 10.1016/j.ymeth.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Pines A, Gibby MG, Waugh JS. Proton-Enhanced Nmr of Dilute Spins in Solids. J Chem Phys. 1973;59:569–590. [Google Scholar]
- 48.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear Decoupling in Rotating Solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 49.Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid-state NMR. J Chem Phys. 2007;126:064506. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- 50.Petkova AT, Tycko R. Sensitivity enhancement in structural measurements by solid state NMR through pulsed spin locking. J Mag Res. 2002;155:293–299. doi: 10.1006/jmre.2002.2519. [DOI] [PubMed] [Google Scholar]
- 51.Morris GA, Freeman R. Enhancement of nuclear magnetic resonance signals by polarization transfer. J Amer Chem Soc. 1979;101:760–762. [Google Scholar]
- 52.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 53.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 55.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Amer Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 57.Siemer AB, Arnold AA, Ritter C, Westfeld T, Ernst M, Riek R, Meier BH. Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc. 2006;128:13224–13228. doi: 10.1021/ja063639x. [DOI] [PubMed] [Google Scholar]
- 58.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. H-1, C-13 and N-15 Random Coil Nmr Chemical-Shifts of the Common Amino-Acids. 1 Investigations of Nearest-Neighbor Effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 59.King CY. Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J Mol Biol. 2001;307:1247–1260. doi: 10.1006/jmbi.2001.4542. [DOI] [PubMed] [Google Scholar]
- 60.Chang H-Y, Lin J-Y, Lee H-C, Wang H-L, King C-Y. Strain-specific sequences required for yeast prion [PSI+] propagation. Proc Natl Acad Sci U S A. 2008;105:13345–13350. doi: 10.1073/pnas.0802215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ross ED, Minton AP, Wickner RB. Prion domains: sequences, structures and interactions. Nat Cell Biol. 2005;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- 62.Wickner RB, Shewmaker F, Kryndushkin D, Edskes HK. Protein inheritance (prions) based on parallel in-register β-sheet amyloid structures. Bioessays. 2008;30:955–964. doi: 10.1002/bies.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan JCC, Oyler NA, Yau W-M, Tycko R. Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10--39 of the yeast prion protein Ure2p. Biochemistry. 2005;44:10669–10680. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.