Abstract
The world-wide explosion of overweight people has been called an epidemic. The inflammatory nature of obesity is widely recognized. Could it really be an epidemic involving an infectious agent? In this climate of concern over the increasing prevalence of overweight conditions in our society, we focus on the possible role of oral bacteria as a potential direct contributor to obesity. To investigate this possibility, we measured salivary bacterial populations of overweight women. Saliva was collected from 313 women with a body mass index between 27 and 32, and bacterial populations were measured by DNA probe analysis. Levels in this group were compared with data from a population of 232 healthy individuals from periodontal disease studies. The median percentage difference of 7 of the 40 bacterial species measured was greater than 2% in the saliva of overweight women. Classification tree analysis of salivary microbiological composition revealed that 98.4% of the overweight women could be identified by the presence of a single bacterial species (Selenomonas noxia) at levels greater than 1.05% of the total salivary bacteria. Analysis of these data suggests that the composition of salivary bacteria changes in overweight women. It seems likely that these bacterial species could serve as biological indicators of a developing overweight condition. Of even greater interest, and the subject of future research, is the possibility that oral bacteria may participate in the pathology that leads to obesity.
Keywords: bacterial physiology, obesity, overweight, infectobesity saliva, body mass index, oral health
Introduction
The bacteria of the oral cavity can be altered in disease conditions such as oral cancer and dental caries. The meanings of these associations can be useful diagnostics and potentially reflect an underlying etiology. With the recognition that there are likely as many as 700 bacterial species that can inhabit the oral cavity of man (Paster et al., 2001), and the development of molecular microbiological assays based on DNA probe arrays (Paster et al., 2001; Socransky et al., 2004), the screening of individuals in various disease categories for changes in oral bacterial populations becomes a practical procedure that can provide insight into the pathological manifestations and potential etiologic consequences of the underlying disease process.
Methods
Overweight individuals were recruited from a Boston population by subway advertisement. After obtaining informed consent, we screened 410 persons, and 313 Caucasian females in good general health, between the ages of 20 and 45 yrs, with a body mass index (BMI) between 27 and 32, were recruited for this study. Women who smoked were not excluded. Individuals accepted into the study were not on long-term medication (e.g., hypertensives, psychiatric therapies) and did not have chronic or acute illnesses requiring chronic medical treatment and/or systemic therapy.
The control population consisted of 232 healthy individuals (58% female), also from Boston, greater than 18 yrs of age (40 ± 16 yrs), with at least 20 teeth (average missing, 2.0 ± 2.3) and no known systemic diseases (e.g., diabetes or AIDS). These individuals had not received systemic antibiotic therapy within the preceding 3 mos and did not require antibiotic prophylaxis. They were healthy controls for periodontal disease studies and had no pocket depth or attachment level measurements > 3 mm.
Salivary bacterial species were identified and enumerated by DNA probe analysis with whole genomic probes (Socransky et al., 2004). Each person provided a 1 to 3 mL sample of whole unstimulated saliva by expectorating into a sterile graduated cylinder. A 0.2-mL sample of whole saliva was mixed by vortex with 1.5 mL filter-sterilized Tris EDTA buffer. A 0.2-mL sample of this diluted saliva was transferred to a new tube, and 0.1 mL of 0.5 M NaOH was added. This sample was heated in a boiling water bath for 10 min, neutralized by the addition of 0.8 mL 5 M ammonium acetate, applied to the surface of a nylon membrane, and evaluated individually for the levels of 40 species of oral bacteria, by means of the “checkerboard” DNA probe set. Numbers of bacteria in each sample were determined by analysis of scanned images of samples compared with standards and computed as 100 x log10 N, with N being the number of bacteria in each of the 40 bacterial species monitored for each of the saliva samples. The DNA probe method was adjusted to detect approximately 104 bacteria (sensitivity), with 93.5% of cross-reactions exhibiting less than 5% of the homologous probe signal (specificity).
For each saliva sample, numbers of each of the 40 bacteria were divided by the sum of the numbers of all 40 bacterial species from that sample and multiplied by 100 to obtain an estimate of the percentage of that species in each sample (DNA percent). This procedure is justified for estimating percentages, since the total counts of these 40 bacteria have been found to represent 55%-60% of the bacteria in subgingival biofilms. Medians were computed as descriptive variables for bacteria in saliva samples. Differences between healthy and overweight individuals were tested by the Mann-Whitney U-test adjusted for the reduction in power due to multiple comparisons. Categorization analysis was accomplished with commercial software (CART, Salford Systems, Sunnyvale, CA, USA), with percentages of bacteria as the predictor and class (healthy or overweight) as the target variable, with the Gini algorithm.
Results
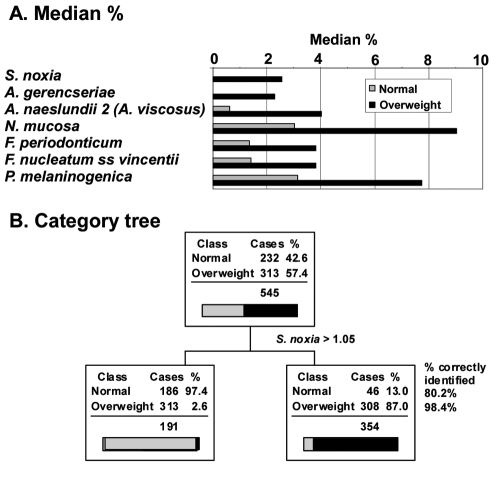
As part of a larger study, we measured salivary bacterial populations of 313 overweight women, targeting 40 bacterial species from 6 bacterial phyla (Euzeby, 2008). The median percentage difference of 7 of the 40 species measured was greater than 2% in the saliva of overweight individuals (Fig. 1A, Table). This included one species of Selenomonas in the phylum Firmicutes (S. noxia), two species of Actinomyces in the phylum Actinobacterium (A. gerencseriae and A. naeslundii 2), one species of Neisseria in the phylum Proteobacteria (N. mucosa), two species of Fusobacterium in the phylum Fusobacteria (F. periodonticum and F. nucleatum ss vincentii), and one species of Prevotella in the phylum Bacteroidetes (P. melaninogenica). All Firmicutes, except Eubacterium sp. and G. morbillorum, had a significantly greater median percent in overweight individuals. Significantly greater median percentages were seen in species of the other 5 phyla as well.
Figure 1.
Median percent of salivary bacteria most prominent in overweight individuals and identification by salivary levels of S. noxia. (A) Median percentage of salivary bacteria increasing with overweight relative to normal weight. Only changes greater than 2% are illustrated and sorted in the order of Normal:Overweight ratio. No reductions in median % of this magnitude were observed. (B). Categorization analysis based on the level of S. noxia greater than 1.05%. When S. noxia was considered as a diagnostic for overweight, it correctly identified 98.4% of individuals who were overweight (sensitivity) and 80.2% of individuals who were normal (specificity).
Table.
Median Percentage of Salivary Bacteria
| Bacteria | Healthy Median % | Overweight Median % | Difference | p < 0.001 |
|---|---|---|---|---|
| Firmicutes | ||||
| Selenomonas noxia | 0.00 | 2.54 | 2.54 | * |
| Veillonella parvula | 4.97 | 6.64 | 1.67 | * |
| Streptococcus mitis | 1.71 | 3.17 | 1.46 | * |
| Streptococcus anginosus | 0.00 | 1.16 | 1.16 | * |
| Streptococcus oralis | 1.31 | 2.40 | 1.09 | * |
| Streptococcus gordonii | 0.53 | 1.55 | 1.01 | * |
| Streptococcus intermedius | 0.00 | 0.63 | 0.63 | * |
| Streptococcus sanguis | 0.71 | 1.18 | 0.47 | * |
| Peptostreptococcus micros | 0.87 | 1.33 | 0.46 | * |
| Streptococcus constellatus | 0.57 | 1.01 | 0.45 | * |
| Eubacterium nodatum | 0.62 | 0.46 | −0.16 | |
| Eubacterium saburreum | 2.01 | 1.56 | −0.45 | |
| Gemella morbillorum | 2.71 | 1.66 | −1.05 | |
| Bacteroidetes | ||||
| Prevotella melaninogenica | 3.13 | 7.75 | 4.63 | * |
| Prevotella intermedia | 1.44 | 3.11 | 1.67 | * |
| Prevotella nigrescens | 1.74 | 3.30 | 1.56 | * |
| Capnocytophaga ochracea | 0.00 | 1.29 | 1.29 | * |
| Porphyromonas gingivalis | 0.39 | 1.57 | 1.18 | * |
| Capnocytophaga sputigena | 0.00 | 0.92 | 0.92 | * |
| Tanerella forsythia | 0.00 | 0.90 | 0.90 | * |
| Capnocytophaga gingivalis | 0.00 | 0.75 | 0.75 | * |
| Proteobacteria | ||||
| Neisseria mucosa | 3.02 | 9.02 | 6.00 | * |
| Aggregatibacter | ||||
| actinomycetemcomitans | 0.38 | 1.52 | 1.14 | * |
| Campylobacter rectus | 0.55 | 1.05 | 0.50 | * |
| Campylobacter showae | 1.41 | 1.75 | 0.34 | |
| Campylobacter gracilis | 1.09 | 1.06 | −0.03 | |
| Eikenella corrodens | 0.89 | 0.58 | −0.32 | * |
| Fusobacteria | ||||
| Fusobacterium periodonticum | 1.35 | 3.82 | 2.47 | * |
| Fusobacterium nucleatum | ||||
| ss nucleatum | 1.41 | 3.83 | 2.42 | * |
| Fusobacterium nucleatum | ||||
| ss vincentii | 1.34 | 3.28 | 1.94 | * |
| Leptotrichia buccalis | 1.38 | 1.61 | 0.23 | |
| Fusobacterium nucleatum | ||||
| ss polymorphum | 1.95 | 1.91 | −0.04 | |
| Actinobacteria | ||||
| Actinomyces naeslundii 2 | ||||
| (A. viscosus) | 0.61 | 4.01 | 3.41 | * |
| Actinomyces gerencseriae | 0.00 | 2.28 | 2.28 | * |
| Actinomyces naeslundii 1 | 0.43 | 1.69 | 1.27 | * |
| Actinomyces israelii | 0.00 | 1.11 | 1.11 | * |
| Propionibacterium acnes | ||||
| (serotypes I & II) | 0.43 | 1.36 | 0.93 | * |
| Actinomyces odontolyticus | ||||
| (serotype I) | 0.50 | 0.73 | 0.23 | |
| Spirochaetae | ||||
| Treponema socranskii | 0.00 | 1.14 | 1.14 | * |
| Treponema denticola | 0.18 | 1.05 | 0.87 | * |
Mann-Whitney U-test for 545 cases. Data are grouped by phyla and sorted by median percent difference between 232 normal and 313 overweight individuals. Probability values are computed from the non-parametric Mann-Whitney U-test. Values significant at p < 0.0013 are indicated with a “*” as an adjustment for multiple comparison for an overall p < 0.05.
Classification tree topology (Steinberg and Colla, 1997), a computer algorithm that seeks from a set of many variables (in this case, % of each of 40 bacteria) the combination that best separates the defined classes (in this case, healthy and overweight individuals), was used to identify the most prevalent component bacteria. The method computes a diagnostic sensitivity and specificity for identification of categories. This approach is not the same as selecting parameters based on the largest differences. Classification tree topology of salivary bacteria data in predicting health or overweight conditions (Fig. 1B) demonstrated that 98.4% of the overweight individuals were correctly identified by the presence of the single bacterium S. noxia > 1.05% to the exclusion of all others. Although all Firmicutes identified overweight individuals with ≥ 95% probability (sensitivity), their identification of healthy individuals was only 40%-60% (specificity). More complex models involving more bacterial species served only to “subset” healthy persons, with no further improvement in recognition of overweight cases. As a diagnostic to identify obesity, the condition of S. noxia > 1.05% had a sensitivity of 98.4 and a specificity of 80.2. The importance of this variable in the prediction model was twice that of the next closest bacterium.
Discussion
What is the possibility that weight gain is directly the result of a change in oral flora? The results of this study suggest that of the 40 species surveyed, levels of many bacteria differed in the saliva of overweight women when compared with levels in the saliva of healthy individuals. In particular, the percentage of the bacterium S. noxia was capable of identifying 98.4% of overweight women from a group of generally healthy persons, a surprisingly high degree of diagnostic power.
S. noxia could be considered a disease candidate. Selenomonads, in general, are motile, crescent-shaped, non-spore-forming, Gram-negative bacteria which actively ferment glucose to produce propionic acid, and are obligate anaerobes found in both the mouth and the gastro-intestinal tract. The type strain for S. noxia (ATCC 43541) was isolated from the gingival crevice of a person with rapidly progressive periodontitis. It was among the species found to increase in experimental gingivitis (Selenomonas D4) in both children and adults (Moore et al., 1984). It was one of the candidate species associated with de novo development of periodontal disease, where the percentage in periodontal plaque increased from 0.9% at healthy sites to 5.9% in disease-active interproximal sites (Tanner et al., 1998). It has also been reported to be elevated in the mouths of mothers giving birth to pre-term low-birthweight babies (Buduneli et al., 2005). Although these studies suggest that S. noxia may be capable of triggering an inflammatory reaction and presumably stimulating release of inflammatory mediators, they do not immediately suggest a link between S. noxia and obesity.
Several findings indirectly support the hypothesis that oral bacteria could be related to obesity. The most direct evidence for a bacterial association with obesity comes from animal studies (DiBaise et al., 2008). In these studies, germ-free C57BL_6 mice were shown to eat more but to gain less weight than littermates infected by fecal samples from wild-type mice. Concomitantly, levels of glucose, insulin, and leptin were higher in conventionalized mice (i.e., germ-free mice exposed to cecal contents of wild-type mice), indicating the development of an insulin-resistant state. These studies demonstrated that gut microbiota promoted absorption of monosaccharides in these mice, which resulted in lipogenesis. Adult mice infected from wild-type mice had 60% more fat than their germ-free counterparts. In subsequent studies, this group demonstrated that bacteria responsible for this effect in mice were in the Firmicutes phylum. It is interesting to note that S. noxia was the only Firmicutes that was significantly elevated in saliva (Table, Fig. 2). Other investigators have shown that this tendency to gain weight in mice is transmissible between mice by coprophagia. Elevated levels of Firmicutes relative to Bacteroidetes were also found in studies of the gene sequences of human gut bacterial species. The authors hypothesized that individuals predisposed to obesity may have gut levels of Firmicutes that promote more efficient extraction and/or storage of energy from a given diet, compared with lean individuals, and that intentional manipulation of gut microbiology may be useful for controlling weight in overweight individuals. The condition of obesity arising from infection has been called “infectobesity”. Although these studies focused on gut bacteria, all gastrointestinal bacteria pass through the oral cavity at some time, and some of those transients might be located in, if not seeded from, the oral cavity. It has been estimated that approximately 1 gram of bacteria (1011) is swallowed with the 500 to 1500 mL of saliva produced daily (Socransky and Haffajee, 2005). If the levels of S. noxia are > 1.05%, this represents approximately 109 cells swallowed each day. It is therefore plausible that salivary microbiology would affect gastrointestinal microbiology. This sentiment is echoed by the great interest in orally administered exogenous bacteria (probiotic therapy).
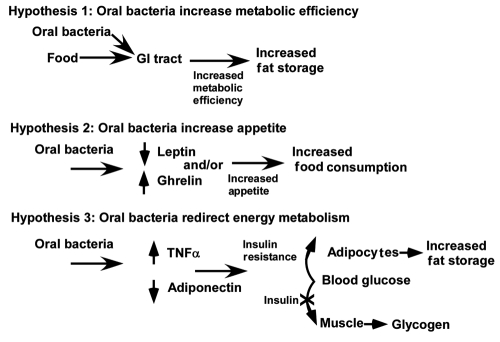
Figure 2.
Possible mechanisms by which oral bacteria could affect body weight and contribute to obesity.
In a completely different context, investigators studying the social interactions between overweight and non-overweight humans (Christakis and Fowler, 2007) found that obesity appears to spread through social associations. Based on a study of social interactions among 12,607 participants in the Framingham Heart Study, a person’s chances of becoming overweight increased by 57% if he or she had a close friend who became overweight. This was true of siblings and spouses, but not of neighbors. This spread is reminiscent of that described for the periodontal pathogen P. gingivalis, where spouses and children of individuals with periodontal disease have higher levels of “pathogens” than those who do not have periodontal disease (Socransky and Haffajee, 2005).
Two prevalent oral diseases are found in most, if not all, people: dental caries and periodontal disease. Dental caries, the most common oral disease, is clearly related to the consumption of fermentable carbohydrates. Since obesity has also been related to carbohydrate consumption, one would think that dental caries and obesity would correlate strongly. Such is not the case. Most authors have not reported a positive association.
In contrast to dental caries, the association between periodontal disease and obesity is subtle, but widely acknowledged. Both periodontal disease and obesity produce increased levels of inflammatory mediators. In a review of this subject (Ritchie, 2007), fat was recognized as a reservoir for inflammatory cytokines, and it was suggested that obesity would likely affect periodontal disease. In the context of this paper, however, we would propose the converse by considering the likelihood that periodontal disease may contribute to the development of obesity. Periodontal disease is an inflammatory condition thought to be caused by the “red complex” bacteria P. gingivalis, T. denticola, and T. forsythia. By community ordination analysis, S. noxia was found as an outlier, not commonly associated with any of the periodontal disease complexes. Studies of early periodontal lesions (Tanner et al., 1998), however, suggested that T. forsythia, C. rectus, and S. noxia were principal species associated with sites converting from periodontal health to disease. Periodontitis has also been significantly correlated with overweight conditions. Analysis of data from a national health survey (NHANES III) indicated that BMI and periodontal attachment loss are positively correlated (Wood et al., 2003). Analysis of populations with and without periodontitis indicate that 70% with periodontitis are overweight or obese, as compared with 37% of the healthy individuals (Socransky and Haffajee, 2005). In addition, overweight individuals appeared to have more severe periodontal disease, as evaluated by plaque, bleeding on probing, pocket depth, attachment loss, and percentage of subgingival T. forsythia.
Tumor necrosis factor-α (TNFα) is one of many pro-inflammatory cytokines produced by diseased periodontal tissues that could be a pivotal inflammatory cytokine encouraging obesity. TNFα increases insulin resistance, induces C-reactive peptide production, and inhibits adiponectin, an important anti-inflammatory adipokine. Increased levels of TNFα in gingival crevice fluid have been shown to correlate with increased body mass index (Lundin et al., 2004). Treatment of periodontitis with locally administered tetracycline and repeated scaling and root planing reduced blood levels of TNFα (Iwamoto et al., (2001) In individuals with diabetes, effective periodontal therapy with antibiotics reduced glycated hemoglobin (Janket et al., 2005). By this mechanism, periodontal bacteria would be seen to stimulate the formation of inflammatory cytokines such as TNFα that divert energy metabolism to lipid synthesis, perhaps contributing to obesity.
Oral bacteria may contribute to the development of obesity by at least 3 mechanisms (Fig. 2). First, the oral bacteria may contribute to increased metabolic efficiency, as suggested by the infectobesity proponents. It is instructive to note that, by this mechanism, even a small excess in calorie consumption, with no change in diet or exercise, might result in unacceptable weight gain. An increase by 5% (100 calories/day) would add approximately 10 pounds of fat per year. A second hypothesis is that oral bacteria could increase weight gain by increasing appetite. Even in the absence of data, the teleology of this proposition is so attractive that it should be mentioned in this context. By stimulating host appetite, the bacteria get more to eat! A third hypothesis is that oral bacteria redirect energy metabolism by facilitating insulin resistance through increasing levels of TNFα or reducing levels of adiponectin.
By any of these mechanisms, even a small excess in calorie consumption with no change in diet or exercise could result in unacceptable weight gain. By the same logic, individuals who are infected may have reduced their caloric intake and/or increased their exercise to compensate and not appear overweight. Hence, any study of infection-related weight gain should include a measure of food consumption and exercise, which was not incorporated into our pilot study. It should also be recognized that an important limitation of the study as conducted is the use of a convenience population as a control, rather than using selected cohorts. This reservation aside, however, rarely have data been reported from saliva in which S. noxia represented such a large percentage of the oral bacterial population (Mager et al., 2003). While it is not justified to suggest that S. noxia infections have an etiologic role in obesity, based on these data, it is reasonable to suggest that S. noxia may be an indicator of change in oral microbial ecology.
The reasons for a relationship between obesity and oral bacteria are undoubtedly complex and varied. The relationship may be circumstantial, as being related to diet. It could be opportunistic, such as proliferation driven by metabolic changes that have occurred in the host. It could also be causal, as participating in initiation or propagation of the disease. Whatever the reasons, it is clear that the parallel microbiological universe that travels with man changes as man changes, and appears to be affected by a tendency to gain weight.
Acknowledgments
Special appreciation to those who aided in this study, including B. Newman, J. Leonel, C. Floros, M. Chvetchkova, M. Sweeney, and E. Regan.
Footnotes
This study was funded by Interleukin Genetics of Waltham, MA, USA, and by DE018184 from the National Institute of Dental and Craniofacial Research.
References
- Buduneli N, Baylas H, Buduneli E, Turkoglu O, Kose T, Dahlén G. (2005). Periodontal infections and pre-term low birth weight: a case-control study. J Clin Periodontol 32:174-181 [DOI] [PubMed] [Google Scholar]
- Christakis NA, Fowler JH. (2007). The spread of obesity in a large social network over 32 years. N Engl J Med 357:370-379 [DOI] [PubMed] [Google Scholar]
- DiBaise JK, Zhang H, Crowell MD, Krajmalnik-Brown R, Decker GA, Rittmann BE. (2008). Gut microbiota and its possible relationship with obesity. Mayo Clin Proc 83:460-469 [DOI] [PubMed] [Google Scholar]
- Euzeby JP. (2008). List of prokaryotic names with standing in nomenclature: a folder available on the Internet. Classifications of domains and phyla, pp. 590-592 http://www.bacterio.cict.fr/ [DOI] [PubMed]
- Iwamoto Y, Nishimura F, Nakagawa M, Sugimoto H, Shikata K, Makino H, et al. (2001). The effect of antimicrobial periodontal treatment on circulating tumor necrosis factor-alpha and glycated hemoglobin level in patients with type 2 diabetes. J Periodontol 72:774-778 [DOI] [PubMed] [Google Scholar]
- Janket SJ, Wightman A, Baird AE, Van Dyke TE, Jones JA. (2005). Does periodontal treatment improve glycemic control in diabetic patients? A meta-analysis of intervention studies. J Dent Res 84:1154-1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin M, Yucel-Lindberg T, Dahllöf G, Marcus C, Modéer T. (2004). Correlation between TNFalpha in gingival crevicular fluid and body mass index in obese subjects. Acta Odontol Scand 62:273-277 [DOI] [PubMed] [Google Scholar]
- Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. (2003). Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol 30:644-654 [DOI] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Smibert RM, Cato EP, Burmeister JA, Palcanis KG, et al. (1984). Bacteriology of experimental gingivitis in children. Infect Immun 46:1-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. (2001). Bacterial diversity in human subgingival plaque. J Bacteriol 183:3770-3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie CS. (2007). Obesity and periodontal disease. Periodontol 2000 44:154-163 [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. (2005). Periodontal microbial ecology. Periodontol 2000 38:135-187 [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Smith C, Martin L, Haffajee JA, Uzel NG, et al. (2004). Use of checkerboard DNA-DNA hybridization to study complex microbial ecosystems. Oral Microbiol Immunol 19:352-362 [DOI] [PubMed] [Google Scholar]
- Steinberg D, Colla P. (1997). CART—classification and regression trees. San Diego, CA:Salford Systems [Google Scholar]
- Tanner A, Maiden MF, Macuch PJ, Murray LL, Kent RL., Jr (1998). Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol 25:85-98 [DOI] [PubMed] [Google Scholar]
- Wood N, Johnson RB, Streckfus CF. (2003). Comparison of body composition and periodontal disease using nutritional assessment techniques: Third National Health and Nutrition Examination Survey (NHANES III). J Clin Periodontol 30:321-327 [DOI] [PubMed] [Google Scholar]