Abstract
Targeted introduction of a double-stranded break (DSB) using designer zinc finger nucleases (ZFNs) in mammalian cells greatly enhances gene targeting - homologous recombination (HR) at a chosen endogenous target gene, which otherwise is limited by low spontaneous rate of HR. Here, we report that efficient ZFN-mediated gene correction occurs at a transduced, transcriptionally active, mutant GFP locus by homology-directed repair, and that efficient mutagenesis by non-homologous end joining (NHEJ) occurs at the endogenous, transcriptionally silent, CCR5 locus in HEK293 Flp-In cells, using designed 3- and 4-finger ZFNs. No mutagenesis by NHEJ was observed at the CCR2 locus, which has ZFN sites that are distantly related to the targeted CCR5 sites. We also observed efficient ZFN-mediated correction of a point mutation at the endogenous mutant tyrosinase chromosomal locus in albino mouse melanocytes, using designed 3-finger ZFNs. Furthermore, re-engineered obligate heterodimer FokI nuclease domain variants appear to completely eliminate or greatly reduce the toxicity of ZFNs to mammalian cells, including human cells.
Keywords: Custom-designed zinc finger nucleases, Homologous recombination, Homology-directed repair, Non-homologous end joining, ZFN-mediated gene targeting
INTRODUCTION
Gene modification by homologous recombination (HR) is a relatively inefficient process mammalian cells, typically occurring at a low frequency of about one event per 105 - 107treated cells as compared to the high rate of random, non-targeted integration of the donor DNA (non-homologous joining; NHEJ), which occurs in about one cell per 102 - 105 treated cells. Thus, in most cells, targeted recombinants (i.e. the intended events’ occurring via HR) are overshadowed by more than 1000-fold higher random, non-targeted integrants (i.e. unintended events, occurring via NHEJ). Several approaches including positive-negative selection have been used to enrich the ratio of targeted recombinants to random integrants for applications in mouse embryonic stem (ES) cells (1). However, positive-negative selection achieves only a low degree of enrichment in mammalian cells other mouse ES cells, and many colonies must be screened to identify the targeted recombinants. approaches that substantially enhance the rate of gene targeting have great potential applications in biotechnology (including agriculture) and biomedical sciences (including human gene therapy).
When a defined chromosomal double-strand break (DSB) is introduced at a unique chromosomal site, local HR is induced at that site in a large fraction of the cells with the damage (2). Therefore, if DSB could be artificially introduced uniquely at a given locus in the genome, then HR would efficiently via normal cellular physiologic processes to repair the DSB. Over the past fifteen years, laboratory has engineered chimeric zinc finger nucleases (ZFNs), which combine the non-specific cleavage domain of the FokI restriction enzyme with zinc finger proteins (ZFPs), as molecular tools to introduce a targeted chromosomal DSB (3-8). In principle, ZFNs offer a general means to deliver a site-specific DSB to any genomic site (3). Since a ZFN requires two copies of the 9-bp or 12-bp recognition sites in a tail-to-tail orientation in order to dimerize and produce a DSB, the pair of ZFNs needed to cut at a given genomic site effectively has a 18-bp or 24-bp recognition site, which is long enough to specify unique address within mammalian genomes (4-6). The two binding sites need not be identical, provided the ZFNs that bind to both sites are delivered into the target cells (7-9). Since the recognition specificities of zinc finger motifs are amenable to easy experimental manipulation, gene targeting using designed ZFNs has been successfully applied for genome modification of cells of several species, including frog oocytes (7), Drosophila (10), C.elegans (11), zebra fish (12, 13), plant (14) and human (15-17).
Here, we have designed and characterized 3- and 4-finger ZFNs that target the human CCR5 gene (CCR5) and the mouse tyrosinase gene (TYR), respectively. We have studied the efficiency and efficacy gene targeting of the designed ZFNs in: (i) HEK293 Flp-In cells with an integrated mutant GFP gene encoding the hCCR5 target sequence or the mTYR target sequence and (ii) mouse albino melanocytes (Melan-c cells) (18) and conducted a detailed genetic analysis. We report that efficient ZFN-mediated gene modification occured at a transcriptionally active, transduced mutant GFP locus by homology-directed repair in HEK293 Flp-In cells, using designer ZFNs. In addition, efficient targeted mutagenesis by NHEJ occured at the transcriptionally silent endogenous CCR5 chromosomal locus in the cells. No mutagenesis by NHEJ was observed at the distantly related CCR2 locus, indicating specificity. We also observed efficient targeted ZFN-mediated gene correction at the endogenous chromosomal mutant tyrosinase gene locus in albino mouse melanocytes.
MATERIALS AND METHODS
The materials and methods are described in detail in Supplementary Information.
RESULTS
Design, engineering and characterization of CCR5 ZFNs
We designed, engineered and characterized several pairs of 3- and 4-finger ZFNs that target specific human CCR5 gene sequences: 5′- GTC CCC TTC TGGGCTCACTAT GCT GCC GCC - 3′; 5′- ACT GTC CCC TTC TGGGCTCACTAT GCT GCC GCC CAG - 3′; 5′- GCT CTC ATT TTC CATACA GTC AGT ATC AAT- 3′; and 5′- GTC ATC CTC ATC CTGAT AAA CTG CAA AAG - 3′ (Table S1). The ZFN designs for the latter CCR5 target site, 5′- GTC ATC CTC ATC CTGAT AAA CTG CAA AAG - 3′, were reported previously by Perez et al. (17). For the other CCR5 target sites, the individual ZFs that had been shown in previous studies to bind to a triplet of the target site, were tethered together to form the 3- and 4-finger zinc finger proteins (ZFPs), which were then converted into ZFN as described elsewhere (5). For the inverted ZFN sites that are separated by 12 bp spacer, the ZFPs were fused to the FokI cleavage domain through a 15 amino acids linker, namely (Gly4Ser)3. Since the other inverted ZFN target sites are separated by spacers of 6 bp or less, and are optimally positioned for efficient cleavage (4, 7), the ZFN fusions contained no linker. The pairs of ZFNs were then screened for their sequence-specific cleavage using rabbit reticulocyte, cell-free transcription/translation system (5). The plasmid encoding CCR5 cDNA, pc.CCR5 (NIH AIDS Research and Reference Reagent Program), was efficiently cleaved by the cognate ZFNs (Supplementary Figure 1S). The ZFNs were then cloned into pIRES under the control of the CMV promoter for expression in mammalian cells.
ZFN-mediated gene correction of the transduced, mutant GFP locus in HEK293 Flp-In cells
In order to reduce toxicity of the 3- and 4-finger ZFNs, we generated two sets of previously reported mutant FokI cleavage domain variants and incorporated them into the CCR5-specific ZFNs (15, 19, 20). We also generated a different set of obligate heterodimer FokI nuclease domain variants at PBPL. The 3 different sets of mutant FokI nuclease domain pairs were fused with each pair of 3- and 4-finger designer ZFNs. We generated HEK293 Flp-In cells containing a chromosomal mutant GFP reporter disabled by insertion of either hCCR5 or mTYR ZFN target sequence (Supplementary Information). We tested the pairs of the designed 3- and 4-finger CCR5 ZFNs fused either to the wild-type FokI nuclease domains or the re-designed FokI nuclease domains, using the GFP gene targeting reporter system to monitor their ability to modify the targeted ZFN sites in human cells (15). Gene targeting and recombination was initiated by the co-transfection of the expression plasmids for the pair of ZFNs and the GFP donor template plasmid. With effective gene targeting and HR, the cell becomes GFP positive, which were detected by flow cytometry. The 3-finger CCR5 ZFNs fused to the wild-type FokI did not yield any GFP positive cells, indicating that these were toxic to cells. The re-designed pairs of 3-finger CCR5-ZFNs and 3-finger TYR-ZFNs, however, yielded GFP positive cells 5 days post-transfection as monitored by microscopy (Figure 1B & 1C).
Figure 1.
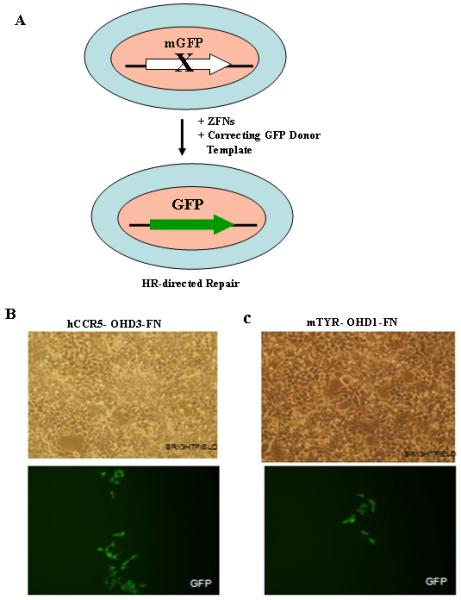
ZFN-mediated gene targeting in HEK293 cells. (A) Schematic representation of ZFN-mediated gene correction of the mutant eGFP locus (disrupted with either the hCCR5 or mTYR ZFN target sites) in HEK293 Flp-In cells. ZFN-mediated gene correction of the mutant eGFP locus using hCCR5-OHD3-FN, 3-finger hCCR5-specific ZFNs containing the FokI nuclease domain variants reported by Miller et al. (19) (B) and mTYR-OHD1-FN, 3-finger mTYR-specific ZFNs containing the FokI nuclease domain variants generated at PBPL (C). GFP positive cells were seen 5 days post-transfection of HEK293 cells with 3-finger ZFNs and donor plasmid. No GFP positive cells were seen by microscopy after transfection of HEK293 cells with donor plasmid alone. OHD depicts obligate heterodimers.
No GFP positive clones were observed post-transfection with the designed 4-finger CCR5 ZFNs (described in this article) fused either to the wild-type FokI domains or the FokI obligate heterodimer nuclease domain variants, indicating that the designed 4-finger ZFNs either did not possess the requisite specificity and affinity to induce targeted cleavage in cells or their target sites were not accessible within human cells, even though they cleaved their cognate substrates in cell-free assays. The results from the experiment with 4-finger CCR5 ZFNs that bind to the target 5′- ACT GTC CCC TTC TGGGCTCACTAT GCT GCC GCC CAG - 3′ was somewhat surprising since this site overlaps the 3-finger site, GTC CCC TTC TGGGCTCACTAT GCT GCC GCC, and the 3-finger CCR5 ZFNs readily stimulate HR to yield GFP positive cells. It appears that the addition of the ZF modules to the corresponding 3-finger CCR5 ZFN designs results in lowered specificity and affinity for the target sites.
We also compared the efficiency and efficacy of the wild-type FokI cleavage domains with different pairs of FokI obligate heterodimer nuclease domain variants (generated by altering amino acid residues at the nuclease domains’ dimer interface) by making fusions to the previously published pair of 4-finger ZFPs that was shown to target CCR5 in human cells (17). ZFN-mediated gene correction at the mutant GFP locus was very efficient in HEK293 Flp-In cells, yielding GFP positive cells upon transduction with the corresponding pairs of ZFNs containing either the wild-type FokI nuclease domains or the FokI obligate heterodimer nuclease domain mutants and correcting plasmid. Transduction with donor alone did not yield any GFP positive cells by microscopy (Figure 2A & 2B). These results indicate that the specificity of the designer ZFNs is a major determinant of the efficiency and efficacy of ZFN-mediated gene targeting in mammalian cells. Quantitative FACS analyses of the GFP positive cells at 3, 5 and 7 days post-transfection with designer ZFNs and donor plasmids are shown in Figure 2C & 2D. The number of surviving GFP positive cells at 7 days post-transfection with ZFNs containing FokI wild type appear to be somewhat lower than those for ZFNs containing the obligate heterodimer FokI nuclease domain variants suggesting that the latter may be less toxic to cells, possibly due to reduced off-target cleavage. Once established, the gene-altered cells are viable and they continued to increase in number for 3 weeks (data not shown). We isolated 4 different individual GFP positive clones by serial dilution of FACS sorted GFP positive cells and grew them for genotypic characterization. Three different loci, namely the mutant GFP locus, the endogenous CCR5 locus and the CCR2 locus, were PCR-amplified using the corresponding locus-specific primers from the isolated genomic DNA of the individual GFP positive clones. The PCR DNA from the mutant GFP locus of the gene-altered clones were all resistant to BstXI digestion indicating that gene correction to the wild-type sequence has occurred in the GFP positive clones (Figure 2E). The PCR DNA from the three different loci was then cloned in the pGEMT vector for transformation into E. coli. DNA sequence analysis of at least 5 recombinant E. coli clones from each individual GFP positive clone confirmed that gene correction indeed has occurred (Table 1). DNA sequence analysis of 5 recombinant bacterial clones generated by cloning the PCR-amplified DNA of the endogenous CCR5 locus from each of the four individual GFP positive clones, as expected, showed simple deletion or insertion mutations at the CCR5 locus resulting from NHEJ. While DNA sequence analysis of the GFP+1, GFP+2 and GFP+3 clones showed identical homozygous mutant alleles, GFP+4 clone indicated the presence of heterozygous mutant alleles at the endogenous CCR5 loci (Table 1). No change in the nucleotide sequence of the CCR2 locus was observed in the limited number of GFP positive clones (four in this case) that were sequenced suggesting that the designed pair of 4-finger CCR5 ZFNs did not cleave at the distantly related site (Table 1).
Figure 2.
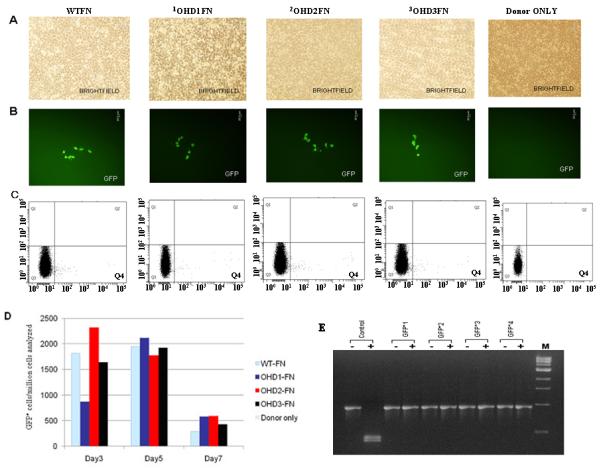
Efficiency and efficacy of ZFN-mediated gene correction in HEK293 cells using 4-finger CCR5-specific ZFNs fused to different FokI nuclease domain variants. Panel (A) brightfield; Panel (B) GFP positive cells seen post-transfection of HEK293 Flp-In cells with 4-finger CCR5-ZFNs and donor plasmids; Panel (C) FACS analyses (Q4 shows GFP+Cells). WT-FN, CCR5-ZFN constructs carrying the wild-type FokI nuclease domains. OHD depicts obligate heterodimers. OHD1-FN, CCR5-ZFN constructs carrying the FokI nuclease domain mutants that were generated at PBPL. OHD2-FN, CCR5-ZFN constructs carrying the FokI nuclease domain mutants reported by Szczepek et al., 2007 (20). OHD3-FN, CCR5-ZFN constructs carrying the FokI nuclease domain mutants reported by Miller et al., 2007 (19). (D) Frequency of gene correction in HEK293 Flp-In cells of a chromosomal mutant GFP reporter disabled by insertion of the CCR5 ZFN target sequence. Quantitative FACS analyses of the GFP positive cells at 3, 5 and 7 days post-transfection with designer ZFNs and donor plasmids. WT-FN, ZFN constructs carrying the wild-type FokI nuclease domains. (E) Analysis of the genotype of four different individual GFP positive clones. Five days post-transfection with ZFNs and the donor plasmids, GFP positive cells were sorted, serially diluted to get individual clones and grown. The genomic DNA was isolated from the GFP positive clones and the eGFP gene at the Flp-In locus was PCR amplified and digested with BstXI. The mutant eGFP gene has two BstXI sites, where the ZFN binding sites are inserted. Correction of the eGFP gene by homology-directed repair results in the loss of the BstXI sites. The PCR product size of the corrected eGFP gene is 930 bp as compared to 990 bp for the mutant gene. BstXI digestion of the mutant eGFP PCR product generates two bands: 450 bp and 540 bp, respectively. Lanes: Control, PCR product of the mutant eGFP gene from untransfected cells before (-) and after (+) digestion with BstXI; GFP+1-4, PCR products of 4 different individual clones obtained from GFP positive sorted cells before (-) and after (+) digestion with BstXI; M, 1 Kb ladder. All GFP positive cells are resistant to BstXI digestion, confirming ZFN-mediated eGFP gene correction in these cells.
Table 1. Genotypic analysis of the eGFP, CCR5 and CCR2 loci in 4 different GFP+ clones.
|
Mutant eGFP locus |
BstXI CCR5 sequence BstXI ...CAGCCGCTACCCATAATAATGGGTCATCCTCATCCTGATAAACTGCAAAAGCCATGATGATGGAAGCAGCACGA... |
| Transduced eGFP locus | .....................CAGCCGCTACCCCGACCACATGAAGCAGCACGA.....................WT .....................CAGCCGCTACCCCGACCACATGAAGCAGCACGA.....................GFP+1 (5) .....................CAGCCGCTACCCCGACCACATGAAGCAGCACGA.....................GFP+2 (5) .....................CAGCCGCTACCCCGACCACATGAAGCAGCACGA.....................GFP+3 (5) .....................CAGCCGCTACCCCGACCACATGAAGCAGCACGA.....................GFP+4 (6) * |
| #Endogenous CCR5 locus | ZFN-L ZFN-R ..................GGTCATCCTCATCCTGATAAACTGCAAAAGG.................................WT ..................GGTCATCCTCATCCTGATctgatAAACTGCAAAAGG.................................GFP+1 (5) ..................GGTCATCCTCATCCTGATctgatAAACTGCAAAAGG.................................GFP+2 (5) ..................GGTCATCCTCATCCTGATctgatAAACTGCAAAAGG.................................GFP+3 (5) ..................GGTCATCCTCAT---------------AACTGCAAAAGG.................................GFP+4 (4) * ..................GGTCATCCTCATCC------------AACTGCAAAAGG.................................GFP+4 (2) * |
| Endogenous CCR2 locus | ZFN-L ZFN-R ..................TGGTCGTCCTCATCTTAATAAACTGCAAAAAGC...........................WT ..................TGGTCGTCCTCATCTTAATAAACTGCAAAAAGC...........................GFP+1 (5) ..................TGGTCGTCCTCATCTTAATAAACTGCAAAAAGC...........................GFP+2 (5) ..................TGGTCGTCCTCATCTTAATAAACTGCAAAAAGC...........................GFP+3 (5) ..................TGGTCGTCCTCATCTTAATAAACTGCAAAAAGC...........................GFP+4 (6) * |
The number of recombinant E. coli clones containing the PCR fragment from each of the GFP+ clone that were sequenced are shown in brackets. Sequences of six recombinant E. coli clones were determined for the GFP+4 clone and 5 for the rest.
NHEJ mutants with bp deletions at the CCR5 locus are depicted by dashes (-), While mutants with bp insertions are shown by bold type and lower case letters.
ZFN-mediated gene correction of the mutant tyrosinase locus in albino mouse melanocytes
Tyrosinase is an essential enzyme in melanin biosynthesis, which is expressed exclusively in melanocytes. Enzymatic activity of tyrosinase is sufficient for the conversion of tyrosine to DOPA and to melanin. Melanocytes derived from albino mice (Melan-C cells) contain a homozygous point mutation (TGT → TCT) in the tyrosinase gene (18). This results in an amino acid sequence change from Cys to Ser at position 85 of the amino acid sequence of the mature tyrosinase, complete inactivation of the tyrosinase enzymatic activity, and absence of cellular pigmentation. Targeted gene correction of this point mutation results in the restoration of tyrosinase activity; melanin biosynthesis and pigmentation change is observed both in vitro and in vivo (18). We used this system to explore the efficacy and efficiency of ZFN-mediated gene targeting of a pre-determined endogenous chromosomal locus within the genome of albino mouse melanocytes.
The design, engineering and characterization of TYR ZFNs that target the site 5′- TTC CCC TTC AAAGGG GTG GAT GAC - 3′ in the first exon of the tyrosinase gene on chromosome 7 of the mouse genome using cell-free assays is described elsewhere (8). The ZFN target site is located ∼ 80 bp distant from the homozygous point mutation (Cys->Ser) site with in the mTYR gene. Pigmented cells were observed by microscopy, as early as 4 days post-nucleofection of Melan-c cells with TYR-specific 3-finger ZFNs containing wild-type FokI cleavage domains and the correcting plasmid. Four different TYR gene-corrected, pigmented Melan-c cells, at 6 days after nucleofection are shown in Figure 3A. The RFLP analysis to determine the genotype of 3 gene-corrected clones (that had originated from individual pigmented Melan-c cells) is shown in Figure 3B. The 179 bp fragment from the DdeI digestion is representative of the wild-type TYR sequence, while the144 bp fragment results from the mutant TYR sequence. The presence of both 179 and 144 fragments in 3 pigmented clones indicates targeted gene-correction of only 1 allele of the 2 chromosomal mutant copies present in (albino) Melan-c cells (Figure 3B). Furthermore, the designed TYR ZFNs containing the obligate heterodimer FokI nuclease domain variants were further tested for mutant eGFP correction using the HEK293 cell line with an integrated copy of mutant eGFP encoding the TYR ZFN target sites. These experiments yielded GFP positive cells (Figure 1C), confirming that gene correction has occurred at the mutant eGFP locus, and that the designed 3-finger TYR ZFNs have the requisite specificity for their cognate sites in cells.
Figure 3.
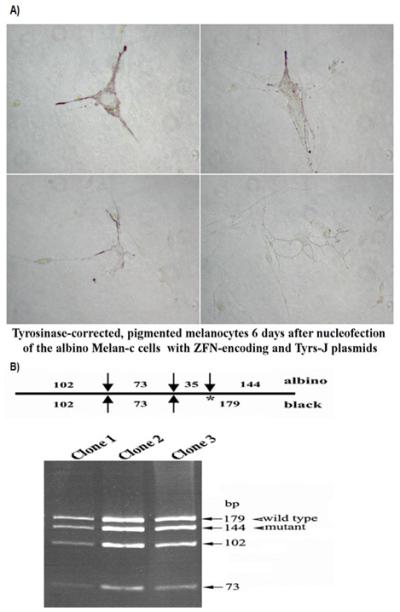
Phenotypic and genotypic changes in the albino mouse melanocytes. A) Four different tyrosinase gene-corrected pigmented melanocytes 6 days post nucleofection of Melan-C cells (albino melanocytes) with the TYR ZFNs and the correcting plasmid (Tyrs J), are shown. B) Three pigmented clones (that had originated from individual pigmented cells) were obtained and correction of the mTYR gene was monitored by RFLP analysis (18). Presence of both 179 bp (a marker for the wild-type) and 144 bp (a marker for the mutant allele) fragments suggest targeted correction of one of the mutant tyrosinase alleles in the pigmented mouse albino melanocytes.
The Melan-c cell culture results indicated that gene correction using 3-finger TYR ZFNs and correcting template plasmid was about ∼200-fold more efficient, compared to the control with template DNA alone at that specific chromosomal locus. We did not detect any pigmented clones that were corrected on both alleles, indicating this occurs at much lower efficiency. By performing another cycle of ZFN-evoked HR using the heterozygous pigmented Melan-c clones, one could enrich for homozygous corrected alleles.
DISCUSSION
The studies on human (HEK293 Flp-In) cells and albino mouse melanocytes reported here provide further evidence that ZFN-based strategies offer a powerful and efficient approach for targeted genetic modification of mammalian cells. While all the designed 3- and 4-finger ZFNs were active in cell-free assays, none of the ZFNs containing the wild-type FokI cleavage domains showed any activity in HEK293 Flp-In cells, when monitored using the GFP gene targeting reporter system. In the case 3-finger ZFNs this is likely due to off-target cleavage, since the CCR5-specific 3-finger ZFNs and the TYR-specific 3-finger ZFNs were active in the GFP gene targeting reporter system, when the corresponding ZFPs were fused to the FokI obligate heterodimer nuclease domain variants. In the case of 4-finger ZFNs, it could be that they lack the requisite high specificity to find their cognate sequences or that the cognate sites in the genome of HEK293 are not accessible to ZFNs (9).
The sequence-specificity of the designed ZFNs appears to be the major determinant of ZFN activity for efficient gene targeting and for greatly reduced cellular toxicity. The potential of ZFN technology applications in human therapeutics depend on the ability to produce ZFNs that cleave the target sequences with exquisite sequence-specificity and high affinity (15). While designed 3- or 4-finger ZFNs with reasonable specificity (generated from the available literature data on ZF designs for various DNA triplets) might be sufficient for the laboratory-based research applications, design and/or selection coupled with further optimization of the designed ZFNs as was done by Urnov et al. (15) will likely be needed for therapeutic applications. Furthermore, the use of the re-engineered FokI obligate heterodimer nuclease domain mutants in the pairs of ZFNs will be critical for eliminating or greatly reducing toxicity to human cells.
With the availability of custom ZFNs from both a commercial vendor (Sigma-Aldrich) as CompoZr™ ZFN Technology and from the Zinc Finger Consortium, an academic OPEN source (21), one can expect to see ever increasing applications of the ZFN-evoked strategies to address many interesting and important research problems in biomedical sciences and biotechnology, including applications in agriculture. Many technical challenges, such as the delivery of these reagents to the desired cells, still remain for ZFN-based strategies to be successfully applied for human therapeutics in the future. We anticipate that the first clinical applications of ZFN technology to treat a monogenic human disease will likely to occur, using ex-vivo gene therapy of human stem cells (9, 16). Here, the desired gene-modified cells could be identified through selection, expanded in culture, their genotype and phenotype carefully characterized and monitored, and then administered to carefully selected patients.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported partly by grants from National Institute of General Medical Sciences (GM077291) and Maryland Stem Cell Research Fund to S.C. (JHU) and partly by a grant from The Bill & Melinda Gates Foundation through the Grand Challenge Explorations Initiative to K.K. (Pondicherry Biotech Private Limited, PBPL) and a National Foundation for Cancer Research fellow Award to C.I.C. We thank Dr. Joseph Margolick’s lab at JHU for assistance with the flow cytometry and FACS studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- (1).Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- (2).Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- (3).Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to FokI cleavage domain. Proc Natl Acad Sci U S A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mani M, Kandavelou K, Dy JF, Durai S, Chandrasegaran S. Design, engineering and characterization of zinc finger nucleases. Biochem Biophys Res Commun. 2005;335:447–457. doi: 10.1016/j.bbrc.2005.07.089. [DOI] [PubMed] [Google Scholar]
- (6).Mani M, Kandavelou K, Dy JF, Durai S, Chandrasegaran S. Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochemical Biophysical Research Communications. 2005;334:1191–1197. doi: 10.1016/j.bbrc.2005.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kandavelou K, Mani M, Durai S, Chandrasegaran S. ‘Magic’ scissors for genome surgery. Nat Biotechnol. 2005;23:686–687. doi: 10.1038/nbt0605-686. [DOI] [PubMed] [Google Scholar]
- (8).Wu J, Kandavelou K, Chandrasegaran S. Custom-designed zinc finger nucleases: what is next? Cellular and Molecular Life Sciences. 2007;64:2933–2944. doi: 10.1007/s00018-007-7206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by a chimeric nuclease. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Morton J, Davis MW, Jorgensen EM, Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci U S A. 2006;103:16370–16375. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Doyon Y, MaCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregary PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Shukla VK, Doyon Y, Miller JC, DeKelver RC, et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- (15).Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–51. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- (16).Lombardo A, Genovese P, Beausejour CM, Colleoni S, et al. Gene editiing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. S. [DOI] [PubMed] [Google Scholar]
- (17).Perez EE, Wang J, Miller JC, Jouvenot Y, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Alexeev V, Yoon K. Stable and inheritable changes in genotype and phenotype of albino melanocytes induced by an RNA-DNA oligonucleotide. Nat Biotechnol. 1998;16:1343–1347. doi: 10.1038/4322. [DOI] [PubMed] [Google Scholar]
- (19).Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. et a. [DOI] [PubMed] [Google Scholar]
- (20).Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- (21).Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, et al. Rapid ‘Open-Source’ engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


