Abstract
We measured T-cell responses to human immunodeficiency virus type 1 (HIV-1) cryptic epitopes encoded by regions of the viral genome not normally translated into viral proteins. T-cell responses to cryptic epitopes and to regions normally spliced out of the HIV-1 viral proteins Rev and Tat were detected in HIV-1-infected subjects.
The immune system responds to invading pathogens by recognizing pathogen-derived peptides. A retrovirus such as human immunodeficiency virus type 1 (HIV-1) utilizes host cellular machinery for synthesis of viral proteins, and an infected cell will present proteasomal cleavage products commonly derived from the breakdown products of the main viral proteins. During protein processing, translational errors can result in bypassing of the main viral protein reading frames, resulting in the production of alternate reading frame or cryptic peptide epitopes (1, 2). Additionally, in the presence of certain cytokines, such as gamma interferon, the composition and function of the proteosome are altered, with resulting changes in the processing of epitopes (4). T-cell responses to the protein products of alternate reading frames can also be induced through vaccination (11).
The functional role of T-cell responses to cryptic epitopes in an experimental model of retroviral disease control has recently been established (5), and an epitope from HIV-1 that was derived from an alternate reading frame has also previously been described (3). Although most HIV-1 structural viral proteins are produced from a single, continuous open reading frame, a subset of the accessory proteins are produced from spatially separated regions of the genome. In these accessory proteins, failure of the normal processes of splicing introduces additional regions of peptide-coding sequence into the mRNA. These regions have been shown to generate immune responses in primates infected with simian immunodeficiency virus (9).
We used in silico T-cell immunogenicity prediction methods to identify peptide epitopes for the HLA-B58, A2, and B7 superfamilies within alternate reading frames in HIV-1 (7). We measured T-cell responses of HIV-1-positive individuals with good immunological control of HIV-1 viral load (controllers), individuals on highly active antiretroviral therapy (HAART) (HAART-suppressed individuals), and untreated individuals with uncontrolled HIV-1 viral loads (noncontrollers).
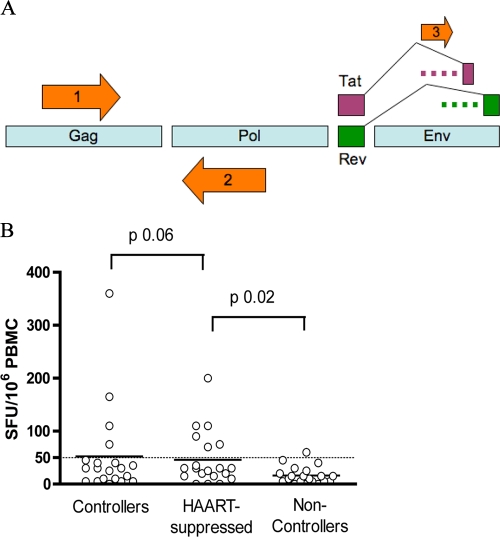
Candidate peptides were selected from the HIV-1 genome by analyzing the HXB-2 DNA sequence. Open reading frames that did not correspond to known protein-coding regions of the HIV-1 genome were identified in the sense direction. Antisense open reading frames were also identified. Alternative splice sites were identified upstream of the main splice sites for the Tat and Rev proteins. A graphical representation of epitope sources is shown in Fig. 1A.
FIG. 1.
(A) Sources of cryptic epitope peptides encoded by the HIV-1 genome. Candidate cryptic epitope peptides were selected from three sources, identified by arrows on the figure: alternate reading frames with Met start codons whose products differed from the main structural or accessory protein coded by that region of the genome (arrow 1), antisense reading frames on the strand opposite from that corresponding to the main structural or accessory protein coded by that region of the genome (arrow 2), and alternate splice sites with intervening sequence that code for additional amino acids potentially added to multiexon proteins coded by the HIV genome (arrow 3) (8). The dashed lines that precede each downstream splice site for each protein represent the additional peptide region incorporated into the proteins by the use of the alternate splice site. (B) T-cell responses to pools of cryptic epitope antigens as measured by an enzyme-linked immunospot assay. The study participants tested were divided into three clinical categories: “controllers,” with less than 5,000 copies/ml HIV-1 plasma viral load without HAART therapy, “HAART-suppressed” individuals, who were on various regimens of HAART, and “noncontrollers,” with uncontrolled viral load (not on HAART). The median is shown by the solid black line and the cutoff threshold of 50 SFU/million cells drawn with a dotted line across the graph. Statistics were performed using the Mann-Whitney test.
Peptides were identified within these open reading frames with NetCTL 1.2 software (http://www.cbs.dtu.dk/services/NetCTL/). Top-scoring epitopes for the HLA-B58, A2, and B7 supertypes were selected for peptide synthesis. The entire regions of amino acids resulting from the use of alternate splice sites in Rev and Tat were synthesized as 9- and 10-mer peptides, respectively. Eight peptides in total were synthesized and tested in a pool or on an individual basis.
Sixty-six HIV-1-infected adults were selected for this study. We focused on a subset of individuals who were able to maintain low to undetectable levels of HIV-1 in the absence of any therapy (“controllers,” with fewer than 5,000 copies/ml HIV-1 plasma viral load without HAART therapy). As a comparison group, we studied 24 individuals who had higher levels of viremia (“noncontrollers”). Finally, we studied 20 HAART-treated individuals with undetectable plasma HIV-1 RNA levels (“HAART-suppressed” individuals).
Peptides were tested in a gamma interferon enzyme-linked immunospot assay by using cryopreserved peripheral blood mononuclear cells (PBMC) (10). Cryptic-peptide-specific responses were assessed for 22 controllers (median CD4+ T-cell count, 629; median HIV RNA level, 299 copies/ml), 24 noncontrollers (median CD4+ T-cell count, 282; median plasma HIV RNA level, 50,625 copies/ml), 20 HAART-suppressed individuals (median CD4+ T-cell count, 580; median plasma HIV RNA level, <50 copies/ml), and 35 HIV-1-negative low-risk volunteers.
No HIV-1-negative volunteer had responses to the pool of HIV-1 cryptic epitope peptides (data not shown). A total of 11/66 HIV-1 positive subjects had responses to the pool of cryptic epitope peptides (>50 spot-forming units [SFU]/106 PBMC) (Fig. 1B). The noncontrollers had the lowest median T-cell response to the cryptic peptide pool, and this was statistically lower than the level for the HAART-suppressed group and trended to be lower than that for the controllers. PBMC from these subjects were then tested with individual peptides which made up the pool (Table 1). Ten subjects made responses to the Rev- or Tat-derived peptides, and one made responses to other cryptic peptides (Table 1). One controller (subject 1133; HLA-A3,31,B57) had a particularly robust response of 305 SFU/106 PBMC to the Rev alternate splice site peptide (Table 1) and also made a response to the Tat-derived peptide (Table 1). In total, 10/11 patients made responses to the alternate splice site peptides (Table 1). These are the first alternate splice site peptide epitopes recognized in humans infected with HIV-1. It is intriguing to note that the noncontrollers had the lowest-magnitude responses to the cryptic peptides, although we cannot tell whether they might have less control of viremia, because they never made cryptic responses or their lack of control led to a failure of cytotoxic T lymphocyte responses to these epitopes.
TABLE 1.
Positive responses to the cryptic peptide poola
| Patient no. | No. of SFU/million PBMC for:
|
|||
|---|---|---|---|---|
| HXB2- ORF18-140 (IAFPTFCHM) | HXB2- ORF19-64 (TSSSARLPF) | REV leader (RIFTIIVSD) | TAT leader (SRDIHHYRFR) | |
| 720 | 325 | |||
| 839 | 130 | |||
| 1133 | 305 | 290 | ||
| 1516 | 105 | 50 | ||
| 2017 | 70 | |||
| 2050 | 60 | |||
| 2056 | 90 | 65 | ||
| 2063 | 55 | |||
| 2089 | 65 | |||
| 2102 | 90 | |||
| 3101 | 230 | |||
For the 11 positive responders to the cryptic peptide pool, responses to the individual cryptic peptides were tested. Values for responses that were positive are shown in the table. The amino acid sequence (in amino acid code) is shown for each of the four cryptic peptides that gave positive responses. No positive responses to HXB2-ORF14-27 (TSWCSLLYW), HXB2-ORF15-17 (LSSSHSFPY), HXB2-ORF28-79 (LAYFPVFRF), or HXB2-ORF36-17 (KTSNSPYHF) were detected.
The HIV-1 accessory proteins Rev and Tat are each produced via the removal of what is effectively an intron sequence in the envelope region of the genome, followed by the joining of two exons coding for the two parts of the functional protein. There is a preferred target splice site for the proper joining of the two component exons into a single functional protein (12). Alternate splice sites also exist upstream of the target splice acceptor site for both the Tat and Rev proteins (6). Use of these alternative splice sites would result in the addition of novel amino acids encoded upstream of the main splice sites for the Tat and Rev mRNA. When the aberrantly spliced mRNA is translated, additional amino acids encoded in the upstream regions of RNA would be included in the protein. It is in this region where immunogenic peptides have been identified in primates (9). Although the peptides that we selected from HIV-1 do not have high levels of sequence homology to the simian immunodeficiency virus sequence, they are located within functionally equivalent areas of the two genomes.
T-cell responses to cryptic epitopes could aid in the control of HIV-1 replication. It is unclear how much variability within these regions can be tolerated in a functional viral genome. Mutations resulting in the loss of the upstream splice site in the Tat or Rev reading frame could adversely affect Env gene function, and the reduced functionality of the Tat or Rev proteins caused by use of the alternate splice site could lead to viral latency. As such, responses against splice variant regions could help to target and eliminate latently infected cells. Further study will be needed to investigate the potential of cryptic epitopes as antigens in a vaccine against HIV-1.
Acknowledgments
We thank Nick Maness and David Watkins, University of Wisconsin, for helpful discussions.
This work was supported in part by the AIDS biology program of the AIDS Research Institute, UCSF. Steven G. Deeks and Jeffrey N. Martin are supported in part by the National Institutes of Health (grants AI069994 and AI071713) and the UCSF Center for AIDS Research (grant AI027763).
Footnotes
Published ahead of print on 1 July 2009.
REFERENCES
- 1.Bullock, T. N., and L. C. Eisenlohr. 1996. Ribosomal scanning past the primary initiation codon as a mechanism for expression of CTL epitopes encoded in alternative reading frames. J. Exp. Med. 184:1319-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullock, T. N., A. E. Patterson, L. L. Franlin, E. Notidis, and L. C. Eisenlohr. 1997. Initiation codon scanthrough versus termination codon readthrough demonstrates strong potential for major histocompatibility complex class I-restricted cryptic epitope expression. J. Exp. Med. 186:1051-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardinaud, S., A. Moris, M. Fevrier, P. S. Rohrlich, L. Weiss, P. Langlade-Demoyen, et al. 2004. Identification of cryptic MHC I-restricted epitopes encoded by HIV-1 alternative reading frames. J. Exp. Med. 199:1053-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffin, T. A., D. Nandi, M. Cruz, H. J. Fehling, L. V. Kaer, J. J. Monaco, and R. A. Colbert. 1998. Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J. Exp. Med. 187:97-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho, O., and W. R. Green. 2006. Cytolytic CD8+ T cells directed against a cryptic epitope derived from a retroviral alternative reading frame confer disease protection. J. Immunol. 176:2470-2475. [DOI] [PubMed] [Google Scholar]
- 6.Kammler, S., M. Otte, I. Hauber, J. Kjems, J. Hauber, and H. Schaal. 2006. The strength of the HIV-1 3′ splice sites affects Rev function. Retrovirology 3:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, et al. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 8.Korber, B. 1998. Numbering positions in HIV relative to HXB2CG, p. 102-111. In B. Korber, C. L. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. W. Mellors, and J. Sodroski (ed.), Human retroviruses and AIDS. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 9.Maness, N. J., L. E. Valentine, G. E. May, J. Reed, S. M. Piaskowski, T. Soma, et al. 2007. AIDS virus specific CD8+ T lymphocytes against an immunodominant cryptic epitope select for viral escape. J. Exp. Med. 204:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meiklejohn, D. A., R. K. Karlsson, A. C. Karlsson, J. M. Chapman, D. F. Nixon, and B. Schweighardt. 2004. ELISPOT cell rescue. J. Immunol. Methods 288:135-147. [DOI] [PubMed] [Google Scholar]
- 11.Schirmbeck, R., P. Riedl, N. Fissolo, F. A. Lemonnier, A. Bertoletti, and J. Reimann. 2005. Translation from cryptic reading frames of DNA vaccines generates an extended repertoire of immunogenic, MHC class I-restricted epitopes. J. Immunol. 174:4647-4656. [DOI] [PubMed] [Google Scholar]
- 12.Staffa, A., and A. Cochrane. 1994. The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3′ splice site. J. Virol. 68:3071-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]