Abstract
A number of experimental studies have shown that natural killer (NK) cells can eliminate cancer cells and the mechanisms involved in this effect have been uncovered during the last two decades. Clinical data from haploidentical haematopoietic stem cell transplantation (haplo-HSCT) revealed that NK cells were responsible for remarkably favourable effects in both adult and paediatric high-risk leukaemias. NK receptors specific for major histocompatibility complex (MHC) class I molecules, including killer immunoglobulin (Ig)-like receptors (KIR) and CD94/NKG2A, play a major role in the anti-leukaemia effect (mediating either inhibitory or activating signals). Haplo- HSCT requires a heavy conditioning regimen for the patient and the use of large numbers of T cell-depleted HSC to be grafted. After transplantation, natural killer cells develop from HSC shortly after engraftment and may include ‘alloreactive’ NK cells that kill leukaemic cells and prevent graft-versus-host disease (GvHD). Alloreactive NK cells are characterized by the expression of KIR that are not engaged by any of the human leucocyte antigen (HLA) class I alleles expressed by the patient. Their generation is dependent upon the existence of a KIR/HLA class I mismatch between donor and recipient. Novel important information on the function and specificity of different KIR has been obtained recently by the analysis of donor-derived alloreactive NK cells in a cohort of paediatric patients given haplo-HSCT to cure acute, high-risk leukaemias.
Keywords: killer Ig-like receptors, leukaemia, NK cells, transplantation
Recognition of major histocompatibility complex (MHC) by human natural killer (NK) cells
NK cells, discovered originally in the early 1970s [1,2], represent a specialized arm of the innate immune system that possess potent perforin- and interferon-γ-dependent effector functions and participate in early defence mechanisms against intracellular microbial infections and different types of tumours [3–5]. Distinct from B and T cells, NK cells forego the process of rearranging genes to produce a repertoire of antigen receptors. Instead, they assemble different combinations of germline-encoded receptors that do not undergo somatic recombination. Some of these NK receptors inhibit, while others trigger, NK cell effector function. A portion of these receptors bind specifically to major histocompatibility complex (MHC) class I or class I-like molecules that were thought to be dedicated uniquely to T cell-mediated adaptive immune responses [6–10].
The ‘missing self’ mode of NK cell recognition, proposed originally by Ljunggren and Karre [11], refers to a functional mechanism by which NK cells keep target cells in check for surface expression of MHC class I. This function is possible thanks to the expression at the NK cell surface of a series of MHC class I-specific inhibitory receptors that prevent NK cell activation, thus protecting potential target cells from NK cell-mediated attack. However, sensing the absence of self-MHC class I molecules is not sufficient for inducing NK cell activation. NK cells also require stimulation by ligands on target cells recognized by activating receptors including natural cytotoxicity receptors (NCRs), DNAX accessory molecule-1 (DNAM-1) and NK group 2 member D (NKG2D) [12–15]. Thus, NK cell recognition of target cells is regulated tightly by a process involving the integration of multiple signals delivered by both activating and inhibitory receptors.
In humans, a relatively large family of NK cell receptors for MHC is represented by killer Ig-like receptors (KIR) [16]. KIR are characterized by two (KIR2D) or three (KIR3D) extracellular immunoglobulin domains. In addition, they have either short (S) or long (L) intracytoplasmic tails which transduce activating or inhibitory signals, respectively [17–20]. The known ligands for inhibitory KIRs are all represented by MHC class I molecules: thus, HLA-C is recognized by KIR2DL1, KIR2DL2 and KIR2DL3 [21–24]; HLA-B by KIR3DL1 [25,26] and HLA-A by KIR3DL2 [27,28], while HLA-G is recognized by KIR2DL4 [29]. The HLA determinants that bind to inhibitory KIRs are known as KIR epitopes. In particular, HLA-C allotypes have either the C1 epitope (shared by HLA-Cw alleles characterized by Ser77Asn80, including HLA-Cw1, -Cw3, -Cw7, -Cw8, -Cw12, -Cw13, -Cw14 and -Cw16 alleles), recognized by KIR2DL2/3, or the C2 epitope (Asn77Lys80 shared by HLA-Cw2, -Cw4, -Cw5, -Cw6, -Cw15, Cw*-1602, -Cw17 and -Cw18), the ligand for KIR2DL1 [30,31]. Similarly, all HLA-B allotypes have either the Bw4 or Bw6 epitope, but only the Bw4 epitope is a ligand for KIR, its cognate inhibitory receptor being KIR3DL1 [32–34].
Regarding the comparison between inhibitory and activating KIRs, they are characterized by highly homologous codon sequences in the extracellular domain, namely KIR2DL2/3-KIR2DS2, KIR2DL1-KIR2DS1 and KIR3DL1-KIR3DS1. In contrast to the inhibitory KIR, in most instances the ligands for the activating KIR are poorly defined despite the fact that activating KIRs possess similar extracellular regions. In addition to KIRs, another mode of MHC recognition by NK cells is mediated by the CD94-NKG2A (lectin-like) inhibitory receptor [35,36], specific for complexes of HLA-E and peptides derived from the leader sequences of many, but not all, HLA-A, HLA-B and HLA-C polypeptides [37–39].
Each individual differs in the number and type of inherited KIR genes and KIR gene products are distributed clonally in the NK cell repertoire [21,40,41]. Importantly, individual NK cells express only one or few inhibitory KIRs, while some NK cells do not express KIR but express CD94-NKG2A. In addition, inhibitory KIR and CD94-NKG2A receptors may be co-expressed by subsets of NK cells. Thus, in most cases, every NK cell expresses one or more inhibitory receptors, which through the interaction with autologous HLA class I molecules prevent NK cells from killing autologous normal cells.
The KIR gene family exhibits extensive variations in gene number and content, as well as allelic polymorphism [42,43]. Fourteen KIR genes, located in the leucocyte receptor complex on human chromosome 19, encode the various inhibitory and activating KIRs. These genes segregate independently of HLA, with the important consequence, in unrelated allogeneic transplantation, that matching for HLA does not match for KIR. The KIR repertoire in NK cells is determined primarily by the KIR genotype, although the HLA genotype also has a subtle, modifying influence [44]. Diverse KIR haplotypes can be simplified into two biologically distinct groups, A and B. Group A haplotypes have a fixed number of genes that encode inhibitory receptors, with the exception of the activating 2DS4, while group B haplotypes have variable gene content including several additional activating receptor genes. All individuals can be categorized as having one of two KIR genotypes: A/A, which is homozygous for group A KIR haplotypes, or B/x, which contains either one (A/B heterozygous) or two (B/ homozygotes) group B haplotypes [16,40,45].
Alloreactive NK cells in haploidentical haemopoietic stem cell transplantation
Loss of HLA molecules is a common event in cells undergoing tumour transformation or infection with some viruses. When NK cells expressing KIR with specificity for a particular allotypic determinant shared by some HLA class I alleles interact with target cells lacking these alleles (i.e. sensing the ‘missing-self’), they are not inhibited and kill the potentially harmful target cell.
In settings of clinical stem cell transplantation (SCT), the term ‘KIR-ligand mismatch’ is often used to describe such ‘missing-self’ recognition. In this case, a fraction of NK cells from the donor may display the ability to kill target cells from the recipient. These NK cells are referred to as ‘alloreactive’[22]. Remarkably, the presence of alloreactive donor-derived natural killer (NK) cells have been correlated with highly improved survival after HSCT for acute myelogenous leukaemia (AML). Indeed, alloreactive NK cells appear to promote engraftment, reduce graft-versus-host disease (GvHD) and, more importantly, decrease leukaemic relapses [46–49]. NK cell alloreactivity occurs when a subset of NK cells express KIR specific for an allelic epitope that is absent on (recipient) allogeneic cells. Under these conditions, these NK cells would kill allogeneic cells (at least those expressing ligands recognized by activating NK receptors). The existence of human ‘alloreactive’ NK cells was discovered originally by our group two decades ago [21–23]. It should be stressed that an HLA class I mismatch between NK cells and target cells does not lead necessarily to NK-mediated target cell killing. As indicated above, KIRs recognize allotypic determinants that are shared by groups of HLA class I alleles. Thus ‘alloreactive’ NK cells must not only express KIR that are not engaged by any of the HLA class I alleles present on allogeneic target cells [50] but also should not express CD94/NKG2A, as HLA-E is present on all HLA class I+ cells [51].
The majority of patients, particularly of paediatric age, have a family member who is identical for one HLA haplotype. One hundred per cent of parents and 50% of siblings fulfil this criterion. In most instances, haploidentical transplants are mismatched at all the HLA class I and II loci, a situation that would lead to severe GvHD and poor clinical outcome. However, a promising approach to HLA-mismatched haploidentical haematopoietic cell transplantation (HCT) has been developed which may prevent graft rejection and eliminate leukaemic cells [52]. First, patients are given an intense conditioning regimen involving total-body irradiation, cytotoxic drugs and polyclonal anti-thymocyte antibodies [46]. Moreover, in order to facilitate engraftment, the numbers of CD34+ haematopoietic stem cells (HSC) infused are 5–20 times greater than in the setting of HLA-matched transplants. The use of such ‘megadoses’ of HSC has been made possible thanks to the use of peripheral blood as a source of mobilized CD34+ HSC [52,53]. Megadoses of CD34+ cells are also important because they can generate much larger numbers of NK cells, thus contributing to the maintenance of donor's KIR repertoire, efficient alloreactivity and anti-leukaemic effect.
In this transplantation setting, alloreactivity is mediated by NK cells provided the existence of KIR–ligand mismatches. Notably, two groups of haploidentical transplants can be identified. The first is represented by donor–recipient combinations in which NK cells of the donors express KIRs that do not recognize HLA class I molecules expressed by the patient (i.e. ‘alloreactive’ NK cells). In the second category of donor–recipient combinations all the KIR expressed by NK cells recognize the HLA class I alleles of the patient (‘NK matched’). Selection of the most appropriate donor is based on the assessment of the degree of ‘alloreactivity’ of NK cells. This can be determined by the phenotypic identification of the alloreactive NK cell subset by flow cytometric analysis (revealing its size) and by evaluation of the ability to kill leukaemic cells [revealing the potential graft-versus-leukaemia (GvL) effect]. The same experimental approach allows identification and evaluation of the functional capability of alloreactive NK cells in the recipient after transplantation [54]. Indeed, in view of the key role of alloreactive NK cells in eradicating leukaemias in haplo-HSCT, it is important to ask whether such NK cells are originated and persist in the recipient for a significant time interval [55].
In the early studies, the presence of alloreactive NK cells in the recipients was documented up to 1 year after transplantation. In some patients, the proportions of alloreactive NK cells were larger in the recipient than in the donor, thus suggesting that they had been selected/expanded preferentially in vivo[46,56]. More recent studies have confirmed and extended these findings, showing that alloreactive NK cells can be detected for a period of more than 3 years after transplantation in several paediatric patients with high-risk leukaemias ([54] and Locatelli et al., in preparation).
Previous studies have shown that patients with AML had greatly improved disease-free survival and reduced leukaemia relapses when a KIR–ligand mistmatch was present [46,56]. Remarkably, patients transplanted from an NK alloreactive donor also benefited from higher rates of engraftment and reduced incidence of GvHD. A recent study on high-risk paediatric acute lymphoblastic leukaemia (ALL) refractory to chemotherapy provided further evidence of the key role of alloreactive NK cells in haploidentical HSC transplantation. This study represents a major breakthrough in the cure of otherwise fatal leukaemias, as the estimated 3-year survival in children with ALL transplanted from an NK alloreactive relative is ∼75% (Locatelli et al., in preparation).
As indicated by a murine model and suggested by clinical results, the GvL effect correlates with the generation of alloreactive NK cells characterized by potent anti-leukaemia activity. The reduced rate of GvHD is also likely to be the result of the NK cell-mediated killing of recipient antigen-presenting cells (APC) [57]. In agreement with this concept, in vitro studies, using selected NK cell clones, showed that alloreactive NK cells are capable of killing both immature and mature monocyte-derived DCs [58]. Conversely, autologous NK cells kill only immature DCs while sparing the mature ones. This, together with the release of proinflammatory cytokines [59], represents a major event in the process (‘DC editing’) of NK cell-mediated control of DC maturation. Indeed, NK cell-mediated editing [60] is necessary for selecting optimal DC, capable of priming naive T cells towards T helper type 1 (Th1) responses within secondary lymphoid compartments [61]. Notably, alloreactive NK cells cannot mediate a DC editing process because they kill all DCs of the host, independent on their stage of maturation. This, in turn, would be essential to prevent donor T cell priming and subsequent generation of GvHD.
Generation and tissue specificity of alloreactive NK cells
It is now well established that, during maturation, NK cells require recognition of self-MHC class I to acquire full effector function, a phenomenon referred to as ‘licensing’ or ‘education’[62–64]. However, NK cells lacking MHC-specific inhibitory receptors can also be generated in the absence of this interaction, but they would remain virtually anergic or hyporesponsive. One may ask how donor NK cell precursors undergoing maturation in the mismatched recipient can give rise to alloreactive NK cells capable of killing leukaemic cells? A probable explanation is that, in haplo-HSCT, the infusion of ‘megadoses’ of CD34+ cells may provide a bone marrow (BM) microenvironment (i.e. where NK cells undergo maturation) predominantly of the donor type. Under these conditions, the process of NK education would be similar to that occurring normally in the donor and would allow the generation of NK cells displaying alloreactivity towards the recipient.
However, why do alloreactive NK cells in vivo not cause GvHD? A number of experimental data have demonstrated clearly that NK cells attack haematopoietic cells of the host while sparing other tissues that are, instead, common targets for T cell-mediated GvHD. For example, in the hybrid resistance phenomenon in mouse, alloreactive NK cells caused rejection of the BM graft but did not attack other tissue [65]. Moreover, Ruggeri et al. obtained direct evidence in mice that alloreactive NK cells do not cause GvHD [46], while infusion of allogeneic T cells killed all the mice. The probably molecular basis of the resistance of recipient normal tissues to the NK cell-mediated attack is the lack of ligands for the activating NK receptors. Indeed, these ligands are expressed by cells of different histotypes only upon cell stress infection or tumour transformation [66]. Thus, interaction with normal resting tissues does not lead to alloreactive NK cell activation.
The function and specificity of KIRs regulates the ability of NK cells to kill leukaemic blasts
Our early studies on the inhibitory KIR specific for HLA-C alleles revealed the existence of a clear-cut discrimination between the specificity of the HLA-class I allele of KIR2DL1 and that of KIR2DL2/3. Thus, as mentioned above, KIR2DL1 recognizes the C2 specificity while KIR2DL2/3 is specific for the C1 specificity [6]. We reached this conclusion by functional analysis of selected NK cell clones characterized by given KIR2DL phenotypes. The C2 specificity of KIR2DL1 was confirmed in many studies, including those assessing the cytolytic activity of phenotypically selected NK cell clones against leukaemia blasts. On the other hand, two recent studies have shown that KIR2DL2/3 can also interact with HLA-C alleles belonging to the C2 specificity [54,67]. Although this interaction is weaker than that of KIR2DL2/3 with C1, it is sufficient to generate inhibitory signals that may prevent efficient NK-mediated killing. The inhibition may or may not be detected depending on the intensity of the activating signals generated during NK cell/target cell interaction. Thus, a strong NK cell stimulation via activating receptors may be sufficient to overcome the inhibition. Remarkably, in the case of NCR, their ligands are expressed in most leukaemic cells (anti-NCR antibodies strongly inhibit killing) [68,69].
Recent experimental evidence has revealed that some activating KIRs (e.g. KIR2DS1) may not allow a precise phenotypic identification and influence the function of alloreactive NK subsets. As shown by our early studies, upon recognition of HLA-Cw4 on Epstein–Barr virus (EBV)-transformed target cells, KIR2DS1 induced potent NK cell-mediated cytotoxicity [17]. Additional reports confirmed further that KIR2DS1 recognizes different alleles belonging to the C2 specificity [70,71]. In a recent study we provided direct evidence that KIR2DS1 may play a crucial role in the lysis of C2/C2 leukaemia blasts (not only of EBV-transformed cell lines) [54]. Thus, a series of donor-derived NK cell clones co-expressing KIR2DS1 together with either NKG2A or KIR2DL2/3 isolated from donor or recipient after HSCT killed patient leukaemia blasts efficiently. Moreover, cytolysis was inhibited by monoclonal antibody (mAb)-mediated blocking of KIR2DS1. In line with our findings that, in C1-mismatched HSCT, most KIR2DL2/3+ NK cells killed C2/C2 target cells poorly, Chewing et al. reported that a ‘missing self’ alloresponse to C2 could rarely be detected in NK cells from donors lacking KIR2DS1 [72]. The molecular basis of this finding is that KIR2DL2/3 can recognize not only C1, but also C2 (see above). Remarkably, among the C1-mismatched pairs of our study (in which the alloreactive subset was KIR2DL2/3+) all patients receiving haplo-HSCT from KIR2DS1+ donors (four of eight) are alive and in complete remission, while fatal relapses occurred in three of four recipients transplanted from donors lacking KIR2DS1 [54]. These data support strongly the evidence of a substantial clinical relevance of KIR2DS1-mediated recognition of leukaemia cells (Fig. 1). On the contrary, we could not document a specific role of KIR2DS2 in the mechanism of recognition and killing of leukaemia blasts.
Fig. 1.
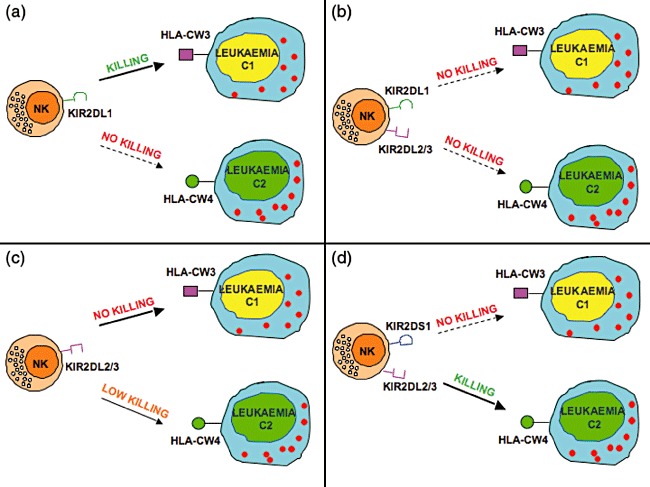
Alloreactivity against leukaemic targets is regulated by both inhibitory and activating killer immunoglobulin (Ig)-like receptors (KIRs). Alloreactive natural killer (NK) cells express KIR and lack CD94/NKG2A [that is specific for human leucocyte antigen (HLA)-E, a class Ib molecule expressed on virtually all HLA class I+ cells]. The most common alloreactivity is mediated by the C2-specific KIR2DL1+ NK cells [21,54], which kill leukaemic targets expressing the C1/C1 specificity (a). NK cells co-expressing [50] both the C2-specific KIRD2L1 and the C1-specific KIR2DL2/3 are not alloreactive (b). As expected, NK cells expressing KIR2DL2/3 [21] do not kill C1/C1+ leukaemias; however, they frequently also display poor cytotoxicity against C2/C2+ leukaemias (c). This has been explained by the recent finding [54,67] that KIR2DL2/3 recognize not only C1 (with high affinity) but also C2 (with low affinity). However, KIR2DL2/3+ NK cells can kill C2/C2+ leukaemias provided that they also express KIR2DS1 (d) or high levels of the natural cytotoxicity receptor (NCR) activating receptors (not shown). The group of C1 alleles include: HLA-Cw1, -Cw3, -Cw7, -Cw8, -Cw12, -Cw13, -Cw14 and -Cw16. The group of C2 alleles include: HLA-Cw2, -Cw4, -Cw5, -Cw6, -Cw15, Cw*-1602, -Cw17 and -Cw18.
Conclusions
The identification of HLA class I-specific NK receptors and the analysis of alloreactive NK cells represent the biological basis to devise a novel translational exploitation capable of bridging basic research with relevant clinical application. Thus, considerable evidence has indicated that alloreactive NK cells developing from haploidentical HSCT represent a most remarkable source of effector cells to fight successfully high-risk leukaemias and possibly other tumours [73,74]. The possibility to assess directly both the size and the cytolytic activity of the alloreactive NK cell populations allows not only identification of donors capable of generating alloreactive NK cells but also selection of the best possible donor [54]. A recent study on high-risk paediatric ALL refractory to chemotherapy extended further the major clinical impact of NK alloreactivity in haploidentical HSC transplantation. This study provided the first clear evidence that the NK alloreactivity against leukaemia blasts is regulated by integrated signals delivered by both inhibitory and activating KIRs ([55] and Locatelli et al., in preparation). Remarkably, the expected alloreactivity against C2/C2 leukaemia blasts mediated by KIR2DL2/3+ NK cells were often impaired because this C1-reactive receptor was found to recognize C2 weakly as well. This affected substantially the ability to kill ‘KIR mismatched’ leukaemic targets unless C2 recognition was counterbalanced by KIR2DS1 functioning as a potent inducer of leukaemic blast-killing [54].
Taken together, these novel findings provide new information that may greatly improve our understanding of the molecular mechanisms at the basis of successful haplo-HSCT in patients with high-risk leukaemias.
Disclosure
A. Moretta is founder and shareholder of Innate-Pharma and hence declares a financial interest. The other authors declare no conflict of interest.
References
- 1.Kiessling R, Klein E, Wigzell H. ‘Natural’ killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol. 1975;5:112–17. doi: 10.1002/eji.1830050208. [DOI] [PubMed] [Google Scholar]
- 2.Herberman RB, Nunn ME, Holden HT, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer. 1975;16:230–9. doi: 10.1002/ijc.2910160205. [DOI] [PubMed] [Google Scholar]
- 3.Moretta A, Bottino C, Mingari MC, Biassoni R, Moretta L. What is a natural killer cell? Nat Immunol. 2002;3:6–8. doi: 10.1038/ni0102-6. [DOI] [PubMed] [Google Scholar]
- 4.Parham P. Innate immunity: the unsung heroes. Nature. 2003;423:20. doi: 10.1038/423020a. [DOI] [PubMed] [Google Scholar]
- 5.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–10. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 6.Moretta A, Bottino C, Vitale M, et al. Receptors for HLA class-I molecules in human natural killer cells. Annu Rev Immunol. 1996;14:619–48. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 7.Colonna M. Specificity and function of immunoglobulin superfamily NK cell inhibitory and stimulatory receptors. Immunol Rev. 1997;155:127–33. doi: 10.1111/j.1600-065x.1997.tb00945.x. [DOI] [PubMed] [Google Scholar]
- 8.Lanier LL. NK cell receptors. Annu Rev Immunol. 1998;16:359–93. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 9.Long EO, Burshtyn DN, Clark WP, et al. Killer cell inhibitory receptors: diversity, specificity, and function. Immunol Rev. 1997;155:135–44. doi: 10.1111/j.1600-065x.1997.tb00946.x. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Botet M, Llano M, Navarro F, Bellon T. NK cell recognition of non-classical HLA class I molecules. Semin Immunol. 2000;12:109–19. doi: 10.1006/smim.2000.0213. [DOI] [PubMed] [Google Scholar]
- 11.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–44. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 12.Moretta A, Biassoni R, Bottino C, Mingari MC, Moretta L. Natural cytotoxicity receptors that trigger human NK-cell-mediated cytolysis. Immunol Today. 2000;21:228–34. doi: 10.1016/s0167-5699(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 13.Moretta A, Bottino C, Vitale M, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 14.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–90. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 15.Bottino C, Castriconi R, Pende D, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. 2003;198:557–67. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–51. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 17.Moretta A, Sivori S, Vitale M, et al. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med. 1995;182:875–84. doi: 10.1084/jem.182.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biassoni R, Cantoni C, Falco M, et al. The human leukocyte antigen (HLA)-C-specific ‘activatory’ or ‘inhibitory’ natural killer cell receptors display highly homologous extracellular domains but differ in their transmembrane and intracytoplasmic portions. J Exp Med. 1996;183:645–50. doi: 10.1084/jem.183.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–8. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 20.Wagtmann N, Biassoni R, Cantoni C, et al. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995;2:439–49. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 21.Moretta A, Bottino C, Pende D, et al. Identification of four subsets of human CD3-CD16+ natural killer (NK) cells by the expression of clonally distributed functional surface molecules: correlation between subset assignment of NK clones and ability to mediate specific alloantigen recognition. J Exp Med. 1990;172:1589–98. doi: 10.1084/jem.172.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciccone E, Pende D, Viale O, et al. Involvement of HLA class I alleles in natural killer (NK) cell-specific functions: expression of HLA-Cw3 confers selective protection from lysis by alloreactive NK clones displaying a defined specificity (specificity 2) J Exp Med. 1992;176:963–71. doi: 10.1084/jem.176.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moretta A, Vitale M, Bottino C, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci USA. 1993;90:12000–4. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Andrea A, Chang C, Franz-Bacon K, McClanahan T, Phillips JH, Lanier LL. Molecular cloning of NKB1. A natural killer cell receptor for HLA-B allotypes. J Immunol. 1995;155:2306–10. [PubMed] [Google Scholar]
- 26.Vitale M, Sivori S, Pende D, et al. Physical and functional independency of p70 and p58 natural killer (NK) cell receptors for HLA class I: their role in the definition of different groups of alloreactive NK cell clones. Proc Natl Acad Sci USA. 1996;93:1453–7. doi: 10.1073/pnas.93.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dohring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A1,2. J Immunol. 1996;156:3098–101. [PubMed] [Google Scholar]
- 28.Pende D, Biassoni R, Cantoni C, et al. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J Exp Med. 1996;184:505–18. doi: 10.1084/jem.184.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189:1093–100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biassoni R, Falco M, Cambiaggi A, et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by ‘group 2’ or ‘group 1’ NK clones. J Exp Med. 1995;182:605–9. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158:4026–8. [PubMed] [Google Scholar]
- 32.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–44. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med. 1994;180:537–43. doi: 10.1084/jem.180.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagtmann N, Rajagopalan S, Winter CC, Peruzzi M, Long EO. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity. 1995;3:801–9. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 35.Sivori S, Vitale M, Bottino C, et al. CD94 functions as a natural killer cell inhibitory receptor for different HLA class I alleles: identification of the inhibitory form of CD94 by the use of novel monoclonal antibodies. Eur J Immunol. 1996;26:2487–92. doi: 10.1002/eji.1830261032. [DOI] [PubMed] [Google Scholar]
- 36.Carretero M, Cantoni C, Bellon T, et al. The CD94 and NKG2-A C-type lectins covalently assemble to form a natural killer cell inhibitory receptor for HLA class I molecules. Eur J Immunol. 1997;27:563–7. doi: 10.1002/eji.1830270230. [DOI] [PubMed] [Google Scholar]
- 37.Lee N, Llano M, Carretero M, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA. 1998;95:5199–204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med. 1998;187:813–18. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braud VM, Allan DS, O'Callaghan CA, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–9. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 40.Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–63. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 41.Valiante NM, Uhrberg M, Shilling HG, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–51. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 42.Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. 1999;10:154–60. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 43.Wilson MJ, Torkar M, Haude A, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. 2000;97:4778–83. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shilling HG, Young N, Guethlein LA, et al. Genetic control of human NK cell repertoire. J Immunol. 2002;169:239–47. doi: 10.4049/jimmunol.169.1.239. [DOI] [PubMed] [Google Scholar]
- 45.Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- 46.Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 47.Velardi A, Ruggeri L, Moretta A, Moretta L. NK cells: a lesson from mismatched hematopoietic transplantation. Trends Immunol. 2002;23:438–44. doi: 10.1016/s1471-4906(02)02284-6. [DOI] [PubMed] [Google Scholar]
- 48.Ruggeri L, Mancusi A, Capanni M, et al. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood. 2007;110:433–40. doi: 10.1182/blood-2006-07-038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stern M, Ruggeri L, Mancusi A, et al. Survival after T cell-depleted haploidentical stem cell transplantation is improved using the mother as donor. Blood. 2008;112:2990–5. doi: 10.1182/blood-2008-01-135285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vitale M, Sivori S, Pende D, Moretta L, Moretta A. Coexpression of two functionally independent p58 inhibitory receptors in human natural killer cell clones results in the inability to kill all normal allogeneic target cells. Proc Natl Acad Sci USA. 1995;92:3536–40. doi: 10.1073/pnas.92.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pende D, Castriconi R, Romagnani P, et al. Expression of the DNAM-1 ligands, Nectin-2 (CD112) and poliovirus receptor (CD155), on dendritic cells: relevance for natural killer–dendritic cell interaction. Blood. 2006;107:2030–6. doi: 10.1182/blood-2005-07-2696. [DOI] [PubMed] [Google Scholar]
- 52.Martelli MF, Reisner Y. Haploidentical ‘megadose’ CD34+ cell transplants for patients with acute leukemia. Leukemia. 2002;16:404–5. doi: 10.1038/sj.leu.2402382. [DOI] [PubMed] [Google Scholar]
- 53.Korbling M, Anderlini P. Peripheral blood stem cell versus bone marrow allotransplantation: does the source of hematopoietic stem cells matter? Blood. 2001;98:2900–8. doi: 10.1182/blood.v98.10.2900. [DOI] [PubMed] [Google Scholar]
- 54.Pende D, Marcenaro S, Falco M, et al. Anti-leukemia activity of alloreactive NK cells in KIR ligand-mismatched haploidentical HSCT for pediatric patients: evaluation of the functional role of activating KIR and re-definition of inhibitory KIR specificity. Blood. 2009;113:3119–29. doi: 10.1182/blood-2008-06-164103. [DOI] [PubMed] [Google Scholar]
- 55.Moretta A, Locatelli F, Moretta L. Human NK cells: from HLA class I-specific killer Ig-like receptors to the therapy of acute leukemias. Immunol Rev. 2008;224:58–69. doi: 10.1111/j.1600-065X.2008.00651.x. [DOI] [PubMed] [Google Scholar]
- 56.Aversa F, Terenzi A, Tabilio A, et al. Full haplotype-mismatched hematopoietic stem-cell transplantation: a phase II study in patients with acute leukemia at high risk of relapse. J Clin Oncol. 2005;23:3447–54. doi: 10.1200/JCO.2005.09.117. [DOI] [PubMed] [Google Scholar]
- 57.Shlomchik WD, Couzens MS, Tang CB, et al. Prevention of graft versus host disease by inactivation of host antigen-presenting cells. Science. 1999;285:412–15. doi: 10.1126/science.285.5426.412. [DOI] [PubMed] [Google Scholar]
- 58.Della Chiesa M, Vitale M, Carlomagno S, Ferlazzo G, Moretta L, Moretta A. The natural killer cell-mediated killing of autologous dendritic cells is confined to a cell subset expressing CD94/NKG2A, but lacking inhibitory killer Ig-like receptors. Eur J Immunol. 2003;33:1657–66. doi: 10.1002/eji.200323986. [DOI] [PubMed] [Google Scholar]
- 59.Marcenaro E, Della Chiesa M, Bellora F, et al. IL-12 or IL-4 prime human NK cells to mediate functionally divergent interactions with dendritic cells or tumors. J Immunol. 2005;174:3992–8. doi: 10.4049/jimmunol.174.7.3992. [DOI] [PubMed] [Google Scholar]
- 60.Moretta A. Natural killer cells and dendritic cells: rendezvous in abused tissues. Nat Rev Immunol. 2002;2:957–64. doi: 10.1038/nri956. [DOI] [PubMed] [Google Scholar]
- 61.Agaugue S, Marcenaro E, Ferranti B, Moretta L, Moretta A. Human natural killer cells exposed to IL-2, IL-12, IL-18, or IL-4 differently modulate priming of naive T cells by monocyte-derived dendritic cells. Blood. 2008;112:1776–83. doi: 10.1182/blood-2008-02-135871. [DOI] [PubMed] [Google Scholar]
- 62.Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105:4416–23. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim S, Poursine-Laurent J, Truscott SM, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–13. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 64.Anfossi N, Andre P, Guia S, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–42. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 65.Bennett M. Biology and genetics of hybrid resistance. Adv Immunol. 1987;41:333–445. doi: 10.1016/s0065-2776(08)60034-6. [DOI] [PubMed] [Google Scholar]
- 66.Bottino C, Castriconi R, Moretta L, Moretta A. Cellular ligands of activating NK receptors. Trends Immunol. 2005;26:221–6. doi: 10.1016/j.it.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 67.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–79. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 68.Costello RT, Sivori S, Marcenaro E, et al. Defective expression and function of natural killer cell-triggering receptors in patients with acute myeloid leukemia. Blood. 2002;99:3661–7. doi: 10.1182/blood.v99.10.3661. [DOI] [PubMed] [Google Scholar]
- 69.Pende D, Spaggiari GM, Marcenaro S, et al. Analysis of the receptor–ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the Poliovirus receptor (CD155) and Nectin-2 (CD112) Blood. 2005;105:2066–73. doi: 10.1182/blood-2004-09-3548. [DOI] [PubMed] [Google Scholar]
- 70.Stewart CA, Laugier-Anfossi F, Vely F, et al. Recognition of peptide–MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci USA. 2005;102:13224–9. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Foley B, De Santis D, Lathbury L, Christiansen F, Witt C. KIR2DS1-mediated activation overrides NKG2A-mediated inhibition in HLA-C C2-negative individuals. Int Immunol. 2008;20:555–63. doi: 10.1093/intimm/dxn013. [DOI] [PubMed] [Google Scholar]
- 72.Chewning JH, Gudme CN, Hsu KC, Selvakumar A, Dupont B. KIR2DS1-positive NK cells mediate alloresponse against the C2 HLA-KIR ligand group in vitro. J Immunol. 2007;179:854–68. doi: 10.4049/jimmunol.179.2.854. [DOI] [PubMed] [Google Scholar]
- 73.Ruggeri L, Capanni M, Casucci M, et al. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94:333–9. [PubMed] [Google Scholar]
- 74.Ruggeri L, Aversa F, Martelli MF, Velardi A. Allogeneic hematopoietic transplantation and natural killer cell recognition of missing self. Immunol Rev. 2006;214:202–18. doi: 10.1111/j.1600-065X.2006.00455.x. [DOI] [PubMed] [Google Scholar]


