Abstract
Base excision repair (BER) is the major pathway for removing mutagenic and cytotoxic oxidative and alkylation DNA modifications. Using a catalytically-inactive, dominant negative protein form of human APE1, termed ED, which binds with high affinity to substrate DNA and blocks subsequent repair steps, we assessed the role of BER in mediating cellular resistance to clinically relevant alkylating drugs and antimetabolites. Colony formation assays revealed that ED expression enhanced cellular sensitivity to melphalan not at all; to decarbazine, thiotepa, busulfan and carmustine moderately (1.2 to 2.4-fold); and to streptozotocin and temozolomide significantly (2.0 to 5.3-fold). The effectiveness of ED to promote enhanced cytotoxicity generally correlated with the agent's (i) monofunctional nature, (ii) capacity to induce N7-guanine and N3-adenine modifications, and (iii) inability to generate O6-guanine adducts or DNA crosslinks. ED also enhanced the cell killing potency of the antimetabolite troxacitabine, apparently by blocking the processing of DNA strand breaks, yet had no effect on the cytotoxicity of gemcitabine, results that agree well with the known efficiency of APE1 to excise these nucleoside analogs from DNA. Most impressively, ED expression produced an ∼5- and 25-fold augmentation of the cell killing effect of 5-fluorouracil (5-FU) and 5-fluorodeoxyuridine, respectively, implicating BER in the cellular response to such antimetabolites; the increased 5-FU sensitivity was associated with an accumulation of abasic sites and active caspase positive staining. Our data suggest that APE1, and BER more broadly, is a potential target for inactivation in anti-cancer treatment paradigms that involve select alkylating agents or antimetabolites.
Keywords: APE1/APEX1, repair inhibitor, DNA-damaging agent, alkylating drug and antifolate, colorectal cancer
Introduction
To cope with the deleterious consequences of endogenous and exogenous DNA-damaging agents, cells have evolved repair systems that maintain genome integrity (1). Defects in DNA repair processes are linked to genomic instability syndromes and cancer predisposition. A significant, yet evolutionarily-unintended role for DNA repair is its involvement in influencing cellular resistance to anti-cancer agents (2, 3). In particular, most drugs employed to eradicate neoplastic disease operate by inducing the formation of complex DNA lesions that ultimately prevent replication and activate cell death responses. The best established demonstration of a role of DNA repair in mitigating therapeutic agent responsiveness is with O6-methylguanine DNA methyltransferase (MGMT), which plays a prominent part in adduct repair that limits the cytotoxic effect of clinical alkylating (methylating or chloroethylating) agents (4).
Most agents employed to treat cancer fall into the following major categories: antimetabolites, DNA-interactive drugs (e.g. alkylators, cross-linking agents, intercalating agents, topoisomerase inhibitors, and DNA cleaving agents), antitubulin agents, molecularly-targeted drugs, hormonal therapies, tumor-targeting strategies, and biological agents (5). Relevant to the studies herein are the alkylating compounds and the antimetabolites. Alkylating agents represent the earliest of anti-cancer therapies and have great utility in both hematological and solid tumor malignancies. The most common of the alkylating agents used in clinical practice include nitrogen mustards, nitrosoureas, platinum complexes, methanesulfonate esters and aziridines. These compounds, or their active metabolites, react with a range of nucleophilic targets, particularly in DNA, to form covalent intermediates that induce cell death (6, 7).
Antimetabolites account for nearly 1/5th of all drugs currently approved by the FDA for the treatment of cancer. These compounds, which are structural analogs of natural compounds, are used primarily in the treatment of hematological malignancies, although some of the more recently developed agents have demonstrated activity against solid tumors. The majority of antimetabolites are analogs of purines or pyrimidines and must be activated by cellular enzymes to nucleotide metabolites, which are incorporated into DNA and/or are direct inhibitors of enzymes required for DNA synthesis, such as DNA polymerases or thymidylate synthase (8). Regardless, nucleoside analogs interfere with normal chromosome replication and thus inhibit cell growth.
While DNA-interactive drugs typically exploit the high replicative capacity of cancerous cells, actively-dividing normal cells (e.g. bone marrow) are also susceptible to the toxic effects of these compounds. Thus, a primary goal of current investigations is to devise combinatorial treatment methods that (a) protect normal cells from and (b) enhance the sensitivity of tumor cells to the toxicity of anti-cancer agents. As noted above, DNA repair systems represent a major protective mechanism against the cytotoxic effects of clinical DNA-interactive drugs (3). Besides MGMT, base excision repair (BER) is another prominent system that eliminates potentially lethal base damage introduced by alkylating agents (6, 7). In addition, 3′ to 5′ DNA exonucleases, which have the capacity to excise chain-terminating nucleoside analogs that have been incorporated into DNA, can determine the efficacy of anti-metabolites (9). Strategic regulation of these repair mechanisms would therefore improve the selectivity and effectiveness of specific anti-cancer treatment paradigms.
Human apurinic/apyrimidinic (AP) endonuclease 1 (APE1) is the major enzyme responsible for the repair of abasic sites in DNA (10). AP sites are common intermediates of alkylation damage to DNA, arising either via spontaneous base loss or through base release by a DNA repair glycosylase. APE1 initiates repair of AP sites by incising the phosphodiester backbone immediately 5′ to the lesion, creating a single-strand break intermediate that is further processed by proteins of the BER pathway. In addition to its AP site incision activity, APE1 also possesses a 3′ to 5′ exonucleolytic function that operates on 3′-obstructive termini, such as mismatched nucleotides, tyrosyl groups, phosphate or phosphoglycolate residues, and certain chain-terminating nucleoside analogs (11-15). Indeed, past studies employing either antisense, RNAi, or small molecule inhibitor strategies have revealed that APE1-deficient cells exhibit hypersensitivity to a number of “DNA-damaging” agents, including the laboratory agents methyl methanesulfonate (MMS), hydrogen peroxide, menadione, and paraquat, and anticancer agents such as ionizing radiation, thiotepa, 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU; a.k.a. carmustine), temozolomide, gemcitabine, and the nucleoside analogue β-l-dioxolane-cytidine (L-oddC; a.k.a. troxacitabine) (16-23). Thus, APE1 represents a sensible target for improved responsiveness to certain therapeutic strategies (24).
Previous work from our laboratory characterized a dominant-negative form of APE1, termed ED, which exhibits a >5.6 × 107-fold reduced AP site incision capacity, but a >10-fold higher binding affinity for substrate DNA (25). This protein was designed strategically to harbor mutations at the catalytic active site residues Glu96 and Asp210. The improved AP-DNA binding affinity of ED likely stems from the neutralization of these acidic, negatively-charged amino acids, which normally would repel the similarly-charged phosphodiester DNA backbone. Significantly, expression of ED in both Chinese hamster ovary (CHO) cells and the human cancer cell line NCI-H1299 was shown to increase cellular sensitivity to the laboratory agent MMS, and the chemotherapeutic agents BCNU and di-deoxycytidine (ddC, a.k.a. zalcitabine); ED had no effect on the cytotoxic effects of the radiomimetic bleomycin, the nucleoside analogue β-d-arabinofuranosylcytosine (araC; a.k.a. cytarabine), the topoisomerase inhibitors camptothecin and etoposide, or the cross-linking agents mitomycin C and cisplatin (25). Using ED as a tool, we explored further the role of APE1 and BER in the survival response to a wide-range of clinical alkylating agents and antimetabolites.
Results
Effect of ED on cellular sensitivity to SN1 and SN2 alkylating agents
Alkylating agents are generally divided into two types based on their reaction mechanism, i.e. either SN1 or SN2 (6, 7). SN2-type, because of their direct biomolecular reaction with DNA, exhibit high nucleophilic selectivity and alkylate almost exclusively the highly nucleophilic nitrogen centers in DNA, producing primarily N7-alkylguanine, lesser amounts of N3-alkyladenine, and only small amounts of O-adducted lesions, such as O6-alkylguanine. SN1-type alkylating agents modify DNA via the intermediate methyldiazonium ion. Because of its high electrophilic reactivity, this intermediate has relatively low selectivity, and therefore modifies not only the highly nucleophilic nitrogen atoms, but also the less nucleophilic oxygen atoms to generate significant, albeit lesser, amounts of O-alkylated nucleotides, such as O6-alkylguanine, O4-alkylthymine and O2-alkylcytosine, as well as alkylphosphates.
We had previously demonstrated that ED expression increases by 4.8 to 6.3-fold cellular sensitivity to the SN2-type alkylating agent MMS, and concurrently leads to a hyper-accumulation of chromosomal AP sites (25). MMS has been routinely used as a classic BER-type DNA-damaging agent, as it creates primarily the base lesions N7-methylguanine and N3-methylguanine, which are either lost spontaneously due to the increased instability of the N-glycosidic bond or removed as substrates of DNA repair glycosylases (6, 7). We explored here the effect of ED on colony formation ability following treatment with two additional laboratory alkylators: the SN1-type methylating agent N-methyl-N-nitrosurea (MNU) and the SN1/SN2-type ethylating agent, ethyl methansulfonate (EMS). Using the previously designed high (ED8), medium (ED5) and low (ED6) ED-expressing CHO clones (25), as well as a parental T-REx control cell line, we found that ED production had a 1.2 to 2.9-fold enhancement of the cell killing effect of MNU, but had a marginal ∼1.2-fold effect on EMS cytotoxicity (Figure 1). The range for the fold increase in sensitivity was derived by determining the fold difference between the LD50s (i.e. the dose at which 50% cell killing is attained) of the low ED-expressing line (ED6, which represents for all practical purposes a vector control) and the medium ED-expressing line (ED5) [i.e. the low end of the range], and the difference between the LD50s of the high ED-expressing line (ED8) and the T-REx control [i.e. the high end of the range].
Figure 1.
Effect of ED on cellular sensitivity to SN1 and SN2 alkylating agents. (A) Colony formation assay with MNU. The T-REx control (T-REx) and the low (ED6), medium (ED5) and high (ED8) ED-expressing CHO cells were plated, exposed to tet for 24 hr, and then challenged with the indicated dose of MNU for 1 hr as described in Materials and Methods. After washing, cells were allowed to form colonies for 10 days in DMEM medium. Colonies were fixed, stained with methylene blue and counted, and the percent survival was determined relative to the appropriate untreated sample. Shown is the average and standard deviation of 6 data points from 3 independent experimental runs. (B) Colony formation assay with EMS. The different CHO cell lines were handled and processed as above, except EMS was used as the cytotoxic agent. Shown is the average and standard deviation of 6 data points from 3 independent experimental runs. Any apparent missing error bars represent standard deviations of <4%.
Effect of ED on clinically relevant alkylating agent sensitivity
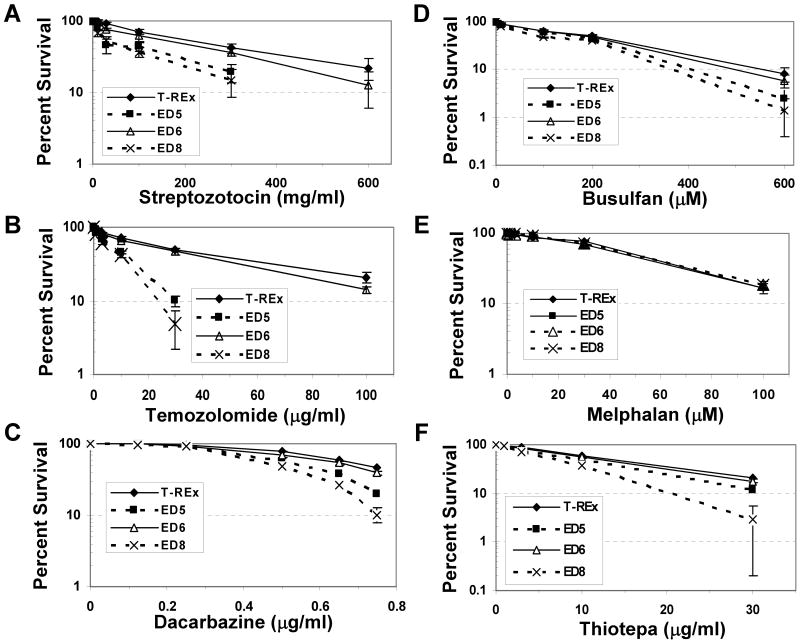
We next used the ED expressing CHO cell lines as a tool to interrogate the role of APE1 (and BER) in clinically relevant alkylating agent resistance, examining specifically the effect of ED on cellular sensitivity to streptozotocin, temozolomide, dacarbazine, busulfan, melphalan and thiotepa. These agents are frequently used in the treatment of a range of malignancies, and span the various sub-classifications of therapeutic alkylating compounds (Table 1). Previous work by our group found that ED expression increased the cell killing potency of carmustine/BCNU, a drug most commonly employed in the management of brain tumors, by 1.4 to 2.2-fold (25). Figure 2 shows the colony formation assays for each of the alkylating agents above, and Table 1 summarizes the quantitative effects of ED on cell survival with all alkylators tested herein. In short, ED had little effect on sensitivity to melphalan, an intermediate, albeit variable, effect with decarbazine, thiotepa, busulfan and carmustine (1.2 to 2.4-fold), and a pronounced effect with streptozotocin and temozolomide (2.0 to 5.3-fold).
Table I. Summary for alkylating agents.
Top three are laboratory chemicals; the remainder are clinical drugs (parentheses denote sub-classification). Unless otherwise indicated, agent is monofunctional. *Adduct profile (where available) was derived from in vitro studies using naked DNA (except for dacarbazine). **Estimates could not be found. Me = methyl; Et = ethyl; PhosTri = phosphotriester. #Based on estimated LD50 values (see Results for explanation). Ψ from (25).
| Alkylator | Reaction Mechanism | Adduct Profile* | Reference(s) | ED Fold Enhancement of Cytotoxicity# |
|---|---|---|---|---|
| MMS | SN2 | 82%, N7-MeG | (50) | 4.8 to 6.3Ψ |
| 11%, N3-MeA | ||||
| 0.3%, O6-MeG | ||||
| 0.8%, PhosTri | ||||
| MNU | SN1 | 68%, N7-MeG | (50) | 1.2 to 2.9 |
| 9%, N3-MeA | ||||
| 7%, O6-MeG | ||||
| 15%, PhosTri | ||||
| EMS | SN2/SN1 | 62%, N7-EtG | (50) | 1.2 |
| 4%, N3-EtA | ||||
| 2%, O6-EtG | ||||
| 13%, PhosTri | ||||
| Carmustine (nitrosourea) | SN1, bifunctional | 93%, N7-G | (51) | 1.4 to 2.2 Ψ |
| 3%, O6-G | ||||
| 3% crosslinks | ||||
| Streptozotocin (nitrosourea) | SN1 | ≥70%, N7-MeG | (42) | 2.0 to 5.3 |
| 5%, N3-MeA | ||||
| 3%, O6-MeG | ||||
| Temozolomide (imidotetrazine) | SN1 | 70%, N7-MeG | (52) | 2.9 to 4.5 |
| 5%, O6-MeG | ||||
| Dacarbazine (triazene) | SN1 | 70%, N7-MeG | (53) | 1.2 to 1.5 |
| 10%, N3-MeA | ||||
| 8%, O6-MeG | ||||
| Busulfan (alkyl sulfonate) | SN2, bifunctional | <10% crosslinks | (54) | 1.2 to 2.4 |
| Melphalan (nitrogen mustard) | SN2, bifunctional | 38%, N7-G | (55) | 1.1 to 1.2 |
| 20%, N3-A | ||||
| 33%, crosslinks | ||||
| Thiotepa (ethylenimine) | trifunctional | **N7-MeG > N3-MeA > O6-MeG and crosslinks | (56) | 1.2 to 1.9 |
Figure 2.
Effect of ED on clinical alkylating agent sensitivity. Colony formation efficiency and percent survival was determined as described in Figure 1 and Materials and Methods, following exposure to the indicated doses of the denoted alkylator: (A) Streptozoticin, (B) Temozolomide, (C) Dacarbazine, (D) Busulfan, (E) Melphalan, (F) Thiotepa. Shown is the average and standard deviation of at least 5 data points from 3 independent experimental runs. Any apparent missing error bars represent standard deviations of <4%.
Effect of ED on sensitivity to chain-terminating nucleoside analogs
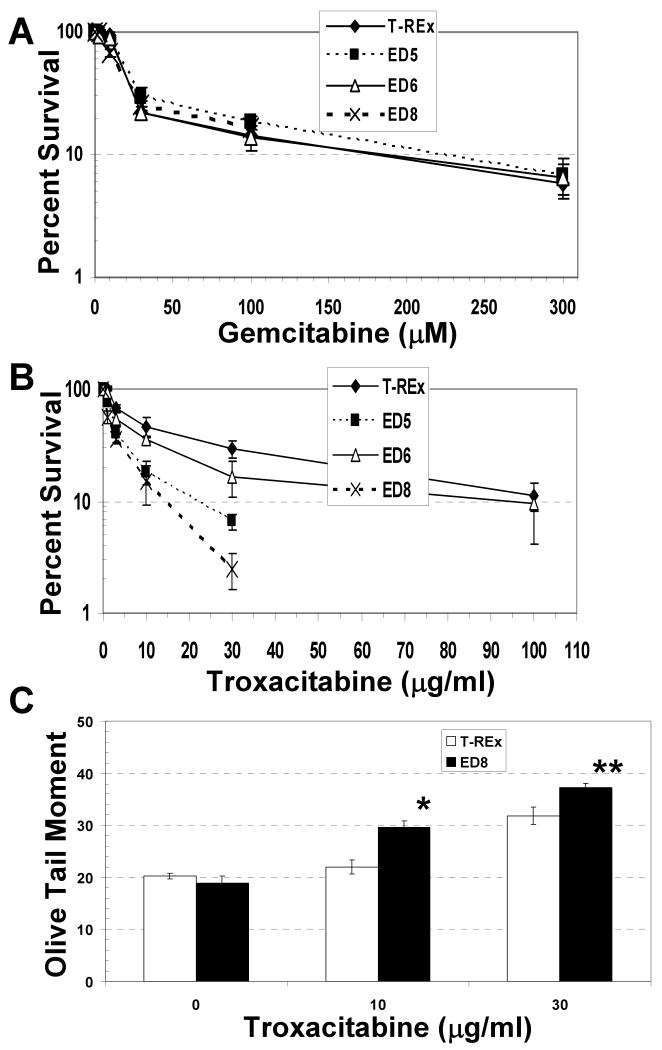
Our prior studies demonstrated that ED production in CHO cells and the human non-small cell lung cancer line NCI-H1299 increased sensitivity to the nucleoside analog zalcitabine, but not to the antimetabolite cytarabine (25). This finding is consistent with the known biochemical properties of APE1, where zalcitabine/ddC is a more favorable substrate for the 3′ to 5′ exonuclease function of APE1 than cytarabine/araC (15). We determined here the effects of ED on the cellular sensitivity to the chain-terminating nucleoside analogs troxacitabine and gemcitabine. Troxacitabine (or L-oddC) is an l-stereoisomeric analog that is a excellent substrate for APE1 excision activity (15). Gemcitabine is a cytidine analog that exerts its cytotoxicity in part through inhibition of DNA synthesis, and is a poor substrate for APE1 exonuclease activity. Impairment of endogenous APE1 function by ED expression resulted in a profound 2 to 3-fold increase in sensitivity to troxacitabine (Figure 3B), but had no effect on cell killing by gemcitabine (Figure 3A), reflective of the excision efficiency of the wild-type enzyme.
Figure 3.
Effect of ED on cellular sensitivity to gemcitabine and troxacitabine. Colony formation efficiency and percent survival was determined as described in Figure 1 and Materials and Methods, following exposure to the indicated doses of gemcitabine (A) or troxacitabine (B). Shown is the average and standard deviation of at least 5 data points from 3 independent experimental runs. (C) DNA strand break levels in ED8 and T-REx cell lines following exposure to troxacitabine. After tet induction, ED8 and T-REx cells were treated with 0, 10 or 30 μg/ml troxacitabine for 24 hr and subsequently processed for Comet analysis (see Materials and Methods). Shown is the average and standard deviation of the OTM determined from a minimum of 50 cells from 3 independent experimental slides. *, P < 0.002; **, P < 0.01. Note: T-Rex cells without tet or troxacitabine treatment were found to have a similar OTM to tet-exposed T-Rex controls (data not shown).
To elucidate the potential mechanism by which ED might induce cell death when combined with troxacitabine, we measured DNA strand breaks using the alkaline single-cell gel electrophoresis (Comet) assay. We postulated that the dominant-negative protein would bind 3′-L-oddC replication intermediates and prevent repair processing, resulting in the hyper-accumulation of blocked termini and genotoxic strand breaks in the chromosomal DNA. As shown in Figure 3C, Comet analysis (see Materials and Methods) indeed found that the high ED expressing cell line ED8 displayed a statistically greater Olive tail moment (OTM; i.e. DNA fragmentation) as compared to the T-Rex control, with an ∼35% and 17% increase in DNA damage at 10 and 30 μM troxacitabine, respectively.
Effect of ED on sensitivity to the antimetabolite 5-fluorouracil (5-FU)
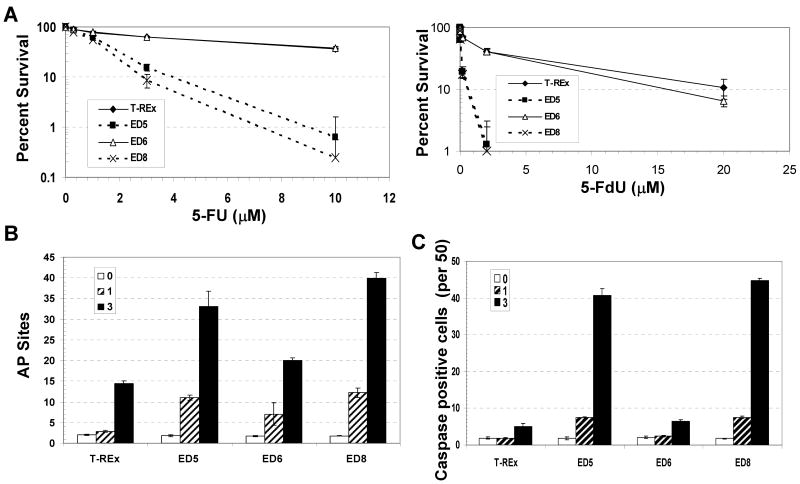
Recent evidence has suggested a role for BER capacity in cellular resistance to the antimetabolite 5-FU, a drug that has been used in the clinic for decades to treat a variety of solid tumors, most notably colorectal cancer (26, 27). First, the 5-FU metabolite, FdUMP, inhibits thymidylate synthase activity, which is responsible for the synthesis of thymidine via reductive methylation of dUMP to dTMP. In the absence of efficient TS function, cellular nucleotide pools become imbalanced with a significant increase in dUTP, resulting in high levels of uracil in chromosomal DNA. Uracil is a substrate of uracil DNA glycosylases, such as UNG, which excise the modified base from DNA to create an AP site as the first step in a BER response (28, 29). Second, the metabolite FdUTP can be directly incorporated into DNA, and recent studies have found that the mammalian DNA glycosylase SMUG1 can remove the abnormal base from DNA and plays a significant role in determining cellular sensitivity to 5-FU exposure (30). Third, studies in yeast have documented a critical role for the principal AP endonuclease, APN1, in protecting cells from the lethality of 5-FU treatment (31, 32). We reasoned that if mammalian APE1 played a major role in dictating responsiveness to 5-FU, ED would increase the potency of 5-FU induced cell killing. Indeed, colony formation assays following ED expression and 5-FU or 5-F-deoxyuridine (5-FdU) treatment resulted in a 4.8 to 5.2-fold and an ∼25-fold increase in drug sensitivity, respectively (Figure 4A).
Figure 4.
Effect of ED on cellular response to 5-FU and 5-FdU. (A) Colony formation assay. Percent survival of ED5, ED6, ED8 and the T-REx control was determined following exposure to the indicated doses of 5-FU (left) or 5-FdU (right) as described in Figure 1 and Materials and Methods. Shown is the average and standard deviation of at least 5 data points from 3 independent experimental runs. (B) AP site levels in the CHO cell lines following 5-FU exposure. After tet induction, the various CHO cell lines were exposed to 0, 1 or 3 μM 5-FU for 24 hr (as indicated), and the cells were collected 24 hr later. Following isolation of chromosomal DNA, AP sites were measured using an aldehyde reactive probe-based colorimetric assay (57). Note: Abasic lesions are expressed as AP sites per 1 × 106 base pairs. (C) Active caspase positive staining following 5-FU exposure. ED5, ED6, ED8 and T-REx control cells were treated with 5-FU for 24 hr (1 or 3 μM, see inset), and subsequently fixed and probed with the capase inhibitor sequence VAD linked to a carboxyfluorescein probe. Shown is the average and standard deviation of the number of green staining, caspase positive cells per 50 from three independent experimental runs. Note: T-Rex cells without tet or 5-FU treatment were found to have a similar AP site level and caspase positive staining in comparison to tet-exposed T-Rex controls (data not shown).
To gain insight into the mechanism of 5-FU induced cell killing, we measured both AP site damage and apoptosis (i.e. active caspase positive cells) in the various ED-expressing and control CHO cell lines. We found that following 1 or 3 μM 5-FU treatment the high (ED8) and medium (ED5) ED expressing cell lines accumulated significantly more abasic damage than the low ED-expressing clone (ED6) (1.6 to 2-fold) or the T-REx control (2.3 to 4.3-fold) (Figure 4B). Notably, this finding suggests that BER DNA substrates/products are indeed formed during the metabolism of 5-FU. In addition, the ED5 and ED8 cell lines exhibited correspondingly greater active caspase staining, presumably reflective of increased apoptotic cell death (Figure 4C).
Chronic ED expression causes G1 arrest and apoptosis
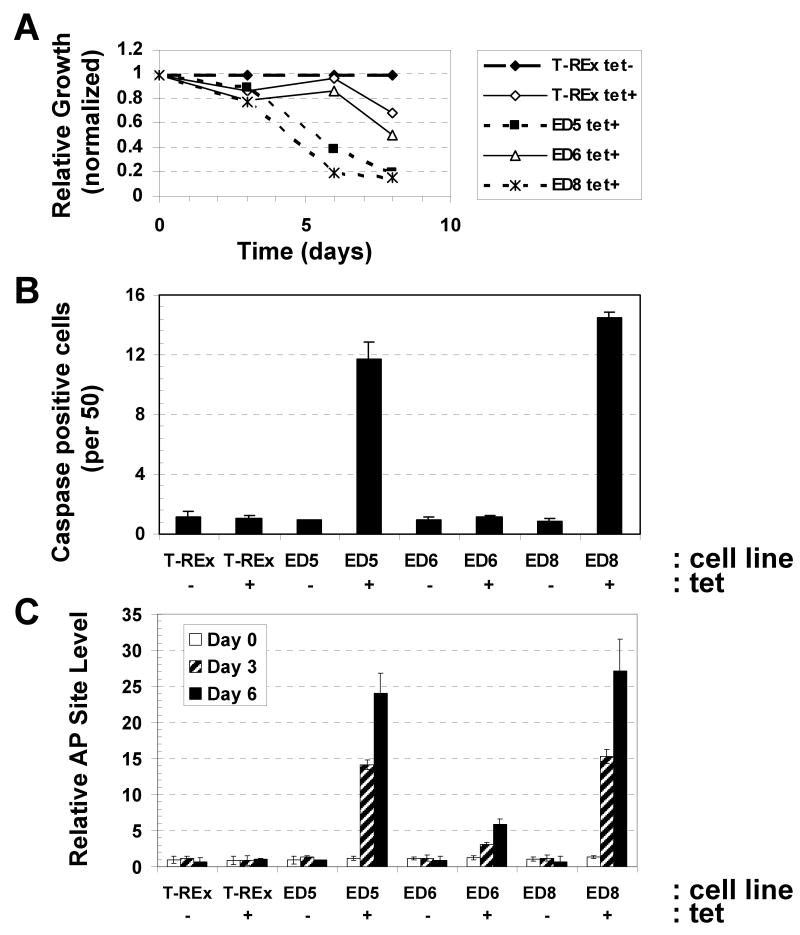
While no obvious cellular changes (such as impaired growth or altered morphology) were observed upon ED expression in the studies above, these experiments were performed with only transient induction periods (∼24 hr). To elucidate the effects of chronic ED production, the low (ED6), medium (ED5) and high (ED8) ED-expressing cell lines, as well as the T-REx parental control, were propagated continuously in the presence of 1 μg/ml tet (with fresh tet-containing media introduced every 24 hr). Cell counts provided us with an initial means of assessing cell growth (i.e. doubling). In these experiments, cell number was measured via standard Coulter counter techniques at days 3, 6 and 8 post initial plating. These studies indicate a clear reduction in cell density (i.e. cells per ml) at day 6 for ED5 and ED8 that is not seen with the ED6 or T-REx lines under conditions of continuous tet exposure (Figure 5A). At day 8, all cell lines began to exhibit reduced proliferative capacity in the presence of tet, presumably due to the cytostatic effects of chronic antibiotic treatment, although impaired growth was more pronounced for ED5 and ED8. Cell cycle analysis using propidium iodide staining and flow cytometry revealed that after 7 days of tet exposure the ED5 and ED8 lines arrested in G1, whereas the low ED expressing ED6 line and the T-REx control maintained a “normal” cell cycle profile with or without tet (Table 2). In addition, studies found that ED5 and ED8 exhibited a tet-dependent 12 to 13-fold increase in the percentage of cells that underwent apoptosis, recorded as active caspase staining (Figure 5B). Finally, consistent with a previous investigation that indicated a causative role for genomic damage in the death of APE1-deficient cells (33), we observed a significantly greater, time-dependent accumulation of abasic sites in the chromosomal DNA of the ED5 and ED8 tet+ clones than in the low ED expressing cell line ED6 or the T-REx control (Figure 5C). These data as a whole indicate that chronic ED production results in AP site accumulation, G1 arrest and apoptotic cell death.
Figure 5.
Effect of chronic ED production in CHO cells on various cellular end-points. (A) Effect of ED on cell growth. The three ED-expressing CHO cell lines, ED5, ED6 and ED8, and the T-REx control were maintained in DMEM with 1 μg/ml tet continuously as described in Materials and Methods. At days 3, 6 or 8, cells from an appointed flask were harvested and counted via a Beckman Coulter counter. Plotted are cells per ml of the tet+ cultures relative to cells per ml of the T-REx control without tet (tet-). (B) Effect of ED on apoptosis. The ED5, ED6, ED8 and T-REx cell lines were maintained as above with or without tet (denoted). At day 7, cells were harvested from each condition, fixed, and probed for active caspases as described in Materials and Methods. Shown is the average and standard deviation of the number of green staining, caspase positive cells per 50 from 3 independent measurements. (C) Effect of ED on AP site accumulation. The ED5, ED6, ED8 and T-REx cell lines were maintained as above with or without tet. At days 0, 3, and 6, cells were harvested from each condition. Following isolation of chromosomal DNA, AP sites were measured using an aldehyde reactive probe-based colorimetric assay (57). Note: AP sites (expressed as AP sites per 1 × 106 base pairs) were all compared to T-Rex control to establish relative AP site level.
Table II. Cell cycle distribution of chronic ED expressing CHO cell lines and the T-Rex CHO control.
The indicated cell line was maintained in DMEM with (+) or without (-) tetracycline (Tet). At day 7, cells were harvested and analyzed for cell cycle profile via flow cytometer. Shown is the percentage of cells in the G1, G2, or S phase. The results are from a representative study of three independent experimental runs.
| Cell Cycle Distribution | T-Rex | ED5 | ED6 | ED8 | ||||
|---|---|---|---|---|---|---|---|---|
| - Tet | + Tet | - Tet | + Tet | - Tet | + Tet | - Tet | + Tet | |
| % G1 | 34.3 | 36.3 | 36.1 | 52.8 | 36.0 | 37.1 | 36.8 | 55.2 |
| % G2 | 7.91 | 8.14 | 9.40 | 11.9 | 7.74 | 8.56 | 8.92 | 10.9 |
| % S | 57.8 | 55.6 | 54.5 | 35.3 | 56.3 | 54.3 | 54.2 | 33.9 |
Discussion
Early studies found that deletion of both alleles of APE1 in mice leads to embryonic lethality, underscoring the essential nature of the protein in animal development and viability (34, 35). More recent work has demonstrated that sufficient depletion of human APE1 via RNAi leads to cell inviability in culture, apparently due to the accumulation of DNA damage such as AP sites (33). As a complementary means of assessing the biological function(s) of APE1, we developed a set of stable, tet-inducible, dominant-negative expressing CHO cell lines (25). The dominant-negative protein, termed ED, exhibits enhanced DNA binding affinity relative to wild-type, yet displays a >56 million-fold reduced nuclease efficiency. Given these properties, we postulated that ED would bind with high affinity to target DNA substrates when produced in cells, and in doing so, block normal APE1 nuclease functions and subsequent repair processing. Indeed, our work found that ED expression rendered cells hypersensitive to agents that generate BER substrates and induced a concomitant hyper-accumulation of AP sites (25). We have employed here the model ED-expressing CHO cell lines to examine more exhaustively the role of APE1 and BER in the survival response to clinical DNA “damaging” drugs, particularly alkylating agents and nucleoside analogs. We also explored the effect of chronic ED production on cell growth and viability.
As for alkylators, we found that ED expression broadly, albeit with some preference, enhanced cellular sensitivity to these agents. In particular, ED had little effect on sensitivity to melphalan; an intermediate effect with decarbazine, thiotepa, busulfan and carmustine; and the most pronounced effect (∼2.0 to 5.3-fold) with streptozotocin and temozolomide (Table 1). At first glance, the features which appear most common among the compounds that experience an ED-dependent enhancement in cytotoxicity (including the SN2 MMS and SN1 MNU laboratory agents) are (i) monofunctionality and (ii) a propensity for N7-guanine, and to a lesser extent, N3-adenine alkylation. Presumably, such DNA adducts undergo spontaneous or glycosylase-mediated base release (7), resulting in the creation of a high number of cytotoxic AP sites, which are “sequestered” by the ED protein (25).
The alkylating agents that appear to escape the “feature-based” prediction outlined above are dacarbazine and busulfan (Table 1). Specifically, the monofunctional alkylator decarbazine would be anticipated to experience a pronounced ED-associated augmentation in cell killing. The lack of notable synergism (only 1.2 to 1.5-fold) could be explained in part by the high degree of O6-guanine alkylation, which is handled by a separate DNA repair response (4). Moreover, dacarbazine may not be effectively metabolized into its reactive form in T-REx CHO cells. As for the bifunctional agent busulfan, the unexpected outcome of a significant ED-dependent enhancement of cytotoxicity (1.2 to 2.4-fold) may stem from its low proclivity to form crosslinks (<10% of total), and possibly, although unsubstantiated, its effectiveness at generating a high level of N7-guanine and/or N3-adenine damage. The absence of an ED-related effect on EMS sensitivity likely stems from the lower frequency of N7-guanine alkylation, and/or a more prominent role for other DNA repair pathways, such as MGMT, nucleotide excision repair, recombination and/or error-prone bypass responses, in resolving the larger ethyl adducts, such as O6-ethylguanine (36, 37). In total, we surmise that (i) monofunctionality, (ii) a propensity to form N7-guanine and N3-adenine adducts, and (iii) a low capacity to generate O6-guanine modifications or DNA inter/intrastrand crosslinks are collectively a predictor for the potential involvement of APE1 and BER in alkylating agent responsiveness.
A role for APE1 in determining cellular sensitivity to thiotepa has been seen previously (22), and a prominent role for APE1 and BER in temozolomide resistance is consistent with the reports of others (20, 22, 38-40). The studies here are the first to suggest a contribution of APE1 and BER in resistance to streptozotocin, and potentially busulfan. The prominent involvement of BER in cellular protection against the cytotoxicity of temozolomide (brand names Temodar and Temodal) and streptozotocin (a.k.a. Zanosar) suggests that this pathway may be a suitable target for improving the therapeutic treatment of certain brain and pancreatic cancer types, respectively (41, 42).
The impact of ED on nucleoside analog sensitivity corresponded well with the known excision efficiency of APE1 for the different 3′-terminal nucleotides once incorporated into DNA. Specifically, the relative efficiency of APE1 3′ to 5′ exonuclease removal of the relevant analogs from deoxyoligonucleotide substrates is as follows: troxacitabine (L-oddC) = 100, zalcitabine (ddC) = 12.3, gemcitabine (dFdC) = 9.0 and cytarabine (araC) = 3.7, although the comparative affinity (i.e. KM) has not been determined (15). The effect of ED on cellular sensitivity was 2 to 3-fold for L-oddC, 1.6 to 2.8-fold for ddC, and essentially zero for both dFdC and araC (results herein and (25)). In addition, as assessed by the Comet assay, ED production increased the level of genotoxic strand breaks when combined with troxacitabine, suggesting that the dominant negative protein prevents normal APE1 processing of 3′-L-oddC DNA intermediates. These findings are by and large in line with past studies showing that overexpression or downregulation of APE1 can correspondingly modify cellular resistance to troxacitabine (21, 43). The major role for APE1 in dictating responsiveness to troxacitabine (brand name Troxatyl) implies that this protein may be an effective target for improving efficacy in the treatment of certain solid tumors and hematologic malignancies.
In the case of gemcitabine, one study found that suppression of APE1 via antisense oligonucleotides augmented the killing of Panc-1 pancreatic cancer cells (19), whereas in a separate study, down-regulation of APE1 by RNAi had no effect on sensitivity of RKO colon cancer cells to this antimetabolite (21). Our results support the latter finding, suggesting that (i) APE1 has no role in excising this nucleoside analog, assuming incorporated into DNA, (ii) gemcitabine induces cell death via a mechanism more related to inhibition of ribonucleoside reductase and depletion of deoxyribonucleotide pools (44), or (iii) the agent's effectiveness is dictated by the array and capacity of the responses specific to the cell type.
The most striking observation within was the pronounced effect that ED had on cell survival following exposure to the antimetabolites, 5-FU and 5-FdU. This enhanced sensitivity was the most dramatic seen for any of the therapeutics explored herein (∼25-fold in the case of 5-FdU). The greater impact of ED on cell killing by 5-FdU relative to 5-FU (∼5-fold) likely stems from the fact that the latter agent affects both DNA and RNA metabolism, whereas the former compound strictly perturbs DNA (26). To our knowledge, this is the first report in a mammalian model system that disruption of endogenous APE1 function is relevant to the mechanism of 5-FU mediated cytotoxicity, and is consistent with the studies in yeast that have found a prominent role for APN1 in protecting cells from the lethality of 5-FU challenges (31, 32). Our studies also insinuate that 5-FU directs a BER response, as we observed an ED-dependent accumulation of AP sites, which likely arise from release of uracil and 5-FU bases from DNA (30, 45). In all, evidence is emerging that implicates BER, as well as other DNA damage response systems, such as mismatch repair, in determining cellular sensitivity to the antimetabolite 5-FU (45, 46), suggesting that these pathways may be reasonable targets for improving the efficacy of treatment for colon, rectal, breast, gastrointestinal, head and neck, and ovarian cancers (27).
Finally, we found that chronic expression of ED in the CHO cell lines leads to impaired cell growth, accumulation of DNA damage, G1 arrest, and eventual apoptosis. This finding is consistent with prior studies that demonstrated that sufficient reduction in APE1 protein leads to cell inviability (33, 47), and further highlights the enormous level of endogenous DNA damage formed spontaneously and the importance of this repair nuclease in genome maintenance. Future studies will continue to dissect out the role of APE1 and BER in clinical agent resistance and more intensely focus on the relative importance of MGMT, MMR and recombinational repair processes in regulating the overall responsiveness to and efficacy of alkylating drugs and antimetabolites.
Materials and Methods
Reagents
All laboratory agents and chemotherapeutics were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise specified. Gemcitabine (NSC# 362856) was obtained from the Developmental Therapeutics Program of the National Cancer Institute (Bethesda, Maryland). Temozolomide and thiotepa were purchased from Schering-Plough Corporation (Kenilworth, NJ) and Bedford Laboratories (Bedford, OH), respectively. Troxacitabine was synthesized as previously described (48). Dulbecco's modified Eagle's media (DMEM) and Minimal Essential media were acquired from Invitrogen Corporation (Carlsbad, CA).
Colony formation survival assays
The T-REx CHO control and ED-expressing cell lines were created and maintained as previously described (25). To evaluate cell survival following a specific chemical exposure, the various ED-expressing CHO cell lines and the T-REx control were grown to confluence, trypsinized and counted. 150 cells of each cell line were transferred to each well of a six well plate. Cells were allowed to adhere for 2 hr before being incubated with 1 μg/ml tetracycline (tet). At the end of the 24 hr tet exposure, cells were treated at the indicated concentrations with one of the following DNA “damaging” agents: EMS (for 1 hr), MNU (1 hr), busulfan (24 hr), dacarbazine (5 hr), melphalan (1 hr), streptozotocin (24 hr), temozolomide (1 hr), thiotepa (1 hr), troxacitabine (24 hr), gemcitabine (4 hr), 5-FU (24 hr), or 5-FdU (24 hr). The cells were then gently washed two times with 1X phosphate buffered saline (PBS), and incubated for 10 days with fresh DMEM to allow individual colonies to form. At that time, colonies were stained with methylene blue and counted, and the percent survival determined relative to the untreated control (25).
DNA damage measurements
Single-cell gel electrophoresis Comet assays were carried out essentially as described in (49). Specifically, after a 24 hr tet treatment, T-Rex and ED8 cells were exposed to 0, 10 or 30 μg/ml troxacitabine for 24 hr under normal growth conditions. The cells were washed twice with 1X PBS, trypsinized, rewashed, and counted using a Beckman Coulter counter. Two million cells from each treatment condition were subsequently isolated and resuspended in 70 μl of 1.2% low melting point agarose (Fisher Scientific, Fair Lawn, NJ) in 1X PBS. The agarose/cell mix was added to a predipped slide coated with 1% normal melting agarose (IBI, Shelton, CT) and spread using a coverslip. After being placed for 5 min on a pre-chilled (iced) aluminum tray, the coverslips were removed and an additional 70 μl of 1.2% low melting point agarose was added, covered with a coverslip, and chilled on the iced aluminum tray. Again, the coverslips were removed, and the slides were then placed in prechilled lysis solution (2.5 M NaCl, 100 mM EDTA, 10 mM Trizma base, pH 10, 1% sodium lauryolsarconsinate, 10% DMSO, and 1% Triton X-100) for 4 hr at 4° C. Slides were washed 3 times for 5 min in 4° C 0.4 M Tris buffer (pH 7.5). Next, the slides were incubated in alkali solution (300 mM NaOH and 1 mM EDTA, pH 13) for 30 min and subsequently electrophoresed horizontally for 30 min at 4° C at 30V. The slides were washed 3 times for 15 min with 4° C 0.4M Tris buffer (pH 7.5), and after staining with ethidium bromide (final concentration 5 μg/ml), were viewed using a Ziess Axiovert 200 M fluorescent microscope (Thornwood, NY). The analysis of the comet tail was carried out using the Komet 5.5 software (Kinetic Imaging, Durham, NC) to determine the OTM. The OTM = [(the mean length of the tail - the mean length of the head) × percentage of DNA in the tail/100]. This experiment was repeated three times for each cell line and experimental condition, and the data shown represent the average and standard deviation of the OTM determined for at least 150 cells (≥50 cells per experiment).
Steady-state AP site levels were measured in purified genomic DNA using the DNA Damage Quantification Kit from Dojindo Molecular Technologies, Inc. (Gaithersburg, MD) as previously described (25).
Cell growth, cell cycle profile and apoptosis measurements
To characterize the response of the cells to chronic tet exposure, samples of the ED-expressing CHO cell lines (i.e. ED5, ED6 and ED8) and the T-REx control were counted using a Beckman Coulter counter. 5,000 cells were then added to a flask and maintained in DMEM media with or without 1 μg/ml tet for the duration of the experiment; fresh media (with or without tet) was added daily. At day 3, 6 and 8, one flask from each cell line under tet+ or tet- growth conditions was trypsinized, counted to determine cells per ml (i.e. cell growth), and frozen for future AP site analysis (see above).
To determine cell cycle distribution, a flask of each cell type with or without 1 μg/ml tet was trypsinized at day 7 and counted using the Beckman Coulter counter. One million cells were then washed with 1X PBS twice, fixed with 70% ice cold ethanol, washed again, and stained with propidium iodide solution (containing RNase A from Bovine pancreas). Cells were subsequently analyzed on a FACSCalibur flow cytometry system (BD Biosciences, San Jose, CA), using the 488-nm excitation to collect forward light scatter and red fluorescence above 600 nm.
Apoptosis was measured using the poly-caspases FLICA apoptosis detection kit from Immunochemistry Technologies, LLC (Bloomington, MN). The kit employs an inhibitor sequence of caspases (VAD, which reacts with all caspases) linked to a green (carboxyfluorescein, FAM) fluorescent probe. In brief, ED5, ED6, ED8 and T-Rex control cells were cultured for 7 days with or without 1 μg/ml tet. Cells were then washed with wash buffer (see detailed procedure provided by manufacturer), exposed to FLICA solution for 1 hr in medium, and washed again. Next, the cells were exposed to propidium iodide, Hoechst stain, and fixed using the standard protocol for adherent cells outlined in the kit manual. 50 plus cells of each reaction condition were visualized using a Zeiss Axiovert microscope and counted for caspase positive (green) staining. Each of the experimental assessments above was repeated at least three times.
Acknowledgments
We thank Robert Wersto, Joe Chrest and Christa Morris from the National Institute on Aging flow laboratory for assistance with the cell cycle distribution analysis.
Grant Information: This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging, and NIH grant CA63477 (Y.-C. Cheng).
Contributor Information
Daniel R. McNeill, Laboratory of Molecular Gerontology, Biomedical Research Center, National Institute on Aging, NIH, Baltimore, MD 21224
Wing Lam, Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06510.
Theodore L. DeWeese, Department of Radiation Oncology and Molecular Radiation Sciences, Johns Hopkins University, Baltimore, Maryland 21231
Yung-Chi Cheng, Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06510.
David M. Wilson, III, Laboratory of Molecular Gerontology, Biomedical Research Center, National Institute on Aging, NIH, Baltimore, MD 21224.
Reference List
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Madhusudan S, Middleton MR. The emerging role of DNA repair proteins as predictive, prognostic and therapeutic targets in cancer. Cancer Treat Rev. 2005;31:603–617. doi: 10.1016/j.ctrv.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 4.Liu L, Gerson SL. Targeted modulation of MGMT: clinical implications. Clin Cancer Res. 2006;12:328–331. doi: 10.1158/1078-0432.CCR-05-2543. [DOI] [PubMed] [Google Scholar]
- 5.Thurston DE. Chemistry and Pharmacology of Anticancer Drugs. CRC Press, Taylor and Francis Group, LLC; Boca Raton, FL: 2007. [Google Scholar]
- 6.Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE. Alkylation damage in DNA and RNA--repair mechanisms and medical significance. DNA Repair (Amst) 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sampath D, Rao VA, Plunkett W. Mechanisms of apoptosis induction by nucleoside analogs. Oncogene. 2003;22:9063–9074. doi: 10.1038/sj.onc.1207229. [DOI] [PubMed] [Google Scholar]
- 9.Wilson DM. III Processing of nonconventional DNA strand break ends. Environ Mol Mutagen. 2007;48:772–782. doi: 10.1002/em.20346. [DOI] [PubMed] [Google Scholar]
- 10.Demple B, Sung JS. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair (Amst) 2005;4:1442–1449. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Chou KM, Cheng YC. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature. 2002;415:655–659. doi: 10.1038/415655a. [DOI] [PubMed] [Google Scholar]
- 12.Hadi MZ, Ginalski K, Nguyen LH, Wilson DM., III Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J Mol Biol. 2002;316:853–866. doi: 10.1006/jmbi.2001.5382. [DOI] [PubMed] [Google Scholar]
- 13.Wilson DM., III Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J Mol Biol. 2003;330:1027–1037. doi: 10.1016/s0022-2836(03)00712-5. [DOI] [PubMed] [Google Scholar]
- 14.Suh D, Wilson DM, III, Povirk LF. 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997;25:2495–2500. doi: 10.1093/nar/25.12.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou KM, Kukhanova M, Cheng YC. A novel action of human apurinic/apyrimidinic endonuclease: excision of L-configuration deoxyribonucleoside analogs from the 3′ termini of DNA. J Biol Chem. 2000;275:31009–31015. doi: 10.1074/jbc.M004082200. [DOI] [PubMed] [Google Scholar]
- 16.Ono Y, Furuta T, Ohmoto T, Akiyama K, Seki S. Stable expression in rat glioma cells of sense and antisense nucleic acids to a human multifunctional DNA repair enzyme, APEX nuclease. Mutat Res. 1994;315:55–63. doi: 10.1016/0921-8777(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 17.Walker LJ, Craig RB, Harris AL, Hickson ID. A role for the human DNA repair enzyme HAP1 in cellular protection against DNA damaging agents and hypoxic stress. Nucleic Acids Res. 1994;22:4884–4889. doi: 10.1093/nar/22.23.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- 19.Lau JP, Weatherdon KL, Skalski V, Hedley DW. Effects of gemcitabine on APE/ref-1 endonuclease activity in pancreatic cancer cells, and the therapeutic potential of antisense oligonucleotides. Br J Cancer. 2004;91:1166–1173. doi: 10.1038/sj.bjc.6602080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bobola MS, Finn LS, Ellenbogen RG, Geyer JR, Berger MS, Braga JM, Meade EH, Gross ME, Silber JR. Apurinic/apyrimidinic endonuclease activity is associated with response to radiation and chemotherapy in medulloblastoma and primitive neuroectodermal tumors. Clin Cancer Res. 2005;11:7405–7414. doi: 10.1158/1078-0432.CCR-05-1068. [DOI] [PubMed] [Google Scholar]
- 21.Lam W, Park SY, Leung CH, Cheng YC. Apurinic/apyrimidinic endonuclease-1 protein level is associated with the cytotoxicity of L-configuration deoxycytidine analogs (troxacitabine and beta-L-2′, 3′-dideoxy-2′, 3′-didehydro-5-fluorocytidine) but not D-configuration deoxycytidine analogs (gemcitabine and beta-D-arabinofuranosylcytosine) Mol Pharmacol. 2006;69:1607–1614. doi: 10.1124/mol.105.021527. [DOI] [PubMed] [Google Scholar]
- 22.Luo M, Kelley MR. Inhibition of the human apurinic/apyrimidinic endonuclease (APE1) repair activity and sensitization of breast cancer cells to DNA alkylating agents with lucanthone. Anticancer Res. 2004;24:2127–2134. [PubMed] [Google Scholar]
- 23.Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, Sternberg MJ, Dianov GL, Hickson ID. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med. 2007;28:375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 25.McNeill DR, Wilson DM., III A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol Cancer Res. 2007;5:61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- 26.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 27.Wyatt MD, Wilson DM., III Participation of DNA repair in the response to 5-fluorouracil. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA--occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 29.Berger SH, Pittman DL, Wyatt MD. Uracil in DNA: consequences for carcinogenesis and chemotherapy. Biochem Pharmacol. 2008;76:697–706. doi: 10.1016/j.bcp.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An Q, Robins P, Lindahl T, Barnes DE. 5-Fluorouracil incorporated into DNA is excised by the Smug1 DNA glycosylase to reduce drug cytotoxicity. Cancer Res. 2007;67:940–945. doi: 10.1158/0008-5472.CAN-06-2960. [DOI] [PubMed] [Google Scholar]
- 31.Seiple L, Jaruga P, Dizdaroglu M, Stivers JT. Linking uracil base excision repair and 5-fluorouracil toxicity in yeast. Nucleic Acids Res. 2006;34:140–151. doi: 10.1093/nar/gkj430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoskins J, Scott BJ. Evidence for distinct DNA- and RNA-based mechanisms of 5-fluorouracil cytotoxicity in Saccharomyces cerevisiae. Yeast. 2007;24:861–870. doi: 10.1002/yea.1516. [DOI] [PubMed] [Google Scholar]
- 33.Fung H, Demple B. A vital role for ape1/ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 34.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ludwig DL, MacInnes MA, Takiguchi Y, Purtymun PE, Henrie M, Flannery M, Meneses J, Pedersen RA, Chen DJ. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat Res. 1998;409:17–29. doi: 10.1016/s0921-8777(98)00039-1. [DOI] [PubMed] [Google Scholar]
- 36.Bronstein SM, Skopek TR, Swenberg JA. Efficient repair of O6-ethylguanine, but not O4-ethylthymine or O2-ethylthymine, is dependent upon O6-alkylguanine-DNA alkyltransferase and nucleotide excision repair activities in human cells. Cancer Res. 1992;52:2008–2011. [PubMed] [Google Scholar]
- 37.Liu N, Lamerdin JE, Tucker JD, Zhou ZQ, Walter CA, Albala JS, Busch DB, Thompson LH. The human XRCC9 gene corrects chromosomal instability and mutagen sensitivities in CHO UV40 cells. Proc Natl Acad Sci USA. 1997;94:9232–9237. doi: 10.1073/pnas.94.17.9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silber JR, Bobola MS, Blank A, Schoeler KD, Haroldson PD, Huynh MB, Kolstoe DD. The apurinic/apyrimidinic endonuclease activity of Ape1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin Cancer Res. 2002;8:3008–3018. [PubMed] [Google Scholar]
- 39.Liu L, Nakatsuru Y, Gerson SL. Base excision repair as a therapeutic target in colon cancer. Clin Cancer Res. 2002;8:2985–2991. [PubMed] [Google Scholar]
- 40.Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Res. 2005;65:6394–6400. doi: 10.1158/0008-5472.CAN-05-0715. [DOI] [PubMed] [Google Scholar]
- 41.Weller M, Steinbach JP, Wick W. Temozolomide: a milestone in the pharmacotherapy of brain tumors. Future Oncol. 2005;1:747–754. doi: 10.2217/14796694.1.6.747. [DOI] [PubMed] [Google Scholar]
- 42.Bolzan AD, Bianchi MS. Genotoxicity of streptozotocin. Mutat Res. 2002;512:121–134. doi: 10.1016/s1383-5742(02)00044-3. [DOI] [PubMed] [Google Scholar]
- 43.Schild LJ, Brookman KW, Thompson LH, Wilson DM., III Effects of Ape1 overexpression on cellular resistance to DNA-damaging and anti-cancer agents. Som Cell & Mol Genet. 2002;25:253–262. doi: 10.1023/a:1019979613989. [DOI] [PubMed] [Google Scholar]
- 44.Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17 5:v7–12. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- 45.Fischer F, Baerenfaller K, Jiricny J. Gastroenterology. Vol. 133. 2007. 5-Fluorouracil is efficiently removed from DNA by the base excision and mismatch repair systems; pp. 1858–1868. [DOI] [PubMed] [Google Scholar]
- 46.Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. J Biol Chem. 2005;280:5516–5526. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- 47.Izumi T, Brown DB, Naidu CV, Bhakat KK, MacInnes MA, Saito H, Chen DJ, Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci USA. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim HO, Shanmuganathan K, Alves AJ, Jeong LS, Beach JW, Schinazi RF, Chang CN, Cheng YC, Chu CK. Potent anti-HIV and anti-HBV activities of (-)-L-β-Dioxolane-C and (+)-L-β-Dioxolane-T and their asymmetric syntheses. Tetrahedron Letters. 1992;33:6899–6902. [Google Scholar]
- 49.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 50.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 51.Eisenbrand G, Muller N, Denkel E, Sterzel W. DNA adducts and DNA damage by antineoplastic and carcinogenic N-nitrosocompounds. J Cancer Res Clin Oncol. 1986;112:196–204. doi: 10.1007/BF00395912. [DOI] [PubMed] [Google Scholar]
- 52.Denny BJ, Wheelhouse RT, Stevens MF, Tsang LL, Slack JA. NMR and molecular modeling investigation of the mechanism of activation of the antitumor drug temozolomide and its interaction with DNA. Biochemistry. 1994;33:9045–9051. doi: 10.1021/bi00197a003. [DOI] [PubMed] [Google Scholar]
- 53.Marchesi F, Turriziani M, Tortorelli G, Avvisati G, Torino F, De VL. Triazene compounds: mechanism of action and related DNA repair systems. Pharmacol Res. 2007;56:275–287. doi: 10.1016/j.phrs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Sanderson BJ, Shield AJ. Mutagenic damage to mammalian cells by therapeutic alkylating agents. Mutat Res. 1996;355:41–57. doi: 10.1016/0027-5107(96)00021-8. [DOI] [PubMed] [Google Scholar]
- 55.Povirk LF, Shuker DE. DNA damage and mutagenesis induced by nitrogen mustards. Mutat Res. 1994;318:205–226. doi: 10.1016/0165-1110(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 56.Maanen MJ, Smeets CJ, Beijnen JH. Chemistry, pharmacology and pharmacokinetics of N, N′, N″ -triethylenethiophosphoramide (ThioTEPA) Cancer Treat Rev. 2000;26:257–268. doi: 10.1053/ctrv.2000.0170. [DOI] [PubMed] [Google Scholar]
- 57.Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 1998;58:222–225. [PubMed] [Google Scholar]