Summary
Two recent theoretical advances have described cargo transport by multiple identical motors and by multiple oppositely directed, but otherwise identical motors [1, 2]. Here we combine a similar theoretical approach with a simple experiment to describe the behavior of a system comprised of slow and fast molecular motors having the same directionality. We observed the movement of microtubules by mixtures of slow and fast kinesin motors attached to a glass coverslip in a classic sliding filament assay. The motors are identical, except that the slow ones contain five point mutations that collectively reduce their velocity ∼15-fold without compromising maximal ATPase activity. Our results indicate that a small fraction of fast motors are able to accelerate the dissociation of slow motors from microtubules. Because of this, a sharp, highly cooperative transition occurs from slow to fast microtubule movement as the relative number of fast motors in the assay is increased. Microtubules move at half-maximal velocity when only 15% of the motors in the assay are fast. Our model indicates that this behavior depends primarily on the relative motor velocities and the asymmetry between their forward and backward dissociation forces. It weakly depends on the number of motors and their processivity. We predict that movement of cargoes bound to two types of motors having very different velocities will be dominated by one or the other motor. Therefore, cargoes can potentially undergo abrupt changes in movement in response to regulatory mechanisms acting on only a small fraction of motors.
Introduction
Intracellular cargoes are often simultaneously attached to multiple types of molecular motors. The motors can have the same or opposite directionality, and they can move on actin, microtubules, or both. Melanosomes [3], lipid droplets [4], peroxisomes [5], mitochondria [6], and intraflagellar transport (IFT) particles [7], are all examples of cargoes that are transported by multiple motors. The questions of to what extent and by what mechanisms different molecular motors coordinate their motility when transporting cargoes together are being explored by both theory and experiments.
Two important theoretical treatments of cargoes attached to multiple motors have recently been published. These papers describe the movement of multiple motor systems using a “transition-rate model” that allows for motor association, dissociation, and stepping with reasonable rates. The first of these two papers modeled systems of multiple motors of the same type [2], while the second extended that treatment to model systems of multiple identical but oppositely directed motors [1]. The latter model demonstrated that experimentally observed discrete stepping behaviors of two-motor systems [8] do not necessitate a coordinating element between the motors. Small changes in the number of actively stepping motors can abruptly switch such a system between forward and backward movements.
The two motors that move cargoes in IFT, kinesin-II and Osm-3, have the same directionality but velocities that differ by a factor of three, as shown by Pan, et al. [7]. In that study, these two motors were combined in a classic in vitro sliding filament motility assay as described [9, 10]. Microtubule movement by mixtures of these two motors in vitro fit well to two models. The first, termed “alternating action”, assumes that the fast and slow motors step completely independently of one another. The second, termed “mechanical competition”, assumes that all motors of both types are always attached and moving, and the forces generated by them are balanced. Subsequent analysis of the behavior of these motors and associated proteins in vivo led these authors to favor the “mechanical competition” model [7]. While both of these models explained the behavior of kinesin-II and Osm-3 motors in vitro, they differ significantly from the approach taken in the aforementioned transition-rate model that allows for motor dissociation from the microtubule.
In both the transition-rate model and the mechanical competition model for multiple-motor systems having two types of motors, the velocity of cargo movement depends on the dissociation forces of the two types of motors involved. If these forces are unequal, either the slow motors decelerate the fast ones or the faster motors accelerate the slow ones. In the latter case, the slow motors would remain microtubule-bound but actually step faster in the mechanical competition model, while the dissociation of slow motors from the microtubule would occur preferentially in the transition-rate model.
Interestingly, single kinesin dimers have asymmetric forward and backward dissociation forces. The forward detachment force is about 45% less than the backward detachment force for a kinesin motor head bound to a microtubule in either a triphosphate nucleotide state or the nucleotide-free state [11]. These asymmetric detachment forces are likely to be critical for the processive mechanism of a single kinesin-1 dimer, and they have been implicated in internal strain mechanisms for kinesin motors (for a discussion of these, see [12]). Some recent results indicate that internal strain acting between the heads of a kinesin dimer can affect the rate of rear head release [13], [14]. In addition, internal strain may affect the rate of ATP binding to the front head, effectively forcing the front head to wait for the rear head to enter a weak binding state before binding ATP [15] [16]. Both of these gating mechanisms may act to increase kinesin processivity, and processive movement is observed even in the surprising case of a kinesin dimer containing one “dead” head that cannot hydrolyze ATP [17]. Abolishing internal strain between the two heads by extending the neck linker compromises processivity [18, 19]. The present work suggests that the same asymmetric forward and backward detachment forces that influence the gating and processive movement of kinesin motors also influence systems of several fast and slow kinesin motors, independent of motor processivity.
The present work extends the transition-rate model to describe a system of two motors having the same directionality, but different velocities. We are able to recapitulate the behavior of the IFT motors described in [7], but also demonstrate that a model that accounts for motor dissociation, like the transition-rate approach used here, is required to describe the behavior of a system of two motors having very different velocities. Our theoretical model uses literature values of dissociation forces, binding and unbinding rates, experimentally measured velocities, and an estimate of the number of motors. Our model is not a fit as there are no free parameters, but it matches our experimental data quite well.
Results
Neck linker mutants generate slow kinesins having reasonable ATPase and microtubule-binding properties
A simplifying aspect of the published transition-rate theoretical treatments of kinesin motors [1, 2] is that the motors are assumed to be identical, except for their directionality [1]. For our experiment, we wished to create a system of two virtually identical kinesin motors with different velocities, but with all other relevant parameters the same (see Table 1 for a list of parameters). These motors could then be combined in a classic sliding filament assay to create the simplest possible experimental system for observing two different motors having different velocities but the same directionality [7, 9, 10] (Figure 1A). To this end, we have used a 576-residue Drosophila kinesin construct as our fast motor, and the same construct containing a total of five point mutations as our slow motor. Details of fast and slow motor DNA constructs and protein purifications can be found in Supplemental methods. The slow motor mutations are found in the neck linker and neck linker interface of kinesin, and they are similar to those described in [20] (Figure 1B). In the theoretical treatment, the binding rates, unbinding rates, forward detachment forces, and backward detachment forces of fast and slow motors were assumed to be identical to simplify our calculation. In reality, these parameters may differ for the two motor types as they have not been directly measured. To address these concerns, we altered the above parameters in our model and compared the output with the experimental data (discussed below).
Table 1. Parameters used in the theoretical model along with literature references.
Parameters shown are for kinesin dimers.
| Symbol | Description | Value | Reference |
|---|---|---|---|
| ε0 | Single motor unbinding rate | 1 sec−1 | [25] |
| π0 | Single motor binding rate | 5 sec−1 | [26] |
| Vf | Single motor velocity, wild type | 0.522 μm/s | this work |
| Vs | Single motor velocity, mutant | 0.034 μm/s | this work |
| Fstall | Stall force | 6 pN | [32] |
| Fd | Dissociation force, minus-end direction | 3 pN | [32] |
| η | Dissociation force asymmetry | 45% | [11] |
| N | Number of motors available to bind | 10 | estimated, this work |
Figure 1. Sliding motility assay using nearly identical fast and slow kinesin motors.
(A), Diagram of a sliding filament assay with fast and slow motors as performed here. The average forces exerted by the motors toward the plus and minus ends of the microtubule balance, but the forward and backward dissociation forces of the motors are asymmetric ([11], see text for discussion). This particular configuration represents N=5 dimeric kinesin motors (Nf=2 and Ns=3) and n=4 (nf=2 and ns=2). See text for definitions. (B), Locations of the five point mutations used to create the slow kinesin motor are indicated usingDrosophila kinesin numbering on the crystal structure of kinesin-1 from PDB ID #1mkj [33]. This figure was generated using PyMol software (www.pymol.org). (C), ATPase activity and velocity of fast and slow motors in the sliding filament assay. Details are described in the Experimental and Supplemental Methods sections.
Figure 1C shows the microtubule-stimulated ATPase activity and microtubule sliding velocities of the fast and slow kinesin motors used in this study. The maximal ATPase rates of the two motors are comparable; 21.8±1.0 ATP/head/s for the fast kinesin and 23.2±0.94 ATP/head/s for slow kinesin. The Km(MT), or the concentration of microtubules at which the motor has half-maximal ATPase activity, of slow kinesin is lower than fast kinesin by a factor of 6.6. The velocity of slow kinesin movement is reduced 15-fold relative to fast kinesin (2 μm/min vs. 34 μm/min).
These measurements are consistent with past data on the properties of kinesin motors containing similar mutations. These mutants have a similar maximal ATPase rate to wild-type kinesin, while their stepping is slow due to defective neck linker docking [20]. Mutations that affect neck linker docking have been shown to result in motors having a lower Km(MT) as we observed, despite the fact that these mutations are not part of the motor's microtubule-binding interface [10, 21]. Kinesin motors that are cross-linked to prevent the neck linker from docking to the core of the motor dissociate from microtubules more slowly than kinesin motors that are cross-linked to force neck linker docking [22]. This is consistent with the fact that a motor head that cannot dock its neck linker “waits” for a longer period of time in a tightly bound apo- (nucleotide-free) state on microtubules [15, 16]. This prolonged, tightly bound apo- state explains why both the cross-link and neck linker mutations result in motors with lower Km(MT)s. While this apo- state is prolonged in the slow motor, it is not rate-limiting, as evidenced by the normal Kcat for the mutant. See supplemental data for a more complete discussion and kinetic model that is consistent with the slow motor's ATPase activity and velocity.
In the theoretical treatment here, we assume that the forward and backward unbinding forces of kinesin differ by the same 45% measured by [11], and we assume that the slow and fast motors are identical in this respect. The fast motors will, on average, experience backward loads. The front heads of the fast motors will either be in a nucleotide-free or ATP state, both of which have ∼2.2-fold higher unbinding forces in backward direction than the forward direction [11]. The slow motors, on the other hand, move at a significantly lower speed and will, on average, experience forward loads. Therefore, the unbinding force of a slow motor in the forward direction will be lower than the unbinding force of a fast motor under backward load, regardless of its nucleotide state.
In vitro motility assays on mixtures of fast and slow kinesins reveal cooperative behavior
Observations of microtubule movement in sliding filament assays containing mixtures of fast and slow motors gave the first clue that fast motors accelerate the dissociation rates of slow motors. Figure 2 shows movement of microtubules over a time period of 1 minute by fast motors (A), slow motors (B), by a mixture containing approximately 40% fast and 60% slow motors with the same total concentration (≈10 motors per microtubule) (C), or the same mixture of motors at near-limiting dilution, with ≈2−3 motors per microtubule (D). These montages were generated from movies of fluorescent microtubules that were contrast-enhanced and inverted to appear black on a white background. A comparison of the montages on the left side of Figure 2 A and C reveals that the average velocity of microtubules in the mixture is not significantly different from fast kinesin alone at the higher density. The velocity at low motor density (Figure 2D, left side) was more variable. In the images on the right side, the same microtubules are shown, except that the microtubule from each movie slice is positioned horizontally, and the left side of the microtubule in each frame is aligned. This repositioning shows movements of different parts of the microtubule relative to the ends. The microtubules have visible light and dark zones throughout their length, which contain different amounts of fluorescent probe. In images of pure fast or slow motors (Figure 2 A and B), the positions of these light or dark zones remain constant relative to the microtubule end, even when the trajectories of the actual microtubules are not straight. In contrast, the images of mixtures of fast and slow motors at both high and low total motor densities show locations where different parts of the microtubule move relative to each other, observable as deviations in the position of light zones relative to the microtubule ends over the timecourse. One such region in the mixture at higher density first slows down, then speeds up, relative to the microtubule end (Figure 2G, *), possibly due to recoiling of the microtubule after motor detachment. This data qualitatively indicates first, that slow motors in the mixture are able to bind to microtubules, and second, that they may be forced to dissociate from microtubules by fast motors. The microtubule recoiling behavior is not explicitly modeled here, as our model assumes that the force stored in the microtubule is shared by all motors and that the averages of those forces balance. Furthermore, our model does not account for any elasticity in the microtubule or stochastic differences in the behavior of individual motors, which would cause the observed recoiling behavior. These properties could be specifically addressed using Monte Carlo methods similar to those used by Kunwar, et al. to describe the movement of organelles by multiple kinesin motors [23]. However, for the purposes of this work, the recoiling of microtubules serves only as a visual demonstration that fast motors may cause slow motors to dissociate.
Figure 2. Movement of microtubules by fast motors, slow motors, and a mixture.
A-D, left side images show three movie frames taken 20 seconds apart showing movement of microtubules. Right side images are kymographs of the same microtubules with one end of the microtubule aligned on the left side to show movements of different regions within the microtubule relative to each other. The * in panel C shows a time period over which a lighter region of a microtubule slows down, then speeds up relative to the microtubule end, consistent with a slow motor dissociation event.
The data shown in Figure 2 indicates that the movement of microtubules by a mixture of fast and slow motors may be positively cooperative, that is, fast motors can accelerate the dissociation rates of slow motors such that the entire system moves at a faster rate than would be expected if the motors did not dissociate. To test whether cooperative dissociation effects significantly alter the behavior of our system, we measured the velocities of different mixtures of fast and slow motors, leaving the total concentration of motors in the assay nearly constant (Figure 3A, Experimental). As the ratio of fast motors to total motors added in the assay (referred to as throughout this paper) was increased, we observed a sharp transition between low and high-velocity microtubule movement. The half-maximal velocity was reached when only about 15% of the motors added to the assay were fast. The microtubule velocities that we measured were more variable in the region of the transition (visible as large error bars in the microtubule velocities measured for between 0.1 and 0.3 in Figure 3A). This is consistent with the fact that slight variations in the number of fast or slow motors moving a particular microtubule can result in large changes in velocity as the system is undergoing the sharp transition from slow to fast movement.
Figure 3. Cooperative behavior in motility assays containing mixtures of fast and slow motors.
A, Microtubule velocity vs. ratio of fast to total motors added to the assay. See Experimental for details on methods and errors. B, Proportion of fast to total motors moving a microtubule , (estimated as described in text) vs. proportion of fast to total motors added to the assay, . Circles show data from the present study, squares from Pan, et al. [7]. The dashed lines in both panels show the results of our model taking motor dissociation into account, described in the text.
In contrast to our data, Pan, et al. observed a smooth concave upward trend in their plot of velocity vs. (Figure 3A, [7]). This is predicted by the alternating action model as a simple consequence of slow motors requiring a longer time to take a single step than fast motors and it does not imply any cooperativity between fast and slow motors. The alternating action model would predict a sharper concave upward trend at a high for our data because of the large difference in velocities of our fast and slow motors. This clearly deviates from our experimental results. Therefore, kinesin-II/Osm-3 motility in vitro can be modeled reasonably well assuming that the motors do not exert any forces on each other whatsoever, while our fast and slow kinesins cannot.
Both the alternating action and mechanical competition models described by Pan, et al. give the relationship between cargo velocity and the number of fast and slow motors bound to a cargo [7]. This can be used to estimate the ratio of fast motors to total motors moving a given microtubule, , from the microtubule velocity in our system (details given in Supplemental data section). A comparison of this estimate to , the ratio of fast to total motors added to the assay can give an idea of whether the system is cooperative or not. We define a positively cooperative system as one in which fast motors cause slow motors to dissociate from the microtubule, such that a larger proportion of fast motors move the microtubule than are added to the assay, in other words, . For our system, in which the stall forces of the two motors are assumed to be the same, the two models described in Pan, et al. [7]yield an identical result, given by equation (1) below.
| (1) |
In the absence of any cooperative behavior, a plot of vs. should yield a straight line. Figure 3B compares the data for our fast and slow kinesins with the data from Pan, et al. on kinesin-II and Osm-3 [7]. Interestingly, the Pan, et al. data (Figure 3B, squares) appears to be mostly non-cooperative, falling very near the straight line. This explains why their data fits reasonably well to the alternating action model that assumes that the motors are completely non-interacting. In contrast, the present data (Figure 3B, circles) can only be explained if the motors alter each others’ dissociation kinetics such that fast motors accelerate the dissocation of slow motors. The dashed lines in Figure 3A and 3B show the results of our theoretical model, described in more detail in the remainder of the paper.
An important property of both the experiments and the theory described here is that they depend on , which can become highly variable at low motor density. This results in more variability in the velocity of moving microtubules observed in experiments (Figure 2 C, D). Our theory assumes that an integer number of motors are available to bind the microtubule. For example, when 5 motors are available to bind to the MT, a total of 4 discrete MT velocities can be predicted by the theory. The extreme case is a single motor, which would lead to a microtubule traveling either fast or slow and never at any intermediate velocity. Both the theory and the experimental measurements of microtubule velocity are more robust at reasonably high motor density, therefore we performed our experiments at approximately 10-fold greater concentration than a limiting dilution (N≈10).
Microtubules moved by fast and slow motors match a transition-rate model
Figure 3B shows that a model in which it is assumed that fast and slow motors do not dissociate from the microtubule can be used to describe the movement of Osm-3 and kinesin-II fairly well [7]. However, it does not reproduce the sharp transitions in the plots of velocity vs. obtained from our experiments on fast and slow motors whose isolated velocities differ by over an order of magnitude. Figure 3B also shows that the solution molar fraction and binding molar fraction of fast motors to total motors can be very different if there is a significant mismatch between motor velocities. We hypothesize that at reasonably high motor density, the velocity difference between fast and slow motors in our system results in a stress that the microtubule distributes across the system, allowing fast motors to accelerate the dissociation of slow motors. To calculate the average number of bound fast motors 〈nf〉 and slow motors 〈ns〉 at each mole fraction, we have adapted the method of Klumpp and Lipowsky [2], replacing their external force term (used in their work to model the behavior of a single group of motors under a shared load such as an optical trap) with internal forces resulting from the difference between the filament velocity and the single-motor velocity. The fast motors produce a force in the plus-ended direction, F+, and the slow motors produce a force in the minus-ended direction, F−:
| (2) |
where Fstall is the maximum force produced by each motor. Eq. (2) is a consequence of the linear force-velocity relationship of the kinesin motor protein at the 3pN dissociation force, and it ensures that internal forces vanish when the velocity mismatch is zero. We note, however, that at high loads the force-velocity relationship may deviate from linearity [24].
We employ a mean-field approximation where F+, which acts to pull off the slow motors, will be distributed over the number of slow motors bound, and F−, which acts to pull off the fast motors, will be distributed over the number of fast motors bound. Following [2], we write the binding rates, π, and unbinding rates, ε, in terms of the number of bound motors of each type:
| (3) |
| (4) |
where the single unloaded motor binding rate π0 and the single unloaded motor unbinding rate ε0 are the same for each type of motor. The unbinding rate is characterized by the dissociation force, Fd. We account for the asymmetry in unbinding between plus-directed and minus-directed forces with the asymmetry parameter η [11]. This asymmetry is such that the forward unbinding force is 45% of the backward unbinding force, so we expect that slow motors, which experience forces in the forward direction (Figure 1A), will be pulled off more readily.
These transition-rates can be inserted into the technique of Klumpp and Lipowsky (see methods) to yield average 〈ns〉 and 〈nf〉 bound to the microtubule for each given molar solution fraction Nf/N. 〈Vmt〉 is calculated from equation (5) below [7] which is derived in the Supplemental data section.
| (5) |
One advantage of our approach is that it has no free parameters. N is roughly estimated (see methods) to be 10. All other parameters are taken from the literature or independently measured in our system. These are summarized in Table 1, and a simulation based on these tabulated parameters is shown as the dashed line in both Figure 3A and B.
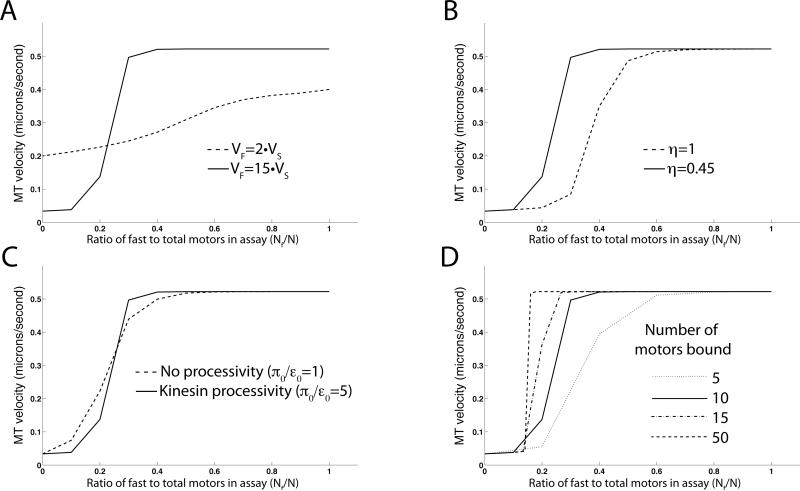
Using this model, several different properties of the system can be explored. First, consider the effects of the velocity mismatch between the two motors. Figure 4A shows the simulation for fast and slow motors having the velocity mismatch that we measured for our fast and slow motors (Figure 1C and Figure 4A, solid line). An abrupt transition between fast and slow microtubule velocities results from the large internal forces generated by the two types of motors. An intermediate velocity is rare, since that would indicate that neither the fast nor the slow motors win the tug-of-war [1]. The dashed line shows the simulation for a motor system where the fast motor is only twice the speed of the slow motor. A smooth transition is seen, as the velocity is averaged between the two different motor types. Even at extremely high motor concentrations, the internal forces are not large enough to drastically affect the binding rates of these two motors (data not shown).
Figure 4. Dependence of transition-rate model on several parameters.
Plots of microtubule velocity vs. mole fraction of fast motor are shown for (A) different fast and slow motor velocities, (B) with and without the asymmetry between forward and backward dissociation forces, (C) for processive and non-processive motors, and (D), for different numbers of motors available to bind the microtubule. Model parameters described in the text are varied as indicated in each panel.
Figure 4B compares the system of fast and slow motors with symmetric forward and backward dissociation forces (η = 1; dashed line) and with the asymmetry found in these forces by [11] (η = 0.45; solid line). The abruptness of the transition from slow to fast movement is not significantly altered for different values of η; however, the location of the transition is shifted to the right for η = 1 as motors are no longer preferentially detached when pulled forward.
The effect of processivity (defined here as π0/ε0 , the ratio of the binding and unbinding rates for single motors) in this multiple motor system is much less pronounced. In our model, motors with the same ratio π0/ε0 exhibit identical behavior, regardless of the particular values of the binding and unbinding rates. Figure 4C shows the system of fast and slow motors with π0/ε0 =5 (solid line) compared to an otherwise identical system having π0/ε0 =1, which would correspond to a nonprocessive motor (dashed line). The motor detachment rate ε0 of 1 s−1 represents the processivity of a single unloaded fast kinesin motor [25], while the motor attachment rate π0 of 5 s−1 is the attachment rate of a single kinesin motor that is part of a multi-motor assembly attached to cargo [26] (Table 1). In reality, motor detachment rates in systems of multiple motors are difficult to estimate due to differences in linkage stiffnesses and the internal forces that the motors exert on one another. It is worth noting that the work of Rogers, et al. in this issue has estimated a lower bound of 4 s−1 for ε2 in a system of two coupled motors and a correspondingly lower than expected single-motor processivity (defined as the ratio π/ε2) for the composite system [27]. This is a minor point for our study, as our model does not change substantially for different reasonable values of π and ε, but in accordance with their result, low processivity (π0/ε0 =1) seems to reflect our data more accurately, as seen in Figure 4C. While in single-molecule assays the processivity of a motor has a significant effect on the observed properties of cargo transport (e.g. run length) [28, 29], at high motor densities processivity is far less critical because the cargo is very likely to be bound by at least one other motor. The effect of reduced unbinding rates at N = 10 is to slightly smooth out the velocity transition.
The final model for several values of N is shown in Figure 4D. The sharp transition between slow and fast microtubule velocity becomes sharper at high motor density, but the difference between N=10 and N=50 is surprisingly subtle. We estimate that N=10 (methods), but concentration estimates at non-limiting dilutions in motility assays are not terribly accurate, as discussed in [30]. Figure 4D shows that an inaccurate estimation of N would not greatly affect our model or its comparison to our experimental data.
Clearly, the velocity mismatch between the slow and fast motors and the asymmetry between the forward and backward dissociation forces are critical parameters for agreement of the model with our data. Assuming a higher unbinding rate and therefore lower motor processivity in our theoretical model (Figure 4C) actually agrees better with the data than using the literature values for binding and unbinding rates shown in Table 1. The accurate representation of the data by this model supports the hypothesis that mechanical competition between different motors modulates the motor unbinding rates as well as the cargo velocity in multiple motor systems.
Discussion
A comparison of mechanical competition and transition-rate models for two-motor systems
Our work shows that in systems of two same-direction motors, motor dissociation becomes more important as the mismatch between the two motors’ velocities increases. The transition-rate model that we use to describe our system of motors with a large velocity mismatch is quite different from the two models used by Pan, et al. that successfully described the movement of Osm-3 and Kinesin-II [7]. The most important distinction between the two models is this: in the Pan, et al. models, slow motors remain microtubule-bound, but they can be induced to effectively step faster by fast motors moving the microtubule at the faster rate. In the transition-rate model, slow motors always travel slowly, but they can be forced to dissociate completely from the microtubule by fast motors. Our transition-rate model recapitulates the behavior of Osm-3 and Kinesin-II shown by Pan, et al. if we make the assumption that the two motors exert very low forces on each other (possibly due to low linkage stiffnesses between motors and glass) and have very high binding rates. This causes both motors to remain microtubule-bound essentially 100% of the time, just as in the Pan, et al. models [7].
In contrast, both our experimental observations and our model indicate that in our system, motor dissociation occurs frequently. We observed that specific regions within a microtubule can slow down, then speed up relative to the microtubule end, which is qualitatively consistent with a slow motor detachment event. These events were observed at both high (N≈10) and low motor density (N≈2−3, Figure 2C and D). Both our model and our experiment are better suited for evaluating cargo movement by >5 motors than by very small numbers of motors. However, a quantitative treatment of a system having N=2 motors appears elsewhere in this issue [27]. We observed a steep transition from slow to fast movement at a small ratio of . In a mechanical competition model, such a transition would indicate that the stall force of the fast motors is far greater than the stall force of the slow motors. This is unlikely in our system because the fast and slow motors are identical except for 5 point mutations. In contrast, the transition-rate model predicts a steep transition from slow to fast movement if the overall force exerted by fast motors on slow motors becomes greater than the force exerted by slow motors on fast motors. Because the fast motors have higher velocity, this transition is predicted to occur at a relatively small ratio of fast to slow motors, as we observed experimentally.
Effects of asymmetry between forward and backward dissociation forces
To keep our model as simple as possible, we assumed that the fast and slow motors used here were identical in their forward and backward dissociation forces as well as their binding and unbinding rates. Because the forward dissociation force of both motors is 45% of the backward dissociation force, this asymmetry gives fast motors pulling toward the weaker forward direction an advantage over the slow motors. Our data matches our model when this asymmetry is taken into account, using the values obtained by [31].
By assuming that our fast and slow motors are identical in every respect except for their velocities, we offer the simplest possible interpretation of our data. However, we have not measured any of the parameters of Table 1 for the slow motors. It may be the case (for example) that the forward and backward dissociation forces of our fast and slow motors are similar but that the slow motor has a significantly lower stall force than fast motor, which would yield essentially the same result. The asymmetry between forward and backward dissociation forces is a parameter that we expect to be highly specific to the motor type used and potentially extremely important for modeling multiple-motor systems.
Experimental
Functional assays
ATPase assays were performed as previously described [10, 20], using purified porcine tubulin and BRB80 buffer in a Beckman DU800 spectrophotometer. For motility assays, coverslips for flow chambers were cleaned with ethanol and acetone, agitated with 1M KOH for 20 minutes at room temperature, then thoroughly rinsed with ddH20. Flow chamber volumes were standardized to a 10 μl volume, then blocked with 0.5mg/ml casein for 5 minutes before each assay. The chambers were then washed with two volumes of BRB80 (80 mM PIPES-KOH, 1 mM EGTA, 2 mM MgCl2, pH 6.9) before a 5-minute incubation with the appropriate motor mixture. Assuming all protein was transferred to the chamber, 70ng of total kinesin was added for each mixture of WT and mutant. This is likely an overestimate because of kinesin motors lost in tubes and tips during the dilution process. A limiting concentration of kinesin in our sliding motility assay was empirically determined to be around 3−7 ng. Movements were rare at that concentration and detachment events occurred at the ends of microtubules. Based on this limiting concentration, we estimate that after adding 70 ng of kinesin to the chamber, we have ∼10 motors bound per microtubule. These measurements are subject to several sources of error (see [30] for a discussion), but our results do not depend strongly on the number of motors (Figure 4D). After incubation, each cell was washed with one volume of BRB80 and loaded with tetramethylrhodamine-labeled microtubules stabilized with 10 μM paclitaxel, diluted 1:500 in an Mg-ATP-containing oxygen-scavenger solution (0.1 mg/ml glucose-oxidase, 0.1 mg/ml catalase, 1 mM BME, 10 mM glucose) with an additional 10 μM paclitaxel. The chamber was then sealed with vacuum grease. All time-lapse movies were conducted on a Nikon TE2000-E microscope through a Photometrics Coolsnap-EZ camera using Metamorph version 7.01 acquisition software. Tracking was done using the MtrackJ plugin for NIH ImageJ. From each slide, microtubules were tracked during 31 consecutive 0.5-second frames. Velocities of individual microtubules were calculated as mean±standard deviation of these 31 measurements, and 8−20 separate measurements were averaged together and their errors propagated for the final measurements shown in Figure 3.
Theoretical methods
Equation (2) couples the calculations for fast and slow motor binding distributions. When motor dissociation is allowed, this equation does not permit an immediate analytic solution for the average microtubule velocity. Instead an iterative approach is followed for each molar solution fraction Nf/N. First, a trial evaluation of Equation (2) is made using guess values for 〈nf〉 and 〈ns〉. This in turn permits calculation of F−(nf,〈ns〉) and F+(〈nf〉,ns). At this point, the method of [2] is followed independently for each motor, as described below. The binding and unbinding rates are evaluated, and these in turn are used to generate a binding probability distribution Pn:
| (6) |
From these distributions, the approximations for 〈nf〉 and 〈ns〉 are updated:
| (7) |
The process is then repeated until the probability distributions are stable to within a tolerance of 1%. The solutions are checked for stability with respect to starting guesses, and also to verify that a stable probability distribution results in zero net force (i.e. the microtubule is not accelerating). Once self-consistent values for for〈nf〉 and 〈ns〉 have been determined, the average effective microtubule velocity Vmt (〈nf〉,〈ns〉) is calculated from Equation (5).
Conclusion
Our major result should caution experimenters performing sliding motility assays on a slow motor in the presence of a contaminating fast motor. Small amounts of a fast contaminant could result in robust fast velocity. Thus, care must be exercised in preparation of slow or reverse-direction motors when using sliding motility assays to determine directionality and/or velocity.
Our results predict that a system of transporters having very different speeds, differently-directed transporters, or transporters moving on differently polarized filaments (e.g. an actin-based motor and a microtubule-based motor that would act in two different directions) could transition very quickly between movement that is dominated by one type of motor and movement that is dominated by another. This is interesting to note, because organelles being moved by multiple different motors can undergo abrupt changes in velocity and directionality of movement even though the number of motors on them is roughly equivalent [8]. The cargo velocity or directionality could be altered by a regulatory switch acting on just one of the components, and this regulatory switch may cause large changes in the system properties by eliciting small changes in . In other words, regulatory mechanisms need not act on a large fraction of motors to alter cargo movement significantly and abruptly.
Supplementary Material
Acknowledgments
SER and AGL are supported by National Institutes of Health 5 R01 GM072656−02. ECL is supported by the College of Liberal Arts and Sciences, DePaul University. The authors wish to acknowledge Michael Diehl, Yao Liang Wong and Mark Alan Seeger for critical commentary on the work, and Park Packing Company for donation of materials for tubulin preparation.
References
- 1.Muller MJ, Klumpp S, Lipowsky R. Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc Natl Acad Sci U S A. 2008;105(12):4609–14. doi: 10.1073/pnas.0706825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klumpp S, Lipowsky R. Cooperative cargo transport by several molecular motors. Proc Natl Acad Sci U S A. 2005;102(48):17284–9. doi: 10.1073/pnas.0507363102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gross SP, et al. Interactions and regulation of molecular motors in Xenopus melanophores. J Cell Biol. 2002;156(5):855–65. doi: 10.1083/jcb.200105055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross SP, et al. Coordination of opposite-polarity microtubule motors. J Cell Biol. 2002;156(4):715–24. doi: 10.1083/jcb.200109047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H, et al. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J Cell Biol. 2007;176(5):641–51. doi: 10.1083/jcb.200608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pilling AD, et al. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell. 2006;17(4):2057–68. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan X, et al. Mechanism of transport of IFT particles in C. elegans cilia by the concerted action of kinesin-II and OSM-3 motors. J Cell Biol. 2006;174(7):1035–45. doi: 10.1083/jcb.200606003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kural C, et al. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science. 2005;308(5727):1469–72. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 9.Cohn SA, Ingold AL, Scholey JM. Quantitative analysis of sea urchin egg kinesin-driven microtubule motility. J Biol Chem. 1989;264(8):4290–7. [PubMed] [Google Scholar]
- 10.Woehlke G, et al. Microtubule Interaction Site of the Kinesin Motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- 11.Uemura S, et al. Kinesin-microtubule binding depends on both nucleotide state and loading direction. Proc Natl Acad Sci U S A. 2002;99(9):5977–81. doi: 10.1073/pnas.092546199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys J. 2007;92(9):2986–95. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crevel IM, et al. What kinesin does at roadblocks: the coordination mechanism for molecular walking. Embo J. 2004;23(1):23–32. doi: 10.1038/sj.emboj.7600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schief WR, et al. Inhibition of kinesin motility by ADP and phosphate supports a hand-over-hand mechanism. Proc Natl Acad Sci U S A. 2004;101(5):1183–8. doi: 10.1073/pnas.0304369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guydosh NR, Block SM. Backsteps induced by nucleotide analogs suggest the front head of kinesin is gated by strain. Proc Natl Acad Sci U S A. 2006;103(21):8054–9. doi: 10.1073/pnas.0600931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenfeld SS, et al. Stepping and stretching. How kinesin uses internal strain to walk processively. Journal of Biological Chemistry. 2003;278(20):18550–6. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thoresen T, Gelles J. Processive movement by a kinesin heterodimer with an inactivating mutation in one head. Biochemistry. 2008;47(36):9514–21. doi: 10.1021/bi800747e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackney DD, et al. Modulation of kinesin half-site ADP release and kinetic processivity by a spacer between the head groups. Biochemistry. 2003;42(41):12011–8. doi: 10.1021/bi0349118. [DOI] [PubMed] [Google Scholar]
- 19.Yildiz A, et al. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134(6):1030–41. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Case RB, et al. Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr. Biol. 2000;10:157–160. doi: 10.1016/s0960-9822(00)00316-x. [DOI] [PubMed] [Google Scholar]
- 21.Sindelar CV, Downing KH. The beginning of kinesin's force-generating cycle visualized at 9-A resolution. J Cell Biol. 2007;177(3):377–85. doi: 10.1083/jcb.200612090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahlen K, et al. Feedback of the kinesin-1 neck-linker position on the catalytic site. J Biol Chem. 2006;281(27):18868–77. doi: 10.1074/jbc.M508019200. [DOI] [PubMed] [Google Scholar]
- 23.Kunwar A, et al. Stepping, strain gating, and an unexpected force-velocity curve for multiple-motor-based transport. Curr Biol. 2008;18(16):1173–83. doi: 10.1016/j.cub.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435(7040):308–12. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 25.Vale RD, et al. Direct observation of single kinesin molecules moving along microtubules. Nature. 1996;380(6573):451–3. doi: 10.1038/380451a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leduc C, et al. Cooperative extraction of membrane nanotubes by molecular motors. Proc Natl Acad Sci U S A. 2004;101(49):17096–101. doi: 10.1073/pnas.0406598101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers AR, et al. Negative Interference Dominates Collective Transport of Kinesin Motors in the Absence of Load. Physical Chemistry Chemical Physics. 2009;(this issue) doi: 10.1039/b900964g. [DOI] [PubMed] [Google Scholar]
- 28.Pierce DW, et al. Single-molecule behavior of monomeric and heteromeric kinesins. Biochemistry. 1999;38(17):5412–21. doi: 10.1021/bi9830009. [DOI] [PubMed] [Google Scholar]
- 29.Romberg L, Pierce DW, Vale RD. Role of the kinesin neck region in processive microtubule-based motility. J Cell Biol. 1998;140(6):1407–16. doi: 10.1083/jcb.140.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hancock WO, Howard J. Processivity of the motor protein kinesin requires two heads. J Cell Biol. 1998;140(6):1395–405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uemura S, Ishiwata S. Loading direction regulates the affinity of ADP for kinesin. Nat Struct Biol. 2003;10(4):308–11. doi: 10.1038/nsb911. [DOI] [PubMed] [Google Scholar]
- 32.Schnitzer MJ, Visscher K, Block SM. Force production by single kinesin motors. Nat Cell Biol. 2000;2(10):718–23. doi: 10.1038/35036345. [DOI] [PubMed] [Google Scholar]
- 33.Sindelar CV, et al. Two conformations in the human kinesin power stroke defined by X-ray crystallography and EPR spectroscopy. Nat Struct Biol. 2002;9(11):844–8. doi: 10.1038/nsb852. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.