SUMMARY
The proliferative lifespan of normal somatic human cells in culture terminates in a permanent growth-arrested state known as replicative senescence. In this study we show that RNA interference-mediated repression of the genes encoding the small ubiquitin-related modifier (SUMO)-specific proteases, Senp1, Senp2, and Senp7, induced low passage primary human fibroblasts to senesce rapidly. Following Senp1 repression, we observed a global increase in sumoylated proteins and in the number and size of nuclear SUMO-containing PML bodies. SUMO/PML bodies also increased during replicative senescence. p53 transcriptional activity was enhanced towards known p53 target genes following repression of Senp1, and inhibition of p53 function prevented senescence after Senp1 repression. These data indicate that Senp1 repression induces p53-mediated premature senescence and that SUMO proteases may thus be required for proliferation of normal human cells.
INTRODUCTION
Replicative senescence is an irreversible growth-arrested state achieved by cells in culture after a finite number of cell divisions (Hayflick et al., 1961). The timing of entry of individual cells into replicative senescence during passage varies greatly, resulting in a slow, gradual decline in the proliferative potential of a heterogeneous cell population (Herbig et al., 2003). Viral oncogenes or constitutive expression of telomerase can allow cultured human cells to bypass senescence and proliferate indefinitely (Sedivy, 1998). Replicative senescence is regarded as a model of cellular aging, and recent results indicate that senescence is an important tumor suppressor mechanism (Campisi, 2005; Chen et al., 2005; Michaloglou et al., 2005). Most notably, mutations that impair senescence result in increased tumor incidence in mouse models of cancer (Braig et al., 2005; Collado et al., 2005a; Collado et al., 2005b).
A state closely resembling replicative senescence can be acutely induced in normal or cancer cells by various stimuli such as activated oncogenes, oxidative stress, chemotherapeutic agents, and the overexpression or reactivation of tumor suppressors, including p53 and p105Rb (Alexander et al., 2001; Cammarano et al., 2005; Chang et al., 1999; Shay et al., 2004; Sugrue et al., 1997; Toussaint et al., 2002; Xu et al., 1997). In contrast to replicative senescence, induced senescence can be rapid and homogeneous, making analysis of key cellular events during senescence more tractable. Furthermore, mRNA expression profiling of induced and replicative senescence demonstrated substantial overlap in the pattern of gene expression in these two senescence systems (Hardy et al., 2005; Johung et al., 2007; Nickoloff et al., 2004).
Sumoylation, the post-translational modification of proteins by conjugation to the small ubiquitin-related protein, SUMO, affects more than 60 proteins involved in numerous cellular processes including transcription, signal transduction, nuclear transport, stress response, and genome integrity (Seeler et al., 2003). Sumoylation occurs in a manner similar to ubiquitination and involves an E1 activating enzyme, an E2 conjugating enzyme, and E3 SUMO ligases (Hochstrasser, 2001), and it may result in increased stability, altered localization, or enhanced transcriptional activity of its targets. Four SUMO paralogues exist in humans. Under normal conditions, most SUMO-1 is present in its conjugated form, while SUMO-2 and SUMO-3 are primarily unconjugated, and increased conjugation of SUMO-2/3 to substrates is believed to be involved in stress responses (Dohmen, 2004). Very little is known about SUMO-4, which has been linked to a number of autoimmune diseases (Pearce et al., 2006; Wang et al., 2006).
Sumoylation is reversible, and the removal of SUMO, desumoylation, is catalyzed by SUMO-specific proteases. Six SUMO proteases have been identified in humans, and they are encoded by the sentrin gene family. As well as desumoylating SUMO-protein conjugates (isopeptidase activity), some SUMO proteases also process SUMO precursors to the mature form that can be conjugated to substrates (endopeptidase activity). SUMO proteases display differential activity in their relative desumoylation abilities. Senp1, a nuclear SUMO-specific protease, can cleave SUMO-1, -2, and -3 conjugates and is involved in the regulation of sumoylation of proteins that mediate proliferation and senescence such as p53, PML, HIPK2, TRF1, TRF2, and Ets-1 (Chen et al., 2003; Gong et al., 2000; Ji et al., 2006; Kim et al., 2005; Potts et al., 2007). Of the six SUMO-specific proteases, Senp1 has the highest endopeptidase and isopeptidase activity in vitro (Mikolajczyk et al., 2007). Knockout of Senp1 in mice is embryonic lethal, and Senp1-/- mouse embryonic fibroblasts (MEFs) accumulate sumoylated proteins and display decreased pools of free SUMO, which is consistent with the notion that Senp1 is the dominant SUMO-specific isopeptidase (Yamaguchi et al., 2005).
Recently, components of the sumoylation pathway have been shown to play a role in senescence. The levels of endogenous E3 SUMO ligase, PIASy, increase during replicative senescence, as do the levels of hypersumoylated proteins (Bischof et al., 2007; Bischof et al., 2006). Overexpression of PIASy in normal human fibroblasts promotes sumoylation and induces premature senescence (Bischof et al., 2006). In addition, overexpression of SUMO-2/3 in HEK293 cells induces premature senescence accompanied by an increase in sumoylated exogenous p53 and p105Rb (Li et al., 2006). Increased sumoylation of specific targets, such as RBP1 or APA-1, have been implicated in playing a role in the regulation of senescence (Benanti et al., 2002; Binda et al., 2006). These data suggest that proper regulation of sumoylation is required for normal growth control and that perturbations of the SUMO pathway that result in increased levels of sumoylated proteins may trigger senescence (Bischof et al., 2007).
Overexpression of promyelocytic leukemia protein (PML) IV can also induce premature senescence, which is mediated by p53 and p105Rb signaling (Bischof et al., 2005; de Stanchina et al., 2004; Ferbeyre et al., 2000; Mallette et al., 2004). PML induces p53 transcriptional activity by recruiting p53 and CREB binding protein (CBP) to PML nuclear bodies where p53 is acetylated and phosphorylated (Bischof et al., 2002; Pearson et al., 2000). Furthermore, following activation of oncogenes such as ras, PML mediates a p53-dependent premature senescence response (Pearson et al., 2000). The presence of SUMO binding motifs in PML are required for PML body formation and may be required for the recruitment or binding of other sumoylated proteins such as p53 (Shen et al., 2006). Taken together, these results suggest that sumoylation or interactions at SUMO binding motifs regulate the formation of PML bodies, thereby initiating interactions between other proteins such as p53 and CBP that are responsible for generating a p53-dependent senescence signal.
To study the role of the nuclear SUMO-specific proteases in cell proliferation, we used RNA interference to repress expression of Senp1, Senp2, or Senp7 in low passage, primary human fibroblasts. We demonstrated that repression of these SUMO proteases rapidly and uniformly induced a phenotype that closely resembled replicative senescence. Repression of these SUMO proteases or extended passage caused a dramatic accumulation of nuclear, SUMO-containing PML bodies. Inactivation of the p53 pathway significantly attenuated the senescence response to Senp1 repression. Together, these results demonstrate a novel role for Senp1 in preventing premature senescence and suggest that SUMO proteases may also play a role in replicative senescence and normal cell proliferation.
RESULTS
Repression of Senp1 expression by RNA interference
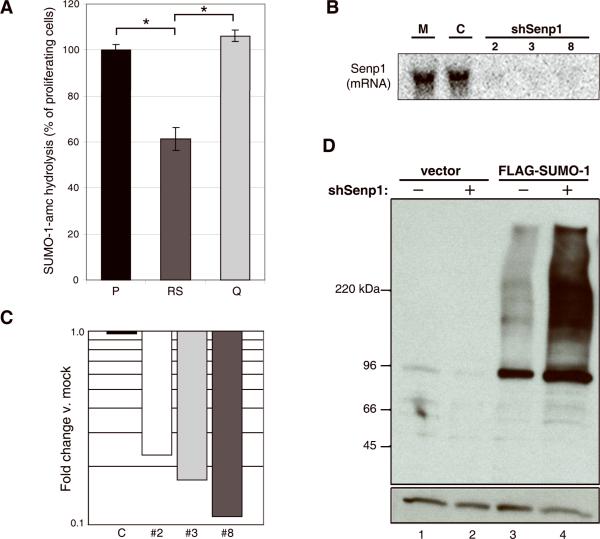
It is not known whether SUMO proteases regulate senescence. To determine if there are differences in SUMO protease activity between proliferating, senescent, and quiescent cells, we compared the relative hydrolytic activity of cell extracts toward SUMO-1-amc, a substrate which generates a fluorescent signal upon cleavage. Of the known SUMO proteases, Senp1 and Senp2 are the dominant isopeptidases for SUMO-1 conjugates. Total lysates from equal numbers of proliferating (passage 9), senescent (passage 34), and contact-inhibited, quiescent (passage 9) primary human foreskin fibroblasts (HFFs) displayed similar levels of SUMO-1-amc hydrolytic activity (data not shown). However, senescent cells are larger and contain more protein on a per-cell basis than proliferating or quiescent cells. Therefore, we measured SUMO-1-amc hydrolysis in reactions containing identical quantities of protein from proliferating, senescent, and quiescent cells. Under these conditions, senescent cells exhibited ~60% of the SUMO-1 protease activity seen in proliferating cells (Fig. 1A). Quiescent cells exhibited similar levels of SUMO-1 protease activity as proliferating cells (Fig. 1A). This decrease in the relative activity of SUMO proteases in senescent cells implies that the same levels of Senp1/Senp2 activity present in young proliferating cells must act in a larger cell volume and on increased protein content in senescent cells. Differences in Senp1 transcription does not account for these activity differences, because Senp1 transcript levels are comparable in proliferating, senescent, and quiescent HFFs, and we have been unable to detect Senp1 protein in proliferating or senescent cells by Western blotting (data not shown). Since the effective SUMO-1 protease activity is thus lower in senescent cells than in proliferating cells, we tested whether reducing SUMO-1 protease activity in cells affected their proliferative status.
Figure 1. Senp1 activity and expression in HFFs.
(A) 30 μg protein from total extracts of proliferating HFFs (passage 9) (P), senescent HFFs (passage 34) (RS), or quiescent HFFs (passage 9, at confluence for two weeks) (Q) were analyzed for SUMO-1-amc hydrolysis (expressed as the percentage of the hydrolytic activity of an extract of proliferating cells). The brackets indicate significant differences in activity (p < 0.0007). (B) Total RNA was harvested from HFFs (passage 9) five days after mock-infection (M) or infection with shControl (C) or one of three shRNAs specific to Senp1 (2, 3, and 8) and analyzed by Northern blot using a Senp1 probe. (C) Total RNA was harvested from HFFs at 24-hours after mock-infection or infection with shControl (C) or shSenp1 hairpins #2, #3, or #8 as indicated, and analyzed by qRT-PCR for Senp1 transcript levels. Expression is shown relative to mock-infected cells after normalization to GAPDH transcript levels. (D) HFFs expressing an empty vector or FLAG-SUMO-1 were mock- (-) or shSenp1-infected (+), and analyzed at three days post-infection by immunoblotting with an anti-FLAG antibody to detect SUMO-1-protein conjugates (top panel) and an anti-actin antibody to detect actin as a loading control (bottom panel). Each lane contained six μg of protein. The band at ~95kDa is probably RanGAP1, one of the most abundant sumoylated proteins in human cells (Matunis et al., 1996).
To study the role of the SUMO protease Senp1 in the regulation of cell proliferation, we used concentrated retrovirus stocks to acutely knockdown Senp1 expression in HFFs by stable expression of short hairpin RNAs (shRNAs). We used HFFs at low passage for all Senp1 repression experiments. By our culture protocol, these cells are approximately 20 passages prior to replicative senescence. We identified three shRNA constructs targeting unique regions of the Senp1 transcript that specifically repressed Senp1 mRNA as detected by Northern blot and quantitative real-time PCR (qRT-PCR) analysis (Fig. 1B and Supplementary Fig. S1). Repression of Senp1 transcript occurred rapidly following shRNA infection (results for 24-hours post-infection are shown in Figure 1C). These hairpins reproducibly knocked down the Senp1 transcript to 5-10% of normal levels in multiple independent experiments, implying that Senp1 expression was inhibited in the vast majority of cells. qRT-PCR demonstrated that expression of the other known closely-related sentrin family members was not affected by shSenp1 expression (Supplementary Fig. S1). Infection with viruses expressing scrambled control shRNA (shControl) did not affect Senp1 mRNA levels compared to mock-infected controls.
Overexpression of Senp1 leads to removal of SUMO-1, SUMO-2, and SUMO-3 from proteins in vivo and results in extensive desumoylation of high molecular weight conjugates (Gong et al., 2000). We investigated whether repression of the endogenous Senp1 gene affected global sumoylation of proteins. To examine changes in SUMO-1 conjugates after Senp1 knockdown, we analyzed HFFs stably expressing FLAG-tagged SUMO-1 from a retroviral vector. Anti-FLAG immunoblotting of whole cell lysates revealed little background signal in cells lacking FLAG-SUMO-1 (Fig. 1D, lanes 1 and 2), and a low basal signal in cells expressing FLAG-SUMO-1 (lane 3). Strikingly, in cells expressing FLAG-tagged SUMO-1, transduction with shSenp1 and Senp1 repression caused the accumulation of high molecular weight SUMO-1 conjugates relative to mock-infected cells (Fig. 1D, lane 4 vs. 3). Similar results were obtained in several independent experiments. The increase in SUMO-1 conjugate levels in HFFs expressing shSenp1 is consistent with compromised SUMO-1 deconjugation directly attributable to diminished Senp1 levels.
Acute repression of Senp1 induced premature senescence
Proliferation of HFFs following infection with shRNA retrovirus was evaluated by BrdU incorporation. Compared to mock- and control shRNA-infected cells, which displayed robust, ongoing DNA synthesis, DNA synthesis in HFFs was significantly inhibited by shSenp1 expression at 24 hours post-infection and decreased to ~8% of mock-infected cells by day three post-infection (Fig. 2A), demonstrating a rapid, growth inhibitory response to loss of Senp1 expression. Similar results were obtained with three independent shRNAs targeting Senp1 (data not shown). In addition, shSenp1 caused substantial inhibition of cell proliferation (see Fig. 4C). Flow cytometry of cells stained with propidium iodide and annexin V demonstrated that repression of Senp1 did not induce apoptosis or necrosis in HFFs (data not shown).
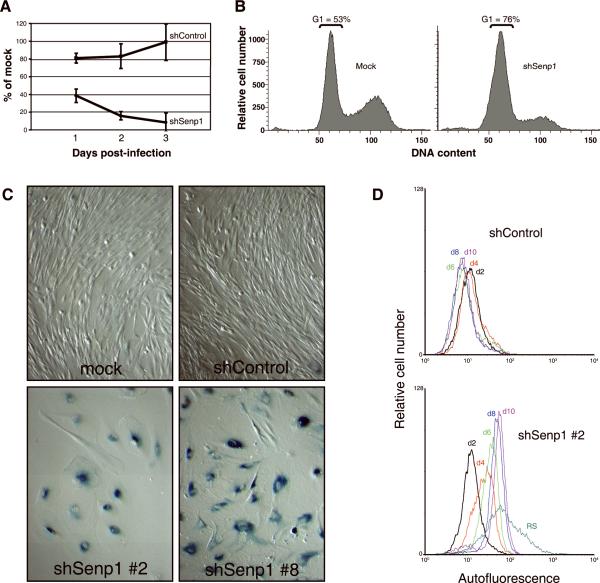
Figure 2. Effect of Senp1 repression on proliferation of HFFs.
(A) HFFs were mock-, shControl-, or shSenp1-infected and analyzed for DNA synthesis at days 1, 2, and 3 post-infection by determining BrdU incorporation (expressed as % of incorporation of mock-infected cells). Error bars indicate standard deviation among four replicates from a single representative experiment. Three independent experiments were conducted with similar results. (B) HFFs at day five after mock-infection or infection with shSenp1 retrovirus were analyzed by FACS for DNA content. The fraction of cells with G1 DNA content is indicated. (C) HFFs were mock-infected or infected with shControl or shSenp1 (construct #2 or #8) retrovirus and fixed at two weeks post-infection and stained for senescence-associated beta-galactosidase activity at pH 6.0. (D) HFFs were infected with shControl- (top panel) or shSenp1 (bottom panel). At the indicated days post-infection, cells were analyzed by FACS for autofluorescence. HFFs serially passaged to replicative senescence (RS) were also analyzed.
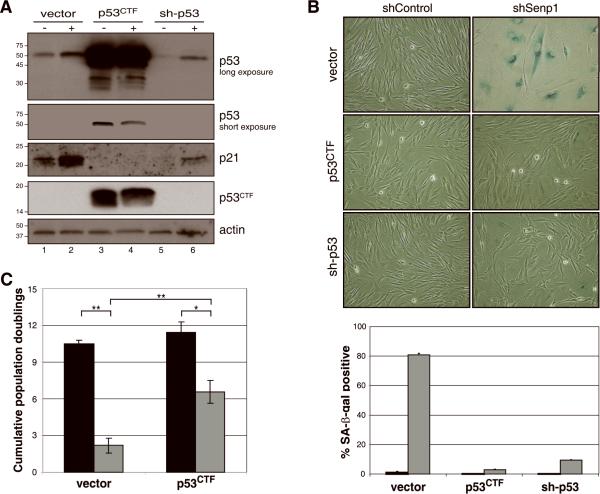
Figure 4. Effect of p53 pathway inhibition on the senescence response to Senp1 repression.
(A) Biochemical analysis of HFFs expressing dominant-negative p53 or sh-p53. HFFs expressing empty vector, dominant-negative p53CTF, or sh-p53 were infected with shControl (-) or shSenp1 (+). Immunoblotting was used to detect truncated p53CTF, full-length p53, p21, or actin as a loading control, as indicated. (B) Effect of inactivation of p53 on senescence-associated β-galactosidase. HFFs expressing empty vector control, p53CTF, or sh-p53 were shControl- or shSenp1-infected and stained for SA-βgal activity after two weeks (top set of panels). SA-βgal-positive cells were quantitated by scoring >200 cells in randomly chosen fields for the presence of blue-green staining (bottom panel). Black bars, shControl; grey bars, shSenp1. Error bars represent standard deviation among three replicates. (C) Effect of p53 pathway inhibition on proliferation following Senp1 repression. HFFs expressing empty vector or p53CTF were shControl- or shSenp1-infected and counted at various time points to determine population doublings. The graph shows cumulative population doublings at day 14-16 after infection with shRNA control (black bars) or shSenp1 (grey bars). The average and standard deviation of three independent experiments is shown. The comparisons shown in brackets are significant as determined by T-tests (**, p<0.003; *, p<0.01).
Cell cycle analysis showed that the fraction of HFFs in the G1 phase of the cell cycle increased from 53% in mock-infected controls to 76% in cells five days after infection with Senp1 shRNA (Fig. 2B). Cells infected with Senp1 shRNAs also exhibited an enlarged, flattened morphology characteristic of HFFs that have undergone replicative senescence (Fig. 2C). We assayed senescence-associated beta-galactosidase (SA-βgal) activity by staining with X-gal at pH 6.0 at day 14 post-infection. SA-βgal, the product of the lysosomal GLB1 β-galactosidase gene, is frequently used as a marker of senescence and can distinguish senescence from other growth arrested states such as quiescence (Dimri et al., 1995; Lee et al., 2006). Greater than 90% of HFFs exposed to any of the three unique Senp1 shRNAs (#2, 3, and 8) exhibited blue-green cytoplasmic staining indicative of SA-βgal activity, whereas the vast majority of mock-infected HFFs or HFFs expressing the scrambled control shRNA were negative for SA-βgal activity and exhibited normal morphology (Fig. 2C and data not shown).
Senescent cells are also characterized by increased cellular autofluorescence due to accumulation of the fluorescent pigment, lipofuscin. Repression of Senp1 induced a gradual increase in cellular autofluorescence compared to shControl-infected HFFs, reaching ~six-fold at ten days after shRNA infection (Fig. 2D). Approximately half of this increase can be attributed to increased cell volume in the senescent population (as assessed by forward scatter; data not shown). Late passage HFFs undergoing replicative senescence displayed on average a >10-fold increase in autofluorescence with a broad fluorescence distribution reflecting the heterogeneous nature of replicative senescence (Fig. 2D). In striking contrast, HFFs exposed to Senp1 shRNA exhibited a narrow autofluorescence distribution, indicating uniform and synchronous entry into senescence. Taken together, these results indicated that acute reduction of Senp1 levels in low passage HFFs induced a growth-arrested state that displays several key features of senescence.
Senescence induced by repression of Senp1 resulted in accumulation of SUMO at PML nuclear bodies
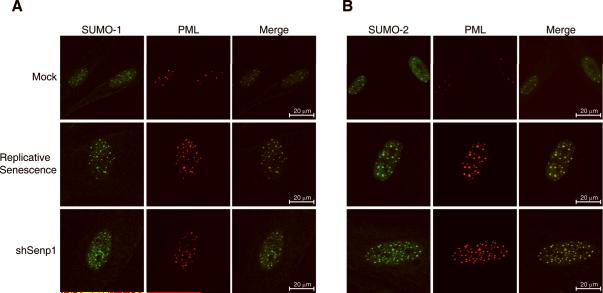
We used immunofluorescence to determine the localization of endogenous SUMO-1 and SUMO-2 in control and senescent HFFs. SUMO-1 and SUMO-2 exhibited predominantly nuclear localization punctuated by discrete foci of high fluorescence intensity (Fig. 3A and 3B). Co-staining with antibody recognizing promyelocytic leukemia (PML) protein identified these foci as PML nuclear bodies (Fig. 3A and 3B). Every PML nuclear body was coincident with both SUMO-1 and SUMO-2 indicating that all detectable PML bodies contain sumoylated proteins (Fig. 3 merged images and data not shown). These bodies also contained p53 and CREB-binding protein (CBP), a component of a p53-PML-CBP prosenescence complex (data not shown) (Pearson et al., 2000).
Figure 3. Accumulation of nuclear SUMO/PML bodies in senescent cells.
Localization of endogenous SUMO-1, SUMO-2, and PML in senescent cells. HFFs at early passage were mock-infected, infected with shSenp1#2, or serially passaged until they reached replicative senescence (passage 30). Immunofluorescence was performed 12 days after infection to detect endogenous SUMO-1 (green, panel A), SUMO-2 (green, panel B), and PML (red). Merged images show co-localization (yellow) of PML and SUMO-1 or SUMO-2 at discreet nuclear foci.
Strikingly, HFFs undergoing replicative senescence and Senp1 knockdown cells displayed a significant increase in the number, size, and staining intensity of SUMO/PML bodies relative to their mock-infected or shControl-infected, proliferating counterparts. Similar results were obtained with three hairpins targeting Senp1 (Fig. 3 and Supplementary Fig. 2). We quantified the differences in the number and size of SUMO/PML bodies with peak fluorescence intensity ≥ four-fold above nuclear background intensity at day 12 post-infection (Table 1). The mean number of PML bodies in shSenp1-infected or replicative senescent cells was 2-3 times greater than in mock-infected cells. In addition, the PML structures were on average 50% larger in replicative senescent and shSenp1-infected cells than in mock-infected or shControl-infected cells (Table 1 and data not shown). All the differences noted above were statistically significant (Table 1). Furthermore, proliferating cells were generally smaller and had smaller nuclei, typically less than one quarter the area of nuclei in the senescent cells. A significant increase in the number and intensity of SUMO/PML bodies was evident in shSenp1-infected cells compared to shControl-infected cells as early as one day post-infection, implying that this increase was a direct effect of Senp1 repression (Supplementary Fig. S3). The increased size and number of SUMO/PML bodies in cells expressing shSenp1 demonstrates that sumoylated proteins accumulate in discrete nuclear structures following Senp1 knockdown, as well as during replicative senescence. These results suggest that accumulation of nuclear SUMO-containing PML bodies may play an important role in both induced and replicative senescence.
Table 1.
Analysis of SUMO/PML bodies
| Numbera | Sizeb | |
|---|---|---|
| Mock | 11±5 | 0.51 |
| Replicative senescence | 27±14 (p<10-6)c | 0.77 (p<10-6) |
| shSenp1 | 28±11 (p<10-6) | 0.79 (p<10-6) |
Number of PML bodies/cell.
Mean size of PML bodies (μm2).
All p values were calculated by two-sided T-test relative to mock-infected cells.
Senescence induced by loss of Senp1 requires p53 function
The tumor suppressor p53 plays an important role in the execution of replicative senescence in normal human cells (Hara et al., 1991; Morris et al., 2002; Shay et al., 1991; Wei et al., 2003). To determine if p53 activity is required for senescence induced by Senp1 repression, we functionally inactivated p53 in HFFs by stably expressing a dominant-negative C-terminal fragment of p53 (p53CTF) or an shRNA targeting p53 (sh-p53#2). We used immunoblotting to confirm inhibition of the p53 pathway. HFFs expressing p53CTF contained markedly elevated levels of full-length p53 (Fig. 4A, lane 3), presumably due to impaired p53-mediated transactivation of the MDM2 gene, which encodes a ubiquitin ligase involved in p53 degradation. Expression of shRNAs targeting p53 resulted in >96% reduction in p53 transcript levels and a reduction of p53 protein levels (Fig. 4A, lane 5, and data not shown). Inhibition of p53 function by either method resulted in >six-fold repression of p21 transcript and substantial reduction in p21 protein levels (Fig. 4A, lanes 3 and 5, and data not shown) due to impaired p53-mediated transactivation of p21 transcription. Taken together, these results demonstrated that the p53 pathway was inactivated by either dominant-negative inhibition or shRNA-mediated knockdown of p53.
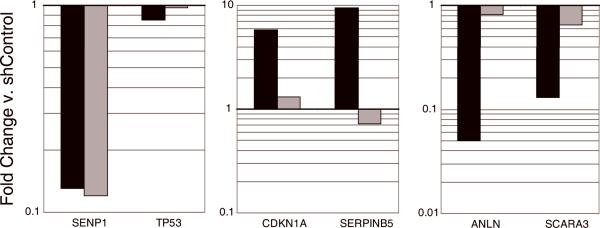
HFFs expressing the empty vector LXSN, p53CTF, or sh-p53 were infected with control or shSenp1 retrovirus. Immunoblot analysis three days after infection revealed a slight upregulation of p53 and p21 levels in control and sh-p53 cells after shSenp1 expression (Fig. 4A, lane 2). After shSenp1 expression, Senp1 mRNA was reduced to a similar extent whether or not p53 function was impaired (Fig. 5, left panel). Although there was no significant change in levels of p53 mRNA (Fig. 5, left panel), repression of Senp1 resulted in enhanced transcriptional activity of p53. qRT-PCR analysis of known p53-responsive genes at day 10 post-infection revealed significant induction of p21 (CDKN1A) and SERPINB5 mRNA, and repression of ANLN, SCARA3, BIRC5 and CAV1 mRNA in cells expressing shSenp1 compared to shControl infected cells (Fig. 5, black bars, and data not shown), consistent with the known effects of p53 on these genes. Similar but less pronounced effects were observed at earlier times after Senp1 repression (data not shown). Expression of dominant-negative p53CTF significantly attenuated these changes in gene expression in response to Senp1 repression (Fig. 5, grey bars, and data not shown), demonstrating that the induction and repression of these genes by Senp1 repression is, in fact, p53-dependent. Despite the increased transcriptional activity of p53, we did not observe phosphorylation of p53 or evidence of a p53-mediated DNA damage response following repression of Senp1 (data not shown).
Figure 5. Expression of p53-dependent genes after Senp1 repression.
HFFs at low passage were infected with LXSN or LXSN-p53CTF, and then infected with shControl or shSenp1. Total RNA was harvested at day 10 post-infection and analyzed by qRT-PCR for expression of SENP1, p53 (TP53), and the indicated p53-responsive genes. Expression of the indicated transcripts in cells expressing shSenp1 is graphed as fold-change compared to shControl-infected cells after normalization to GAPDH expression. The bold horizontal line designates no change in expression. Black bars show results from HFFs expressing empty vector, and the grey bars show results from HFFs expressing dominant-negative p53. Gene names are given as the official symbol in the Entrez Gene database (NCBI). These data are representative of two independent experiments analyzed in triplicate.
As described above, Senp1 repression in unmodified HFFs induced senescence. In contrast, Senp1 repression in HFFs expressing p53CTF or shRNAs targeting p53 did not induce cell flattening, and these cultures contained very few SA-βgal-positive cells and exhibited low autofluorescence (Fig. 4B and data not shown). In addition, although Senp1 repression caused a dramatic proliferative block in control HFFs, p53CTF and sh-p53 cells continued to proliferate after Senp1 repression, albeit to a reduced extent compared to control cells. Similar results were obtained with three different shRNAs targeting different regions of p53 (Fig. 4C, and data not shown). These results showed that inactivation of p53 significantly attenuated the senescence response, indicating that p53 signaling is required for senescence in this system.
Repression of SUMO proteases Senp2 or Senp7 induced accumulation of SUMO/PML bodies and senescence
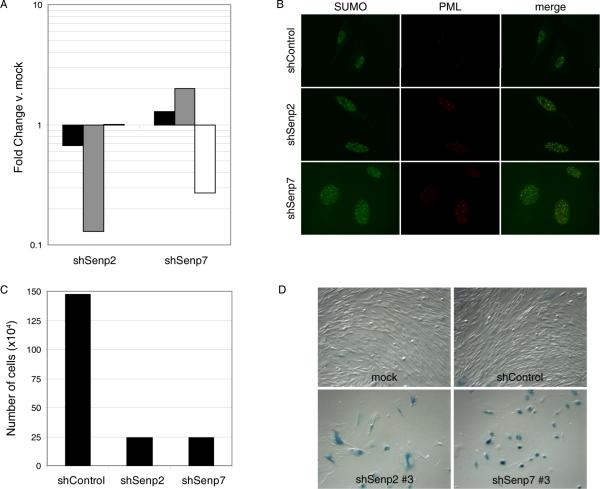
Localization of SUMO proteases is thought to determine their specificity for substrates (Kim et al., 2005). Senp2 shuttles between the cytoplasm and the nucleus, and Senp7, like Senp1, localizes to the nucleoplasm (Mukhopadhyay et al., 2007). To assess the roles of these SUMO proteases in senescence, shRNAs targeting Senp2 or Senp7 were used to infect HFFs at passage 8. By qRT-PCR analysis at day four post-infection, Senp2 and Senp7 transcripts were specifically repressed in HFFs by shSenp2 and shSenp7 expression, respectively (Fig. 6A). Expression of Senp1 was not altered by either shSenp2 or shSenp7 expression. After infection with either shSenp2 or shSenp7, the number of nuclear bodies containing endogenous SUMO-2 and PML was significantly increased, compared to shControl-infected cells (Fig. 6B). In addition, repression of Senp2 or Senp7 resulted in dramatic growth inhibition, and the cells adopted a senescent morphology, and stained positive for SA-βgal activity, indicating that repression of Senp2 or Senp7 induced senescence in HFFs (Figs. 6C and 6D). However, Senp2 and Senp7 repression induced fewer SUMO/PML bodies than did Senp1 repression, and the senescence response took longer to develop (data not shown). As was the case with Senp1 repression, senescence induced by Senp7 repression was significantly attenuated by dominant-negative p53 (Supplementary Fig. S4 and data not shown).
Figure 6. Repression of nuclear SUMO-protease Senp2 and Senp7 induces senescence.
(A) HFFs at early passage (passage 8) were infected with shSenp2 or shSenp7, as indicated. At day four post-infection, total RNA was harvested and analyzed by qRT-PCR for expression of Senp1 (black bars), Senp2 (grey bars), and Senp7 (white bars) transcripts. Expression is shown relative to shControl-infected cells after normalization to GAPDH transcript levels. qRT-PCR analysis was performed in triplicate. (B) HFFs were infected with shControl, shSenp2, or shSenp7 and analyzed at day 12 post-infection by immunofluorescence with an antibody that recognized endogenous SUMO-1, -2, and -3 and an antibody that recognized endogenous PML. (C) 3×104 HFFs were infected with shControl, shSenp2, or shSenp7 and counted at day seven post-infection. (D) HFFs were infected with shControl, shSenp2, or shSenp7 and stained for SA-βgal activity at two weeks post-infection. Images were acquired by phase microscopy.
DISCUSSION
In this study, we demonstrated that repression of the endogenous SUMO proteases Senp1, Senp2, and Senp7 acutely induced senescence in low passage primary human fibroblasts. A growing body of evidence points to a key role for sumoylation in the regulation of proliferation and in the execution of tumor suppressor responses, such as senescence. Li et al. demonstrated that overexpression of SUMO-2/3 induces senescence through p53 and retinoblastoma pathways (Li et al., 2006), and the changes in SUMO-2/3 conjugation they observed were similar to those seen in cells undergoing a hydrogen peroxide-induced stress response. Overexpression of the SUMO E3 ligase, PIASy, also induces premature senescence (Bischof et al., 2006). These published results indicate that factors that promote sumoylation can trigger senescence. In addition, cells that have undergone replicative senescence accumulate hypersumoylated proteins compared to proliferating or quiescent cells, further suggesting that persistent sumoylation of proteins may play a role in the execution and maintenance of senescence (Bischof et al., 2007). In agreement with this notion, we demonstrated that repression of desumoylating factors can stimulate sumoylation of endogenous (Figs. 3 and 6B) and exogenous (Fig. 1C) proteins and trigger a rapid senescence response.
Senp1, Senp2, and Senp7 share conserved C-terminal catalytic domains and have divergent N-terminal domains. In vitro, all three proteases display isopeptidase activity responsible for desumoylation, but Senp7 lacks endopeptidase activity (Drag et al., 2008; Mikolajczyk et al., 2007). SUMO/PML bodies accumulate after repression of Senp1, Senp2, or Senp7, and of these three proteases, repression of Senp1 was the most effective at causing the accumulation of SUMO/PML bodies and at inducing senescence. Taken together, these data suggest that the desumoylation activities of these proteases, not the SUMO precursor processing activities or other potential activities, is rate-limiting in normal human fibroblasts and that reduced desumoylation is responsible for senescence.
The rapidity of the response to repression of SUMO proteases implies that there is a dynamic equilibrium between sumoylation and desumoylation and highlights the sensitivity of cells to perturbation of these processes. The increased SUMO/PML body formation and induction of senescence by repression of any of three different SUMO proteases indicates that these three proteins cannot acutely compensate for loss of each other, either biochemically or in terms of their effects of cell growth. We infer that senescence is induced either by increases in the sumoylation of targets in common to these proteases or by increases in the total level of sumoylated proteins.
Early evidence for a role for SUMO proteases in cell growth was obtained from studies in yeast, where the yeast Senp1 homologue, Ulp1, is required for progression through the G2/M phase of the cell cycle (Li et al., 1999). The functions of SUMO proteases in the growth of mammalian cells have also been investigated. Depletion of Senp1 by RNA interference has no effect on proliferation of androgen-sensitive LNCaP prostate cancer cells in the absence of androgen, but Senp1 repression reduced androgen-stimulated proliferation by 40% (Bawa-Khalfe et al., 2007). The nature of this growth-inhibitory effect was not further investigated. DiBacco and colleagues used siRNA transfection to knockdown Senp5 in HeLa cells (Di Bacco et al., 2006). This resulted in growth arrest, cell enlargement, and the generation of cells containing multiple, aberrant nuclei. These effects were attributed to a defect in cytokinesis, and markers of senescence were not examined. Furthermore, Senp5 repression resulted in accumulation of SUMO-2/3 conjugates, suggesting that Senp5 is required for cell division and loss of Senp5 triggers a stress response.
Senescence following Senp1 repression exhibited remarkable overlap with phenotypic changes observed in replicative senescence, including flattened and granular cell morphology, enlarged cell size, G1 cell cycle arrest, autofluorescence, elevated SA-βgal activity, and accumulation of SUMO-PML bodies. PML and PML bodies accumulate in various induced and replicative senescence systems (Fang et al., 2002; Ferbeyre et al., 2000; Pearson et al., 2000). The formation of PML bodies is dependent on sumoylation of PML, and PML can be desumoylated by Senp1, which has been observed at the periphery of PML bodies (Bailey et al., 2002; Gong et al., 2000; Kang et al., 2006; Shen et al., 2006; Zhong et al., 2000). We showed that endogenous SUMO-1 and SUMO-2 protein conjugates co-localize with PML bodies in induced and replicative senescence. Together, these data suggest that ongoing desumoylation of proteins, possibly including PML itself, by SUMO proteases inhibits the formation and persistence of PML bodies and that inhibition of desumoylation can stimulate rapid PML body formation and initiate senescence.
Senescent cells have decreased free ubiquitin, increased pools of ubiquitin conjugates, and decreased proteasome activity (Grillari et al., 2006). These ubiquitin conjugates accumulate in lysosomes and are believed to contribute to the senescent phenotype and perhaps aging-associated diseases (Dice, 1989; Vernace et al., 2007). In addition, the SUMO ligase, PIASy, is induced, and hypersumoylated proteins accumulate in cells that had undergone replicative senescence (Bischof et al., 2007; Bischof et al., 2006). We showed here that relative SUMO-protease activity declines during replicative senescence and that nuclear SUMO/PML bodies increase during this process. The altered activities of the SUMO and ubiquitin-proteasome pathways in aging cells may permit the accumulation or mislocalization of damaged or oxidized proteins that would otherwise be eliminated from healthy, proliferating cells and may directly contribute to senescence and diseases of aging. Similarly, the specific proteins targeted for sumoylation or ubiquitination may differ as cells age, contributing to cellular pathology.
Our experiments demonstrated that p53 is activated and accumulates in PML bodies and that p53 activity is required for senescence induced by repression of Senp1 or Senp7. Senescence induced by overexpression of PIASy is accompanied by increased sumoylation of exogenous p53 (Bischof et al., 2006). Although sumoylation of p53 has been suggested to enhance its transcriptional activity (Gostissa et al., 1999; Rodriguez et al., 1999), a situation consistent with the increased p53 activity we observed following Senp1 repression, the role of sumoylated p53 in senescence has yet to be elucidated (Melchior et al., 2002; Rodriguez et al., 1999). Following Senp1 knockdown, sumoylation of p53 itself may initiate a senescence program, p53 may mediate a senescence signal initiated by sumoylation of another protein, or a combination of these effects may occur. We have not been able to detect sumoylated p53 following Senp1 repression, which is not surprising because sumoylated p53 accounts for less than 5% of p53 even when both p53 and SUMO are overexpressed (Chen et al., 2003). It is worth noting that significant sumoylation-dependent activities have been observed when sumoylation is below the level of detection, especially for sumoylated transcription factors. It has been proposed that sumoylation of transcription factors may be required for the formation of stable transcriptional complexes, but not for their persistence {reviewed in (Geiss-Friedlander et al., 2007; Hay, 2005)}.
Senp1 is overexpressed or disregulated in some cancers including prostate cancer, infantile teratoma, and thyroid oncocytomas, indicating that abnormally high Senp1 levels may play an important role in tumorigenesis (Cheng et al., 2006; Jacques et al., 2005; Veltman et al., 2005). In addition, the SUMO E2 conjugating enzyme, Ubc9, is upregulated in multiple cancers (Mo et al., 2005). Our results suggest that targeted inhibition of Senp1 enzymatic activity, or the activity of other SUMO proteases such as Senp2 and Senp7, may induce senescence and provide a new approach to prevent or treat cancer.
EXPERIMENTAL PROCEDURES
Vectors, viruses, and cell culture
Normal primary human foreskin fibroblasts (HFFs) obtained from the Yale Skin Disease Research Center were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Gemini Bio-Products, West Sacramento, CA, USA), 2 mM L-glutamine (Gibco, Carlsbad, CA, USA), and 10 mM HEPES [pH 7.3] at 37°C with 5% CO2. HFFs were used at low passage (less than 15 passages) or serially passaged to replicative senescence (approximately 30 passages). Ten μg of the retroviral vector DNA of interest was packaged into vesicular stomatitis virus (VSV) G-protein-pseudotyped recombinant retrovirus by calcium phosphate transfection of 2.5×106 293T/17 cells (ATCC, Manassas, VA, USA) with 4 μg pVSV-G (Clontech, Mountain View, CA, USA) and 6 μg pCL-Eco. VSV-G-pseudotyped recombinant retroviral stocks were concentrated as described previously (Johung et al., 2007) with Centricon Plus-20 columns (Millipore, Billerica, MA, USA) and used to infect HFFs at a multiplicity of infection (MOI) of approximately 50. A dominant-negative C-terminal fragment of p53 (p53CTF (Gottlieb et al., 1994); gift of K. Münger, Harvard University) was cloned into retroviral vector pLXSN. A gene encoding the mature form of SUMO-1 with an N-terminal FLAG tag was cloned into the retroviral vector pBabe-puro. After infection, HFFs were selected with 0.6 μg/mL puromycin or 1 mg/mL G418 as appropriate. For replicative senescence, cells were passaged 1:4 when 80% confluence was reached.
SUMO-1 protease activity assay
Young, proliferating (passage 9), replicative senescence (passage 34) HFFs, or quiescent HFFs (passage 9, cultured at confluence for two weeks) were lysed in buffer A (10 mM Tris, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5% NP-40) at a density of 1000 cells/μl. For assays comparing equal cell number, 100 μl of these lysates were added to 100 μl of buffer B (10 mM Tris, pH 7.4 containing 1 mM DTT) in black 96-well plates (Nunc, Rochester, NY, USA). For assays comparing equal protein content, protein concentration was measured by Bradford assay. Lysates were adjusted with buffer A to a concentration of 0.30 mg protein/ml, and 100 μl of these lysates were added to 100 μl of buffer B in black 96-well plates. One μl of a 50 μM stock of SUMO-1-amc (Boston Biochem, Cambridge, MA, USA) in 10 mM Tris, pH 7.4 (250 nM final concentration) was added to wells. The plate was incubated for 40 minutes at 37°C and fluorescence was measured using a SPECTRAmax Gemini (Molecular Devices, Sunnyvale, CA, USA) at 380 nm excitation and 460 nm emission wavelengths, with a 455 nm cutoff. Individual wells were read as the average of 30 independent measurements and all data points were obtained in triplicate. Data were exported and normalized to activity of proliferating HFF lysate at equal cell number.
Short Hairpin RNAs
Short hairpin RNA constructs (shRNAs) were cloned into the RNAi-Ready pSiren-RetroQ retroviral vector (Clontech, Mountain View, CA, USA) as described previously (Johung et al., 2007) to generate pSiren-shRNA. The shRNAs targeting Senp1 were: shSenp1#2 (1577): 5'-GCGGGAACATTCAGTACATGACGAATCATGTACTGAATGTTCCCGC-3'; shSenp1#3 (2125): 5'-GCAGAATACTCTTGCAATACCCGAAGGTATTGCAAGAGTATTCTGC-3'; and shSenp1#8 (1519): 5'-GCCAGATTGAAGAACAGAAGGCGAACCTTCTGTTCTTCAATCTGGC-3'. Unless indicated otherwise, shSenp1#2 was used for the experiments shown in this paper. The shRNAs targeting p53 were: sh-p53#1 (1097): 5'-GCGCACAGAGGAAGAGAATCTCGAAAGATTCTCTTCCTCTGTGCGC-3'; sh-p53#2 (1022): 5'-GGAAGACTCCAGTGGTAATCTCGAAAGATTACCACTGGAGTCTTCC-3'; and sh-p53#3 (827): 5'-GCATCTTATCCGAGTGGAAGGCGAACCTTCCACTCGGATAAGATGC-3'. The numbers in parentheses refer to the 5' base pair of the Senp1 gene (accession number NM_014554.2) and the 5' base pair of the p53 gene (accession number NM_000546.2) that is targeted by the hairpin, respectively. An shRNA control (shControl) was obtained from Clontech: a scrambled sequence that does not form a hairpin.
Real-time PCR
Quantitative real-time PCR (qRT-PCR) and primer design were performed as described previously (Johung et al., 2007). Briefly, total RNA was harvested using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) or QiaShredder, RNeasy Mini and RNase-free DNase kits (Qiagen, Valencia, CA, USA), and 1 μg RNA was used as template for cDNA synthesis using an iScript™ cDNA synthesis kit (BioRad, Hercules, CA, USA). qRT-PCR was performed by using iQ™ SYBR Green Supermix (BioRad, Hercules, CA, USA) and 40 ng cDNA per 20 μl reaction and the BioRad MyiQ Single-color Real-time PCR detection system. GAPDH transcripts were detected using primers 5'-CTGCACCACCAACTGCTTAG-3' and 5'-GTCTTCTGGGTGGCAGTGAT-3'. Senp1 transcripts were detected using primers 5'-ACTGATAGTGAAGATGAATTTCCTGA-3' and 5'-CATCCTGATTCCCATTACGAA-3'. p53 transcripts were detected using primers 5'-AGGCCTTGGAACTCAAGGAT-3' and 5'-CCCTTTTTGGACTTCAGGTG-3'. qRT-PCR primer sequences for all genes analyzed and the reaction conditions are shown in Supplementary Tables 1 and 2, respectively.
Northern Blot
Total RNA was harvested using Qiashredder and RNeasy Mini kits (Qiagen, Valencia, CA, USA). Five μg of total RNA was electrophoresed on a 1% formaldehyde-agarose gel, transferred to a Nytran Supercharge membrane (Schleicher and Schuell, St. Louis, MO, USA) by downward capillary transfer in SSC and UV-crosslinked with a Stratalinker (Stratagene, La Jolla, CA, USA). Primers 5'-CTGGAGAAGTGACTTTAGTGAACC-3' and 5'-TAGGAAAAGATTTTTCATATGCAGAT-3' were used to PCR amplify a specific probe for Senp1 that was random prime-labeled with [α-32P]-ATP and hybridized to the membrane overnight. The membrane was washed and visualized by exposure to a Phosphor Screen and detection by a Storm 840 PhosphorImager (Molecular Dynamics, Piscataway, NJ, USA).
Senescence Assays
DNA synthesis activity was determined by BrdU incorporation and detection using a Cell Proliferation ELISA kit (Roche, Basel, Switzerland). Briefly, low passage HFFs were plated at 5×103 per well in a 96-well plate and infected the next day with concentrated shSenp1 or control retroviruses. At various time points post-infection, 10 μM 5'-bromo-2'-deoxyuridine (BrdU) was added to the cells for 4-6 hours and incorporation was quantitated by ELISA. For cell cycle analysis, HFFs were trypsinized five days after infection, washed, and resuspended to 1×106 cells/ml in OptiMEM (Gibco, Carlsbad, CA, USA) with 10 μM Vybrant DyeCycle Violet stain (Invitrogen, Carlsbad, CA, USA) and incubated at 37°C for 30 minutes immediately prior to analysis by fluorescence activated cell sorting (FACS) on an LSRII (BD BioSciences, San Jose, CA, USA). Senescence-associated beta-galactosidase staining was performed as described previously (Lee et al., 2006). Quantitation of SA-βgal-positive cells was performed by scoring >200 cells in randomly chosen fields for presence or absence of staining. Autofluorescence was determined by FACS of unstained cells using 488 nm excitation and a 661 nm emission filter on a FACS Caliber flow cytometer (BD Biosciences, San Jose, CA, USA). Forward scatter (FSC) was used to determine cell volume.
Growth Curves
HFFs expressing the pLXSN empty vector or pLXSN-p53CTF were seeded at 2×105 cells/10 cm dish in triplicate for each infection. The next day, the cells were shControl- or shSenp1-infected at MOI ~50. At various days post-infection, cells were trypsinized, resuspended, counted, and replated at 2×105 cells/10 cm dish. Population doubling (PD) was calculated using the equation: PD = log(N/N0)/log(2) where N is the number of cells recovered after a period of proliferation and N0 is the initial number of cells seeded.
Immunofluorescence
For immunofluorescence at day 12 post-infection, cells were plated in 8-well chamber slides at ~3,000/well nine days post-infection. Cells were cultured for three days, fixed in buffered Formalde-Fresh (Fisher, Pittsburgh, PA, USA), permeabilized with 1% Triton-X/Formalde-Fresh, and blocked with 0.5% BSA/0.1% Triton-X/PBS. For immunofluorescence at day one post-infection, cells were plated at 5,000/well of an 8-well chamber slide and infected the next day with shControl or shSenp1 at MOI ~50. The next day, cells were stained as described above. Primary antibodies were diluted in blocking buffer to 2 μg/ml for anti-PML (mouse monoclonal PG-M3, Santa Cruz Biotechnology, Santa Cruz, CA, USA), 2.5 μg/ml for anti-SUMO-1 (rabbit polyclonal, Abgent, San Diego, CA, USA), 2.5 μg/ml for anti-SUMO-2 (rabbit polyclonal, Abgent, San Diego, CA, USA), or 2.5 μg/ml for anti-pan SUMO to detect SUMO-1, -2, and -3 (rabbit polyclonal, Abgent, San Diego, CA, USA). Secondary antibodies were goat anti-mouse AlexaFluor-594 or goat anti-rabbit AlexaFluor-488 (Invitrogen, Carlsbad, CA, USA). Background staining with the secondary antibodies in the absence of primary antibody was undetectable. Images were acquired using a Zeiss LSM510 confocal microscope under a 40X water-immersion objective using the average of 8 scans (1024 × 1024 resolution; 8-bit data depth; scan speed 9). Settings for detector gain, amplifier offset, amplifier gain, laser power and pinhole size were as follows: green/488 nm channel (833; 0; 1; 4; 70.5 μm) and red/594 nm channel (660, -0.005; 1; 66; 70.5 μm). Data were exported as TIFF files and imported into Photoshop CS2 for overlay of the two channels to create merged images. Images used for quantitation were acquired using a 63X water-immersion objective using the average of 8 scans (1024 × 1024 resolution; 16 bit data depth; scan speed 7). Settings for detector gain, amplifier offset, amplifier gain, laser power and pinhole size were as follows: green/488 channel (1025; -0.045; 1.3; 4; 116.4 μm) and red/594 channel (590; 0; 1; 55; 116.4 μm). Images were analyzed using ImageJ software (NIH, Bethesda, MD, USA) to identify PML bodies with fluorescence intensity greater than four-fold above background levels and of size greater than 0.1 μm2. Statistical analysis of the resulting data was performed using Microsoft Excel. Images shown in Supplementary Material and Figure 6 were acquired using a Zeiss Axiovert 200 spinning disk confocal microscope with a 63X water-immersion objective (Zeiss, Thornwood, NY, USA) and a CoolSNAP HQ2 camera (Photometrics, Tucson, AZ, USA).
Western Blots
For immunoblotting of p53CTF, cells were lysed on ice in buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% NP-40, 5 mM dithiothreitol, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, 5 μg/ml aprotinin) for 30 minutes with occasional vortexing. For immunoblotting of p53 and p21, cells were lysed in TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) and total protein was prepared according to manufacturer's instructions. For immunoblotting of FLAG-SUMO-1, cells were lysed by vortexing in buffer (50 mM Tris-HCl [pH 6.8], 1% glycerol, 10 mM dithiothreitol, 25 mM N-ethylmaleimide, 0.2% SDS) for 10 seconds and boiling for 10 minutes. Six μg of supernatant was electrophoresed on a 4-20% Tris-HCl ReadyGel (BioRad, Hercules, CA, USA). After electrophoresis, proteins were transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA) in buffer (12.5 mM Tris, 0.1 M glycine, 20% methanol) and blocked in 5% milk/TBST buffer (5% nonfat dry milk, 25 mM Tris-HCl [pH 8.0], 125 mM NaCl, 0.1% Tween-20). The membranes were probed with the following specific antibodies: mouse monoclonal anti-FLAG (M2, Sigma, St. Louis, MO, USA), mouse monoclonal anti-p21 (F-5, Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-p53 to detect wild-type p53 (OP43, Calbiochem, Darmstadt, Germany), mouse monoclonal anti-p53 to detect p53CTF (OP03, Calbiochem, Darmstadt, Germany), and a broadly reactive goat polyclonal anti-actin raised to human β-actin (C-11, Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Supplementary Material
Acknowledgments
Primary human foreskin fibroblasts were obtained from the tissue culture core of the Yale Skin Disease Research Center (NIH P30 AR41942). We thank Mark Hochstrasser for helpful discussions, David Van Goor for advice and assistance with microscopy, Lynn Cooley and Antonio Giraldez for use of their microscopes, and Jan Zulkeski for assistance in preparing this manuscript. K.E.Y. was supported by a training grant from the NIH (AI55403). G.K. was supported in part by an individual National Research Service Award from the NIH. G.K. thanks Hidde Ploegh for support. This work was supported by grants from the NIH to D.D. (CA16038) and to I.B.R. (CA62099, AG17921, CA95996).
Abbreviations
- SUMO
small ubiquitin-related modifier
- HFF
human foreskin fibroblast
- shRNA
short hairpin RNA
- SA-βgal
senescence-associated beta-galactosidase
- PML
promyelocytic leukemia protein
- MEF
mouse embryonic fibroblast
- MOI
multiplicity of infection
References
- Alexander K, Hinds PW. Requirement for p27KIP1 in retinoblastoma protein-mediated senescence. Mol. Cell. Biol. 2001;21:3616–3631. doi: 10.1128/MCB.21.11.3616-3631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D, O'Hare P. Herpes simplex virus 1 ICPO co-localizes with a SUMO-specific protease. J. Gen. Virol. 2002;83:2951–2964. doi: 10.1099/0022-1317-83-12-2951. [DOI] [PubMed] [Google Scholar]
- Bawa-Khalfe T, Cheng J, Wang Z, Yeh ET. Induction of the sumo-specific protease 1 transcription by the androgen receptor in prostate cancer cells. J. Biol. Chem. 2007;282:37341–37349. doi: 10.1074/jbc.M706978200. [DOI] [PubMed] [Google Scholar]
- Benanti JA, Williams DK, Robinson KL, Ozer HL, Galloway DA. Induction of extracellular matrix-remodeling genes by the senescence-associated protein APA-1. Mol. Cell. Biol. 2002;22:7385–7397. doi: 10.1128/MCB.22.21.7385-7397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda O, Roy JS, Branton PE. RBP1 family proteins exhibit SUMOylation-dependent transcriptional repression and induce cell growth inhibition reminiscent of senescence. Mol. Cell Biol. 2006;26:1917–1931. doi: 10.1128/MCB.26.5.1917-1931.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof O, Dejean A. SUMO is growing senescent. Cell Cycle. 2007;6:677–681. doi: 10.4161/cc.6.6.4021. [DOI] [PubMed] [Google Scholar]
- Bischof O, Kirsh O, Pearson M, Itahana K, Pelicci PG, Dejean A. Deconstructing PML-induced premature senescence. EMBO J. 2002;21:3358–3369. doi: 10.1093/emboj/cdf341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof O, Nacerddine K, Dejean A. Human papillomavirus oncoprotein E7 targets the promyelocytic leukemia protein and circumvents cellular senescence via the Rb and p53 tumor suppressor pathways. Mol. Cell. Biol. 2005;25:1013–1024. doi: 10.1128/MCB.25.3.1013-1024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bischof O, Schwamborn K, Martin N, Werner A, Sustmann C, Grosschedl R, Dejean A. The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol. Cell. 2006;22:783–794. doi: 10.1016/j.molcel.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Braig M, Lee SJ, Loddenkemper C, Rudolph C, Peters AHFM, Schlegelberger B, Stein H, Dorken B, Jenuwein J, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- Cammarano MS, Nekrasova T, Noel B, Minden A. Pak4 induces premature senescence via a pathway requiring p16INK4/p19ARF and mitogen-activated protein kinase signaling. Mol. Cell. Biol. 2005;25:9532–9542. doi: 10.1128/MCB.25.21.9532-9542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Chang BD, Broude EV, Dokmanovic M, Zhu H, Ruth A, Xuan Y, Kandel ES, Lausch E, Christov K, Roninson IB. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59:3761–3767. [PubMed] [Google Scholar]
- Chen L, Chen J. MDM2-ARF complex regulates p53 sumoylation. Oncogene. 2003;22:5348–5357. doi: 10.1038/sj.onc.1206851. [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Schaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig Chen Z, Trotman LC, Schaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:636–637. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Bawa T, Lee P, Gong L, Yeh ET. Role of desumoylation in the development of prostate cancer. Neoplasia. 2006;8:667–676. doi: 10.1593/neo.06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, Beach D, Serrano M. Tumour biology: senescence in premalignant tumours. Nature. 2005a;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Collado M, Serrano M. The senescent side of tumor suppression. Cell Cycle. 2005b;4:1722–1724. doi: 10.4161/cc.4.12.2260. [DOI] [PubMed] [Google Scholar]
- de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell. 2004;13:523–535. doi: 10.1016/s1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol. Cell. Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice JF. Altered intracellular protein degradation in aging: a possible cause of proliferative arrest. Exp. Gerontol. 1989;24:451–459. doi: 10.1016/0531-5565(89)90051-x. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ. SUMO protein modification. Biochim. Biophys. Acta. 2004;1695:113–131. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Drag M, Mikolajczyk J, Krishnakumar IM, Huang Z, Salvesen GS. Activity profiling of human deSUMOylating enzymes (SENPs) with synthetic substrates suggests an unexpected specificity of two newly characterized members of the family. Biochem. J. 2008;409:461–469. doi: 10.1042/BJ20070940. [DOI] [PubMed] [Google Scholar]
- Fang W, Mori T, Cobrinik D. Regulation of PML-dependent transcriptional repression by pRB and low penetrance pRB mutants. Oncogene. 2002;21:5557–5565. doi: 10.1038/sj.onc.1205666. [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000;14:2015–2027. [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nature Rev. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Gong L, Millas S, Maul GG, Yeh ET. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Haffner R, von Ruden T, Wagner EF, Oren M. Down-regulation of wild-type p53 activity interferes with apoptosis of IL-3-dependent hematopoietic cells following IL-3 withdrawl. EMBO J. 1994;13:1368–1374. doi: 10.1002/j.1460-2075.1994.tb06390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillari J, Katinger H, Voglauer R. Aging and the ubiquitinome: Traditional and non-traditional functions of ubiquitin in aging cells and tissues. Exp. Gerontol. 2006;41:1067–1079. doi: 10.1016/j.exger.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Hara E, Tsuri H, Shinozaki S, Oda K. Cooperative effect of antisense-Rb and antisense p53 oligomers on the extension of lifespan in human diploid fibroblasts. Biochem. Biophys. Res. Comm. 1991;179:528–534. doi: 10.1016/0006-291x(91)91403-y. [DOI] [PubMed] [Google Scholar]
- Hardy K, Mansfield L, Mackay A, Benvenuti S, Ismail S, Arora P, O'Hare MJ, Jat PS. Transcriptional networks and cellular senescence in human mammary fibroblasts. Mol. Biol. Cell. 2005;16:943–953. doi: 10.1091/mbc.E04-05-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT. SUMO: A history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Herbig U, Wei W, Dutriaux A, Jobling WA, Sedivy JM. Real-time imaging of transcriptional activation in live cells reveals rapid up-regulation of the cyclin-dependent kinase inhibitor gene CDKN1A in replicative cellular senescence. Aging Cell. 2003;2:295–304. doi: 10.1046/j.1474-9728.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell. 2001;107:5–8. doi: 10.1016/s0092-8674(01)00519-0. [DOI] [PubMed] [Google Scholar]
- Jacques C, Baris O, Prunier-Mirebeau D, Savagner F, Rodien P, Rohmer V, Franc B, Guyetant S, Malthiery Y, Reynier P. Two-step differential expression analysis reveals a new set of genes involved in thyroid oncocytic tumors. J. Clin. Endocrinol. Metab. 2005;90:2314–2320. doi: 10.1210/jc.2004-1337. [DOI] [PubMed] [Google Scholar]
- Ji Z, Degerny C, Vintonenko N, Deheuninck J, Foveau B, Leroy C, Coll J, Tulasne D, Baert JL, Fafeur V. Regulation of the Ets-1 transcription factor by sumoylation and ubiquitinylation. Oncogene. 2006;26:395–406. doi: 10.1038/sj.onc.1209789. [DOI] [PubMed] [Google Scholar]
- Johung K, Goodwin EC, DiMaio D. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J. Virol. 2007;81:2102–2116. doi: 10.1128/JVI.02348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HT, Kim ET, Lee HR, Park JJ, Go YY, Choi CY, Ahn JH. Inhibition of SUMO-independent PML oligomerization by the human cytomegalovirus IE1 protein. J. Gen. Virol. 2006;87:2181–2190. doi: 10.1099/vir.0.81787-0. [DOI] [PubMed] [Google Scholar]
- Kim YH, Sung KS, Lee SJ, Kim YO, Choi CY, Kim Y. Desumoylation of homeodomain-interacting protein kinase 2 (HIPK2) through the cytoplasmic-nuclear shuttling of the SUMO-specific protease SENP1. FEBS Lett. 2005;579:6272–6278. doi: 10.1016/j.febslet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, Kleijer WJ, DiMaio D, Hwang ES. Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- Li T, Santockyte R, Shen RF, Tekle E, Wang G, Yang DC, Chock PB. Expression of SUMO-2/3 induced senescence through p53- and pRB-mediated pathways. J. Biol. Chem. 2006;281:36221–36227. doi: 10.1074/jbc.M608236200. [DOI] [PubMed] [Google Scholar]
- Mallette FA, Goumard S, Gaumont-Leclerc MF, Moiseeva O, Ferbeyre G. Human fibroblasts require the Rb family of tumor suppressors, but not p53, for PML-induced senescence. 2004. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Hengst L. SUMO-1 and p53. Cell Cycle. 2002;1:245–249. [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk J, Drag M, Bekes M, Cao JT, Ronai Z, Salvesen GS. Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J. Biol. Chem. 2007;282:26217–26224. doi: 10.1074/jbc.M702444200. [DOI] [PubMed] [Google Scholar]
- Mo YY, Moschos SJ. Targeting UBC9 for cancer therapy. Expert Opin. Ther. Targets. 2005;9:1203–1216. doi: 10.1517/14728222.9.6.1203. [DOI] [PubMed] [Google Scholar]
- Morris M, Hepburn P, Wynford-Thomas D. Sequential extension of proliferative lifespan in human fibroblasts induced by over-expression of CDK4 or 6 and loss of p53 function. Oncogene. 2002;21:4277–4288. doi: 10.1038/sj.onc.1205492. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Nickoloff BJ, Lingen MW, Chang BD, Shen M, Swift M, Curry J, Bacon P, Bodner B, Roninson IB. Tumor suppressor maspin is up-regulated during keratinocyte senescence, exerting a paracrine antiangiogenic activity. Cancer Res. 2004;64:2956–2961. doi: 10.1158/0008-5472.can-03-2388. [DOI] [PubMed] [Google Scholar]
- Pearce SH, Merriman TR. Genetic progress towards the molecular basis of autoimmunity. Trends Mol. Med. 2006;12:90–98. doi: 10.1016/j.molmed.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy JM. Can ends justify the means?: Telomeres and the mechanisms of replicative senescence and immortalization in mammalian cells. Proc. Natl. Acad. Sci. USA. 1998;95:9078–9081. doi: 10.1073/pnas.95.16.9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- Shay JW, Roninson IB. Hallmarks of senescence in carcinogenesis and cancer therapy. Oncogene. 2004;23:2919–2933. doi: 10.1038/sj.onc.1207518. [DOI] [PubMed] [Google Scholar]
- Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugrue MM, Shin DY, Lee SW, Aaronson SA. Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53. Proc. Natl. Acad. Sci. USA. 1997;97:9648–9653. doi: 10.1073/pnas.94.18.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint O, Remacle J, Dierick JF, Pascal T, Frippiat C, Royer V, Magalhacs JP, Zdanov S, Chainiaux F. Stress-induced premature senescence: from biomarkers to likeliness of in vivo occurrence. Biogerontology. 2002;3:13–17. doi: 10.1023/a:1015226524335. [DOI] [PubMed] [Google Scholar]
- Veltman IM, Vreede LA, Cheng J, Looijenga LH, Janssen B, Schoenmakers EF, Yeh ET, van Kessel AG. Fusion of the SUMO/Sentrin-specific protease 1 gene SENP1 and the embryonic polarity-related mesoderm development gene MESDC2 in a patient with an infantile teratoma and a constitutional t(12;15)(q13;q25) Hum. Mol. Genet. 2005;14:1955–1963. doi: 10.1093/hmg/ddi200. [DOI] [PubMed] [Google Scholar]
- Vernace VA, Schmidt-Glenewinkel T, Figueiredo-Pereira ME. Aging and related protein degradation: who has the UPPer hand? Aging Cell. 2007;6:599–606. doi: 10.1111/j.1474-9726.2007.00329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Podolsky R, She JX. Genetic and functional evidence supporting SUMO4 as a type 1 diabetes susceptibility gene. Ann. N.Y. Acad. Sci. 2006;1079:257–267. doi: 10.1196/annals.1375.039. [DOI] [PubMed] [Google Scholar]
- Wei W, Herbig U, Wei S, Dutriaux A, Sedivy JM. Loss of retinoblastoma but not p16 function allows bypass of replicative senescence in human fibroblasts. EMBO Rep. 2003;4:1061–1066. doi: 10.1038/sj.embor.7400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H-J, Zhou Y, Ji W, et al. Reexpression of the retinoblastoma protein in tumor cells induces senescence and telomerase inhibition. Oncogene. 1997;15:2589–2596. doi: 10.1038/sj.onc.1201446. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Sharma P, Athanasiou M, Kumar A, Yamada S, Kuehn MR. Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol. Cell. Biol. 2005;25:5171–5182. doi: 10.1128/MCB.25.12.5171-5182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.