Abstract
Complex traits and disease comorbidity in humans and in model organisms are the result of naturally occurring polymorphisms that interact with each other and with the environment. To ensure the availability of resources needed to investigate biomolecular networks and systems-level phenotypes underlying complex traits, we have initiated breeding of a new genetic reference population of mice, the Collaborative Cross. This population has been designed to optimally support systems genetics analysis. Its novel and important features include a high level of genetic diversity, a large population size to ensure sufficient power in high-dimensional studies, and high mapping precision through accumulation of independent recombination events. Implementation of the Collaborative Cross has been ongoing at the Oak Ridge National Laboratory (ORNL) since May 2005. Production has been systematically managed using a software-assisted breeding program with fully traceable lineages, performed in a controlled environment. Currently, there are 650 lines in production, and close to 200 lines are now beyond their seventh generation of inbreeding. Retired breeders enter a high-throughput phenotyping protocol and DNA samples are banked for analyses of recombination history, allele drift and loss, and population structure. Herein we present a progress report of the Collaborative Cross breeding program at ORNL and a description of the kinds of investigations that this resource will support.
History of the Collaborative Cross
The Collaborative Cross (CC) is a large, multiparental, recombinant inbred (RI) strain panel that was motivated by the need among the mouse genetics community for a high-precision genetic resource that could serve as a common integration point for the multitude of mouse genetic studies that were sure to follow in the wake of the complete sequencing of the mouse and human genomes. The concept for this common mouse genetic reference population was first proposed at the Edinburgh meeting of the International Mouse Genome Conference in October of 2001 and in print by founding members (Threadgill et al. 2002) of the Complex Trait Consortium (CTC).
An RI strain panel provides significant advantages as a resource because it is a reproducible population for cumulative data integration. Existing RI panels have limited statistical power due to their small size and capture only limited allelic diversity because all current RI sets originate from only two inbred progenitor strains. A large RI panel derived from multiple strains could capture significantly more genetic diversity and would provide sufficient power and resolution for genetic dissection of polygenic traits and construction of systems genetic networks. The CC breeding design was proposed as a strategy to rapidly and randomly mix the genomes of eight founder strains to create independent breeding lines (Churchill et al. 2004). Five classical inbred strains (A/J, C57BL/6 J, 129S1/SvImJ, NOD/LtJ, NZO/H1LtJ) and three wild-derived strains (CAST/EiJ, PWK/PhJ, and WSB/EiJ) were selected to be the eight founders of the CC. Analysis of the allelic variation in mouse inbred strains demonstrates that the eight CC founder strains capture on average 90% of the known allelic diversity across all 1-Mb intervals spanning the entire mouse genome (Roberts et al. 2007). Simulations of the power (Valdar et al. 2006a) and precision (Broman 2005) of genetic mapping with the CC population indicated superior performance to alternative strategies and provided guidelines for sample sizes.
Initial proposals for implementing the CC called for a widely distributed breeding effort. However, logistical and scientific advantages exist for breeding and distributing the CC from a small number of locations that have well-defined and consistent husbandry practices that will minimize confounding effects of environment-specific selective effects. Due to the proposed CC size, attaining the ideals originally envisioned for the CC design requires a large, dedicated facility capable of consistent randomized matings. In May 2005, breeding began at the Oak Ridge National laboratory (ORNL) supported through major funding from The Ellison Medical Foundation and The Department of Energy. Additional support for production and expansion of the CC resource at ORNL has been provided by the National Institutes of Health. Here we report on the current status of the CC lines established at the ORNL William R. and Liane B. Russell Vivarium. To date, 650 lines have been initiated in two cohorts. The first set of 474 CC lines was initiated in 2005 with a second set of 176 lines initiated in 2007 (Fig. 1).
Fig. 1.

Birth records of all CC mice born in the ORNL colony. Each mouse is represented by a single bar. Two groups of lines have been started at ORNL. The first, consisting of 474 started lines, is approaching G2:F12. The second is in the initiation phases. Time to fertility is gradually slowing to a typical pace for inbred lines. This display can be regenerated at http://mouse.ornl.gov/projects/ cc_breeding_progress.html
Construction of the Collaborative Cross at ORNL
To initiate construction of the CC, the eight progenitor strains, referred to as the G0 generation, were first intercrossed to generate 56 possible G1 hybrid combinations. G1 progeny are crossed to create the four-way G2 generation and a G2 × G2 cross yields the first eight-way progeny, the G2:F1 s. G2:F1 s are then propagated by sib-mating through the G2:Fn generations until they are fully inbred, at approximately G2:F22 (Broman 2005). Each resulting independent CC breeding funnel will be a unique and independent combination of the eight founder genomes. A major goal of the ORNL implementation of this breeding scheme has been to minimize clustering of recombination sites that result from strain pair-specific hotspots found in most mating designs (Kelmenson et al. 2005), and selection for single or multiple loci associated with viability, behavioral, and fertility traits by using the breeding software described below. The G0 progenitor strains were obtained from The Jackson Laboratory (TJL), and the G1 animals were produced either at TJL or at ORNL from stock obtained directly from TJL. The colony is restocked periodically to avoid drift in the progenitor lines.
Design for balance
An eight-way CC line can be defined by the order of the initial G0 through G2:F1 matings. Thus, if we represent the eight founder strains by letters A–H, one possible breeding scheme is
((A × B) × (C × D)) × ((E × F) × (G × H)),
where the strain on the left of each pair is the female parent. This notation summarizes the four crosses yielding four two-way hybrids (G1) followed by two crosses yielding two four-way hybrids (G2) followed by a cross yielding an eight-way hybrid (G2F1). There are more than 40,000 possible unique orderings (8!), so a subset must be chosen for production. Each line is initiated with a unique breeding order, which we refer to as a breeding “funnel”.
G0 or G1 animals from the same strain or mating, respectively, are genetically identical. Therefore, two independent reciprocal funnels can be established from the same litters of G1 hybrids, doubling the number of lines obtained from a given set of G1 hybrids without introducing shared recombination sites. Thus, the funnels are set up as reciprocal pairs:
((A × B) × (C × D)) × ((E × F) × (G × H))
and
((E × F) × (G × H)) × ((A × B) × (C × D))
With this systematic breeding design, the genetic contribution of each of the eight CC founder strains to each line is equivalent and when averaged across all CC lines, the allele frequency at each locus will ideally be 0.125 (1/8). While this is automatically true for autosomal loci, it is not necessarily true for mitochondrial genomes and sex chromosomes. Therefore, of the possible funnels, a balanced set is chosen to produce equal contributions for each single factor (X, Y, and mitochondria) as well as for all pairwise combinations of factors across all the breeding funnels.
The location occupied by any particular strain in the initial crosses impacts the genetic composition of the resultant CC line (Fig. 2). One reason for this is that the female parent of a cross contributes mitochondrial DNA through cytoplasm and the male parent of each cross contributes the Y chromosome. In the cross denoted A × B, the strain in the A position contributes mitochondrial DNA, and the strain in the B position contributes the Y chromosome to offspring. When these offspring are crossed to offspring of the cross denoted C × D, mitochondria from the strain in the A position and the Y chromosome from the strain in the D position are retained.
Fig. 2.
A depiction of the Collaborative Cross funnel design showing the progenitor source of autosomal, sex chromosomal, and mitochondrial DNA. The strain in the A position contributes the mitochondrial DNA, and the strain in the H position contributes the Y chromosome. X chromosomal DNA in finished line comes from strains in the A, B, C, E, and F positions
A second way in which progenitor position determines genetic contribution is through the contribution of X chromosomal material. Males contribute their X chromosome to female offspring and their Y chromosome to male offspring. Females contribute their X chromosomes. In the cross denoted above, a female is chosen from the A × B cross, and a male is chosen from the C × D cross. The female has X chromosomal material from parents in both the A and B position, but the male only inherits an X chromosome from the parent in the C position.
Finally, the pairing of progenitors in the earliest generations may confer specific biases in the location and density of recombinations. Meiotic recombination events are cumulative. Those recombinations that occur in early generations are retained in subsequent generations. Additional recombinations occur in each subsequent generation and have the potential to accumulate detectably in each generation prior to inbreeding. In a particular segment of DNA, if alleles are similar in the two progenitors of a cross (identical by state) or fixed through inbreeding (identical by descent), recombination events are not detectable because identical strands of DNA are being broken and recombined. The accumulation of genetic recombination is influenced by genetic background (Kelmenson et al. 2005), and recombinations do not occur with equal probability across the genome. Consequently, each of the eight progenitor strains should occupy the A through H positions an approximately equal number of times when the entire CC panel is considered. Furthermore, it is desirable to balance higher-order combinations of mitochondrial and Y-chromosomal material by balancing all two-way combinations of these factors. Lastly, balance of the combinations of parents in the first generation of crosses will minimize potential bias in recombination accumulation.
Pairwise balance of the founder strain combinations is achieved when all 56 pairwise combinations occur at equal frequency across the set of funnels. This will reduce the impact of systematic allele incompatibilities within and across loci. The variance of the number of lines across 56 pairwise combinations, therefore, gives a single number by which to evaluate pairwise balance in a set of funnels. The controlled pairwise combinations are discussed in the following subsections.
Chr Y-mitochondria combinations
Progenitors A and H, respectively, contribute mitochondria and Y chromosome to the funnel line, and progenitors E and D, respectively, contribute mitochondria and Y chromosome to the reciprocal line. Y-mitochondria combinations will be balanced if all 56 pairwise combinations of strains appear with equal frequency in the progenitor pairs (A:H) and (D:E).
Chr X-Chr Y combinations
Since five progenitors (A, B, C, E, or F in the funnel) may contribute X chromosomes to a line, strain frequency must be averaged over ten two-strain progenitor combinations, with double weight for two combinations (C:E and C:F in the funnel), to evaluate the balance of X-Y combinations.
Autosomal Combinations
Genomes of strains paired in the first generation have an early opportunity for recombination (Broman 2005). To balance any effect of this opportunity, pairwise strain combinations can be balanced over the four matings in the first generation involving progenitor pairs (A:B), (C:D), (E:F), and (G:H).
Balanced CC design schemes are created using customized software (CC8scheme) which systematically tests all available funnel pairs to optimize over the above parameters. CC8scheme builds a design scheme by stepwise addition of the funnel pair that would most improve the balance of the existing set of funnel pairs. The resulting designs avoid using strain combinations known to be infertile or unproductive while still achieving the best possible balance. The current design avoids (NZO × CAST) and (NZO × PWK) hybrids, which are reproductively incompatible, and (PWK × 129) males, which are infertile.
At each generation, two to seven matings are started from a randomly selected litter from the previous generation within each funnel. Normally, a litter from the first-priority mating will provide the next generation. If the first-priority mating fails to produce a litter by the time that one of the other matings has produced a second litter, then the lower-priority litter will be used for the next generation. Thus, litter selection is pseudorandom, since selection for fecundity is allowed only if the survival of the line is threatened. Once a litter is chosen, mice within the litter are randomly assigned mates from available siblings, avoiding inadvertent selection for docility or other behavioral characteristics.
Breeding of the Collaborative Cross is controlled and documented by custom software called Collaborative Cross Database (CCDB), which supports the task of maintaining a randomized mating scheme in the breeding colony based on a specified balanced mating design. CCDB is a three-tier Web application comprising a MySQL database and a Python application that provides a browser-based user interface. Husbandry technicians access CCDB directly from laptops in the mouse handling rooms. The user interface is tightly integrated with the workflow of weaning one generation of mice and setting up matings to create the next generation. CCDB was designed with the following goals: (1) to ensure the randomization of progeny selection and mate choice at each generation, (2) to allow data entry as mice are weaned and mated, (3) to minimize data entry time and data entry errors, (4) to monitor and maintain data integrity, and (5) to allow data entry, monitoring, and reporting from multiple locations. The result is a fully traceable breeding history for each mouse (Fig. 3). These custom software tools are available at http://sourceforge.net/projects/cc8works/.
Fig. 3.

Visual representation of a completely traceable funnel in CCDB. This display shows funnel #55 and its reciprocal #232. The gray highlight indicates the mating cage that was randomly chosen to produce the next generation
Status of the Collaborative Cross at ORNL
A total of 650 CC funnels were initiated from a balanced set of funnels identified by CC8scheme. As of this writing, 452 funnels are extant, with the remaining 198 lost during the breeding process. These include 41 funnels that required crosses (NZO/H1LtJ × CAST/EiJ) and (NZO/H1LtJ × PWK/PhJ) that are not easily obtained or that are often infertile. Because these combinations are now avoided in the first-generation crosses, the breeding success rate should improve. The majority of funnels (126 of 198) were lost during generations G2:F4-G2:F6 (i.e., after three to five generations of inbreeding) (Fig. 4). Currently, the most advanced lines have reached 12 generations of inbreeding, with the bulk of funnels distributed through generations G2:F6–G2:F8. Therefore, the population is theoretically inbred at approximately 75% of all loci (Fig. 4), based on a simulation of allelic identity-by-descent performed using the R/ricalc package (Broman 2005). In reality, the proportion of loci with alleles identical by state will be higher due to shared haplotypes among the founder strains.
Fig. 4.
Summary of ORNL funnels generated to date. The solid line indicates the total number of funnels that have reached the given generation, and the dotted line reveals the maximum number of funnels that can potentially be reached at this time based on a projection of the current number of extant funnels. Some loss is anticipated. The two subgroups of lines have been separated. Infertile G1 s are not represented
Characterizing the Collaborative Cross
During generation of the CC lines, many measurements are routinely collected to support intermediate phenotypic analysis. Breeding records allow analysis of time to fertility, gestation period, sex ratio, and litter size (Fig. 5). Each retired breeder is also characterized through a phenotyping protocol, which includes dissection and storage of tissue samples. A panel of phenotypes selected to broadly index morphology, organ function, and behavior is collected from the retired breeders of each generation and from each line. Tail samples for DNA have been obtained for all breeders in all lines, and a DNA bank has been made for many of the breeders, including the entire G2:F7 generation. Phenotype data and imported husbandry records from CCDB are stored and integrated in the MouseTrack system (Baker et al. 2004), which consists of an ORACLE database and SAS client software for genetic analysis. Software for QTL mapping of interim generations (Mott et al. 2000; Valdar et al. 2006b) is being incorporated into this system.
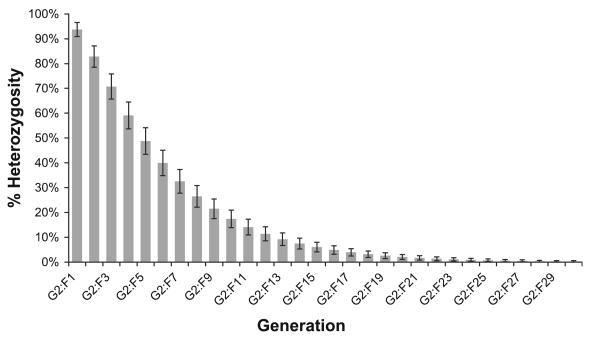
Fig. 5.
Estimate of heterozygosity within the CC. This figure shows the mean estimated identical by descent allele frequency at each generation in 10,000 simulated CC lines
Phenotype distributions and heritability
Breeding and phenotype data are used to monitor heritabilities and phenotypic diversity as inbreeding progresses (Fig. 6). The female and male breeding pair that serves as parents for the subsequent generation within each line are removed from breeding and used in the phenotyping screen after two generations of offspring have been produced for that line. A variety of biological systems is under current investigation, including morphologic, behavioral, and physiologic phenotypes. The panel of phenotypes currently collected includes behavioral wildness, anxiety, activity, sleep, nociception, body weight, tail length, bone density, gastrointestinal microflora, chromosomal aging, fasting plasma glucose, blood chemistry, and the weights of kidney, heart, and gonadal fat pads. For each transition across generations, heritability is computed in the MouseTrack system using parent-offspring regression on either mid-parent values or single-parent values. Heritability analysis tests the utility of the CC as a genetic mapping tool for a given trait, and the dispersion of phenotypes in interim generations tests its validity as a population-based model system. These ongoing studies will provide a wealth of parametric data for simulation, power analysis, and computational tool development (Fig. 7).
Fig. 6.

CC funnel litter size by generation. Litter sizes have reached an asymptote at approximately 4, which is typical for RI lines
Fig. 7.

MouseTrack calculation and display of heritability of two complex phenotypes, fasting plasma glucose, and behavioral wildness (Wahlsten et al. 2003) through seven generations of inbreeding of the Collaborative Cross
Genotyping the Collaborative Cross
Several genotyping efforts are underway, including the Tennessee Mouse Genome Consortium and DOE-funded effort to genotype and characterize the G2:F7 generation. In this project one female and one male of each line will be genotyped on a custom array of 13,000 SNPs that uniquely identify all eight progenitor haplotypes at over 1200 regions of the genome. This single-generation cross section will enable the analysis of population structure, recombination rate, and detection of systematically linked loci. Any such loci that are identified should be the result of actual biological selection, because the CCDB software-assisted breeding has eliminated many effects of human selection for docility and reproductive behavioral effects. A second effort underway will entail in-depth genotyping and phenotyping of individuals from all extant strains at various generations.
The Future Of The Collaborative Cross at Ornl
The CC mice and derivatives of their breeders are currently available through collaborative arrangements with the ORNL Mouse Genetics Research Facility. Plans are being made to ensure that finished lines will be available from a network of phenotyping centers, each of which will have all inbred CC lines on site. CC funnels will continue to be initiated and maintained until the target population size of 1000 CC lines has been met or exceeded. Archiving of strains through cryopreservation will be performed as inbreeding is advanced. The early studies performed on intermediate generations will provide a valuable resource for integrative genetics and genomics, and will yield a demonstration of the utility of the CC. As the CC genotypes are generated and regions of residual heterozygosity identified, mapping studies can be undertaken in their progeny.
Acknowledgments
The Collaborative Cross at ORNL is supported by grants from The Ellison Medical Foundation, the Department of Energy (Field Work Proposal ERKP804 “Mouse Genetics and Mutatgenesis for Functional Genomics”), and the National Institutes of Health (U01CA134240). CCWorks development was supported by P41 HG001656, U01 CA105417, P20 DA021131, and the Center of Genomics and Bioinformatics at UTHSC. The MouseTrack System was originally developed with support from the National Institutes of Health (5U01MH061971). The authors gratefully acknowledge the superb technical efforts of the ORNL staff, including Sarah Shinpock, Lori Easter, Ginger Shaw, Carmen Foster, Jason Spence, Melissa Beckmann, K. T. Cain, and Patricia R. Hunsicker, all at the ORNL, without whose diligence this project would not be possible.
Footnotes
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Elissa J. Chesler, Email: cheslerej@ornl.gov, Systems Genetics Group, Biosciences Division, Oak Ridge National Laboratory, Bldg 1059 MS-6420, P.O. Box 2008, Oak Ridge, TN 37831-6420, USA.
Darla R. Miller, Systems Genetics Group, Biosciences Division, Oak Ridge National Laboratory, Bldg 1059 MS-6420, P.O. Box 2008, Oak Ridge, TN 37831-6420, USA
Lisa R. Branstetter, Systems Genetics Group, Biosciences Division, Oak Ridge National Laboratory, Bldg 1059 MS-6420, P.O. Box 2008, Oak Ridge, TN 37831-6420, USA
Leslie D. Galloway, Systems Genetics Group, Biosciences Division, Oak Ridge National Laboratory, Bldg 1059 MS-6420, P.O. Box 2008, Oak Ridge, TN 37831-6420, USA
Barbara L. Jackson, Systems Genetics Group, Biosciences Division, Oak Ridge National Laboratory, Bldg 1059 MS-6420, P.O. Box 2008, Oak Ridge, TN 37831-6420, USA
Vivek M. Philip, Genome Science and Technology Program, University of Tennessee, Knoxville, TN 37996, USA
Brynn H. Voy, Systems Genetics Group, Biosciences Division, Oak Ridge National Laboratory, Bldg 1059 MS-6420, P.O. Box 2008, Oak Ridge, TN 37831-6420, USA
Cymbeline T. Culiat, Systems Genetics Group, Biosciences Division, Oak Ridge National Laboratory, Bldg 1059 MS-6420, P.O. Box 2008, Oak Ridge, TN 37831-6420, USA
David W. Threadgill, Department of Genetics, University of North Carolina, Chapel Hill, NC 27599, USA
Robert W. Williams, Department of Anatomy and Neurobiology, University of Tennessee Health Science Center, Memphis, TN 38163, USA
Gary A. Churchill, The Jackson Laboratory, Bar Harbor, ME 04609, USA
Dabney K. Johnson, Systems Genetics Group, Biosciences Division, Oak Ridge National Laboratory, Bldg 1059 MS-6420, P.O. Box 2008, Oak Ridge, TN 37831-6420, USA
Kenneth F. Manly, Department of Biostatistics, University at Buffalo, Buffalo, NY 14260, USA
References
- Baker EJ, Galloway L, Jackson B, Schmoyer D, Snoddy J. MuTrack: a genome analysis system for large-scale mutagenesis in the mouse. BMC Bioinformatics. 2004;5:11. doi: 10.1186/1471-2105-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW. The genomes of recombinant inbred lines. Genetics. 2005;169(2):1133–1146. doi: 10.1534/genetics.104.035212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36(11):1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Kelmenson PM, Petkov P, Wang X, Higgins DC, Paigen BJ, et al. A torrid zone on mouse chromosome 1 containing a cluster of recombinational hotspots. Genetics. 2005;169(2):833–841. doi: 10.1534/genetics.104.035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA. 2000;97(23):12649–12654. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel de Villena F, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome. 2007;18(6–7):473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Hunter KW, Williams RW. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm Genome. 2002;13(4):175–178. doi: 10.1007/s00335-001-4001-Y. [DOI] [PubMed] [Google Scholar]
- Valdar W, Flint J, Mott R. Simulating the collaborative cross: power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics. 2006a;172(3):1783–1797. doi: 10.1534/genetics.104.039313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006b;38(8):879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- Wahlsten D, Metten P, Crabbe JC. A rating scale for wildness and ease of handling laboratory mice: results for 21 inbred strains tested in two laboratories. Genes Brain Behav. 2003;2(2):71–79. doi: 10.1034/j.1601-183x.2003.00012.x. [DOI] [PubMed] [Google Scholar]