Abstract
The intrabody technology has become a promising therapeutic avenue for a variety of incurable diseases. This technology is an intracellular application of gene-engineered antibodies, aimed at ablating the abnormal function of intracellular molecules. Parkinson’s disease (PD) is a common neurodegenerative disease with no cure. Recent studies have explored possible intrabody applications against alpha-synuclein (α-syn), whose misfolding is believed to cause a familiar form of PD. Here, we review the origin, production, and therapeutic mechanisms of intrabodies and the potential of intrabody protection against α-syn toxicity. Furthermore, we propose possible intrabody applications against leucine-rich repeat kinase 2 (LRRK2), whose mutations are the most frequent known cause of familial and sporadic PD.
Introduction
Recently, the intrabody technology has emerged as a novel tool to modulate the function of intracellular proteins [1]. Moreover, this technology has been applied in various research settings that model complex human pathological conditions such as Parkinson’s disease (PD), the second most common neurodegenerative disorder after Alzheimer’s dementia [2]. The pathological features of PD include a loss of dopaminergic neurons in the substantia nigra pars compacta [3] and the formation of the Lewy body (LB), a cytoplasmic proteinaceous inclusion composed principally of alpha-synuclein (α-syn) [4]. Currently, while medicines such as Levodopa may help to manage symptoms, to date, no treatment has been shown to slow or stop the progression of PD. Although PD arises essentially as a sporadic condition, it is occasionally inherited [5]. In the past decade, studies on various proteins whose genetic mutations cause familial PD, such as α-syn, parkin, DJ-1, PTEN-induced putative kinase 1 (PINK1), and leucine-rich repeat kinase 2 (LRRK2), have largely improved our understanding of possible pathogenic mechanisms of PD and have provided us with potential therapeutic targets for PD.
Based on a paucity of available intrabody studies in PD, the intrabody therapy for this common disorder has unambiguously a long path to explore. However, it holds one of the promises of a cure for PD since it aims to remedy the pathogenic abnormalities at the molecular level. In this review, to provide a general background, we will first discuss the origin, production and therapeutic mechanisms of intrabody. Then, we will discuss intrabody studies in PD, with focus on α-syn. Finally, we will propose potential intrabody applications to prevent the cellular toxicities caused by mutant LRRK2, shedding light on intrabody studies in new PD-related scenario.
Antibody and scFv
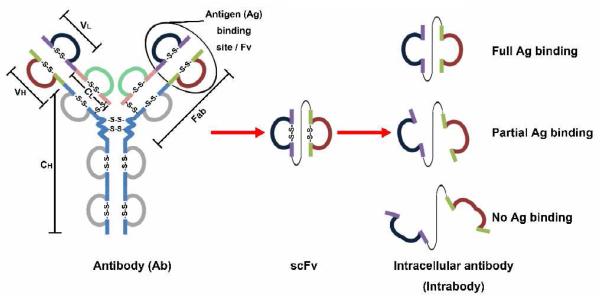
Antibodies, or immunoglobulins, are one of the major protein effectors of an immune system. A typical antibody molecule consists of two heavy chains and two light chains, connected by disulfide bonds. Compared to the constant regions (CH and CL), which share high homology among different antibodies, the variable regions (VH and VL) are extremely variable, forming a unique three-dimensional structure, the antigen-binding site or Fv (Fig. 1). The Fv binds a structurally complementary molecule, known as an antigen (Ag). The high variability of Fv makes it possible for a natural antibody pool to recognize a broad range of antigens.
Figure 1.
Schematic representation of antibody, scFv, and intrabody molecules. A typical antibody structure consists of two heavy chains and two light chains, which contain disulfide bond linked domains. Each chain has a variable region (VH or VL) and a constant region (CH or CL). While the antigen binding site, Fv, refers to the structure formed by VH and VL regions, a Fab molecule contains a light chain (VL and CL), a VH region and the first domain of a CH region, linked by disulfide bond. The VH and VL, linked by a flexible linker peptide, form scFv, a single valent antigen-binding molecule. In the cytosol of mammalian cells, based on its intrinsic folding property, scFvs have various stabilities in terms of forming functional antigen-binding molecules.
Antibodies have been broadly used for disease diagnosis and therapy mainly due to their high specificity and high affinity. An antigen determinant, also termed epitope, is the portion of an antigen that binds to an antibody. Antibody specificity is the property that enables antibodies to discriminate between two different antigen determinants. The strength with which an antibody binds to an antigen determinant is called affinity, usually described by the dissociation constant, KD (mol/L, M). Since Kohler & Milstein described the monoclonal antibody (MAb) technology in 1975 [6], an ideal MAb with both high specificity and high affinity has been believed to be a “magic bullet” for target therapy. Although initial clinical trials with MAbs showed some promising antitumor effects [7], MAb therapy is hindered by the following major obstacles: human antimouse antibody (HAMA) responses generated by the immunogenicity of mouse MAbs, difficulties in identifying specific targets such as tumor specific antigens, and the low efficacy of antibody enrichment at affected tissues [8].
To overcome the above issues, in vitro antibody display technologies have been developed to display a large repertoire of gene-engineered antibodies (>109 members) on the surfaces of phage, yeast, or others, for rapid screening of a specific antibody of interest [9-14]. Gene-engineered antibodies refer to recombinant antibody molecules that are generated by expressing the cDNA of engineered antibody fragments, such as “VH-linker-VL” (Fig. 1). Usually, the engineered antibody fragments contain at least one complete or partial antigen binding site to retain antigen-binding function. Gene-engineered antibodies consist of many types of antibody molecules, such as single-chain Fv (scFv, ∼25 kDa) and Fabs (antigen binding fragments, ∼55 kDa) (Fig. 1). Here, throughout this review, for the simplicity of demonstration, we use scFv to represent all other gene engineered antibodies.
ScFv, a key molecule of gene engineered antibodies, is composed of the VH and VL regions, which are connected by a flexible short linker with a major form of (Gly4Ser)3 (Fig. 1) [15]. ScFv is attractive due to the following advantages. First, it has a small size of ∼25 kDa (vs ∼150 kDa of a typical antibody), yielding rapid renal elimination and better tumor tissue penetration [16-18]. Yet, it retains good antigen-binding specificity because the antigen-binding site is unaltered. Second, to generate a scFv, the coding gene segments of VH and VL regions are isolated either from a hybridoma expressing the MAb with desired specificity [19], or from a large antibody library with more than 109 members of diverse scFvs; the latter gives the potential for screening scFvs against virtually any possible antigen in a short time [20]. Third, most antibody libraries contain scFvs as human proteins, which do not induce HAMA responses, because these scFvs are made from the coding genes of VH and VL regions from human lymphocytes. Fourth, optimized scFv functional characteristics, such as high affinity, long retention time (the capability to stay in antigen-binding status), and enhanced protein stability, can be achieved by further mutagenesis of the scFv cDNAs and by further selection from sub-libraries containing these mutated scFvs [21]. Clearly, scFvs and other gene-engineered antibodies represent a type of promising reagent for targeted therapy. Furthermore, innovative technologies have largely improved the likelihood of an antibody being a “magic bullet” and are still undergoing further development, especially in the improvement of minimizing immunogenicity, in increasing specificity, affinity, and stability, and in employing new application scenarios. For example, recently, the application of scFv inside of human cells has become a completely novel type of molecular therapy, intrabody therapy.
Intrabody
Intrabodies, or intracellular antibodies, here refer to gene-engineered antibodies (mainly scFvs) that are expressed intracellularly to modulate the function of their intracellular targets. Usually, cell transfections or infections with scFv cDNA-containing plasmids or viruses are the means to express scFvs intracellularly. Clearly, the intrabody technology is an intracellular application of gene-engineered antibodies.
Challenges
To use scFvs intracellularly, a major obstacle is the fact that the cytoplasmic reducing environment prevents the formation of disulfide bonds. As shown in Fig. 1, disulfide bonds are necessary for optimized folding of the antigen binding site of scFv [22]. Thus, for most scFvs isolated from antibody libraries or from hybridomas, the antigen binding capacities are more or less decreased or even completely lost when these scFvs are expressed as intrabodies in the cytosol (Fig. 1) [23,24]. For this reason, to select the scFvs that can bind their antigens inside cells, researchers have developed the following two strategies: a) in vitro screening of scFvs from antibody libraries followed by further intracellular validation in mammalian cells [25,26]; b) intracellular scFv screening to identify intracellular functional scFvs directly [27-29]. As a result, the most critical parameter of a successful intrabody is its intrinsic folding capacity under a reducing environment, rather than its in vitro affinity [30]. In addition, there are other important challenges for intrabody therapy, such as the efficacy of gene delivery, which are not in the scope of this review.
Production of intrabodies
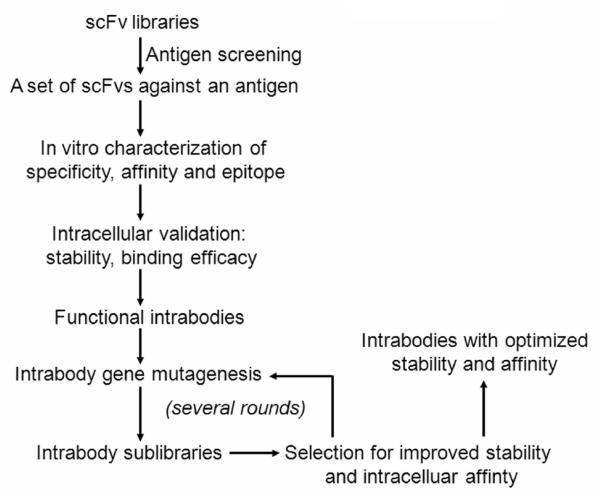
As mentioned above, intrabodies can be generated by using direct intracellular scFv screening methods [27-29]. However, while these methods are waiting to be widely applied and tested, the following strategy remains as a feasible and widely used method to generate functional intrabodies (Fig. 2).
Figure 2.
Schematic representation of the production of scFvs of interest, identification of functional intrabodies, and further optimization of intracellular stability and affinity. This approach applies with various antibody display system, including phage display and yeast display antibody technologies, toward yielding robust intrabodies as therapeutics.
First, antigens are usually produced as recombinant proteins by expressing the antigen cDNAs in bacteria, followed by protein purification. Of note, some eukaryotic proteins require post-translational modifications for maximal folding/activity, whereas bacteria do not have the machinery required to accomplish complex modifications. Therefore, when these modifications are necessary, eukaryotic cells, such as yeast cells and insect cells, can be used for the production of recombinant proteins. In the cases that recombinant proteins are not readily available, or scFvs against specific epitopes are needed, synthesized peptides are often used as antigens. Numerous antibody libraries are available for scientific studies, such as synthetic phage libraries (the Tomlinson I and J antibody libraries, MRC Center for Protein Engineering, Cambridge, England) and naïve yeast libraries [11]. With the antigen and antibody libraries available, one can screen a set of scFvs specifically against an antigen according to standard protocols [9,11]. Once a set of binders are selected, it is important to characterize their in vitro features, including specificity, affinity, and epitopes. From this in vitro characterization, a set of scFvs with good antibody features usually can be acquired. However, only the scFvs with functionally intracellular binding to their specific targets can be used as intrabodies (see later discussion in the section of in vitro versus in situ characterization of scFvs).
Therapeutic strategies
Since 1988, when a study showed that intracellular expression of the cDNAs of antibody heavy and light chains against yeast alcohol dehydrogenase I successfully inactivated the enzyme in vivo [31], the intrabody technology has been broadly employed in the studies involving human diseases, mainly AIDS and cancer. The following strategies are used for intrabodies to ablate or restore the function of intracellular molecules. First, intrabodies can block the interaction between their target proteins and other molecules, such as proteins and nucleic acids. For example, human immunodeficiency virus type1 (HIV-1) protein Tat functions through binding with human cyclinT1 to promote the transcription elongation of HIV-1. By disrupting the molecular interaction between HIV-1 Tat and human cyclinT1, anti-human cyclinT1 intrabodies effectively block HIV-1 replication without causing cellular toxicity [32]. Second, by retaining their targets in a particular subcellular compartment, such as endoplasmic reticulum (ER), intrabodies can prevent their target molecules from being at their physiological locations to function [33]. In fact, by attaching an organelle-targeting sequence to an intrabody, the expression of intrabodies at specific subcelluar compartments can be achieved, such as in nuclei, ER and lysosomes [34]. For instance, tagged with the ER-retention signal (SEKDEL), an anti-gp120 intrabody has been used to trap gp120, an envelope protein of HIV-1, inside the lumen of ER to prevent the assembly of new infectious virions [35]. Third, intrabodies can restore normal protein function that has been lost due to mutagenesis. For instance, p53 is a tumor suppressor gene that is mutated in ∼ 50% of cancers. Mutation of p53 results in a loss of function of p53 protein by diminishing its DNA binding activity, which is required for its tumor suppressor function. An anti-p53 intrabody has been shown to restore the normal conformation and DNA binding activity of p53 and thus to suppress tumor progression [36]. Fourth, intrabody can prevent aberrant protein conformation and protein aggregation. This aspect will be discussed later in the section related to α-syn.
Advantages
Obviously, intrabodies are valuable for both functional studies of a given intracellular molecule and the development of novel therapeutics. In fact, compared to other similar technologies aimed at blocking protein function, such as RNA interference (RNAi), intrabody technology has the advantage of modifying certain domains of a protein without affecting other domains because intrabodies have domain-specific binding capabilities. In addition, intrabodies can also modify the three-dimensional structure of a protein and thus are important for both stabilizing active protein conformations and correcting aberrant protein conformations. The abnormal protein conformations are closely related to the pathogenic mechanisms of neurodegenerative diseases, such as PD. Moreover, for proteins with relatively long half-lives or for proteins whose mRNAs have short half-lives, RNAi may be less effective while intrabodies can still function effectively through direct binding to the proteins.
Efficacy in animal studies and clinical trails
Geared toward a molecular therapy for human diseases, the intrabody technology has been applied in animal studies and clinical trials. For instance, the neutralizing anti-Ras intrabody Y13-259, which induces in vitro transient phenotypic reversion of rodent fibroblasts transformed by ras, an oncogene, has been shown to invoke sustained tumor regression in nude mice [37]. Moreover, an ER-targeted intrabody against Tie-2, a receptor that is up-regulated in capillaries during neovascularization processes produced in tumors, has been shown to significantly inhibit tumor growth in mouse models [38]. More recently, the intrabody EM48 has been shown to suppress neuropil aggregates in a mouse model of Huntington’s disease (HD) [39], thus indicating promising applications of intrabodies in neurodegenerative diseases. In the Phase I clinical trial using an ER-targeted anti-erbB-2 intrabody, which had been previously shown to prolong survival in human ovarian cancer xenograft models [40], of 13 ovarian cancer patients with erbB-2 overexpression, 5 (38%) had stable disease and 8 (61%) had evidence of progressive disease [41]. Except for showing that the intrabody approach is feasible for human application with very limited side effects, this clinical study indicates that compared to cellular and animal studies, the intrabody technology, including gene delivery methods, needs to be further developed to achieve a dramatic effect for the therapy of human diseases.
Intrabody and PD
In light of the initial successful application of intrabodies in the studies of AIDS and cancer, the intrabody technology has been employed in the studies of neurodegenerative diseases with the hopes of preventing abnormal protein aggregation. For instance, in HD, the polyglutamine expansion in the huntingtin (htt) protein causes htt aggregation, which is thought to be toxic to cells. It has been shown that anti-htt intrabodies can inhibit mutant htt aggregation and thus rescue cells from death [42,43] and the anti-htt intrabody C4 shows in vivo efficacy of protection in a Drosophila model of HD [44]. Moreover, the intrabody EM48 has been shown to suppress neuropil aggregates in a HD mouse model [39]. Another example in the study of Alzheimer’s disease is the use of intrabodies to inhibit the generation of the toxic amyloid beta-peptide, by blocking the beta-secretase cleavage site or by retaining the beta-amyloid precursor protein in ER [45].
In the following sections, we will discuss the intrabody application in PD. Although the intrabody technology can be used for studying all PD-related proteins, here, we focus on α-syn and LRRK2, two proteins that cause autosomal dominant PD, to demonstrate a proof of principle of existing and future intrabody applications in PD.
α-syn
α-syn is a 140 amino acid protein that belongs to the synuclein protein family, which also includes β-syn and γ-syn. Previous studies using both optical methods and NMR have shown that α-syn exhibits a random-coil structure (or natively unfolded structure) under physiological conditions [46,47]. Mutations in α-syn cause a rare form of PD. The fact that α-syn is the major component of LB, a hallmark of PD, has put α-syn under intense study that eventually led to the consensus that protein aggregation is one of the major pathogenic mechanisms of PD. Although it is debatable whether LBs are toxic or protective and which forms of aggregated α-syn such as oligomers versus fibrils mediate toxicity, it is clear that α-syn is a natively unfolded protein that may acquire a gain of toxic function due to self aggregation [48-51]. Therefore, the development of anti-α-syn intrabodies may be potentially useful in blocking α-syn aggregation and therefore protect cells from toxic α-syn aggregates.
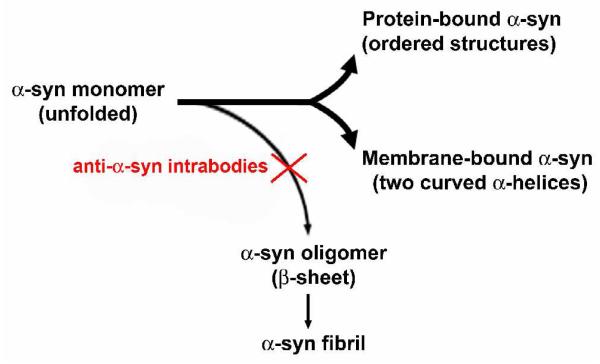
Natively unfolded proteins have been categorized as a novel protein family — naturally unfolded (or intrinsically unstructured) protein family. It is hypothesized that these natively unfolded proteins often acquire ordered three-dimensional structures upon binding to their partners, such as proteins and nucleic acids, and modify the function of their partner molecules [52,53]. Based on the available knowledge of natively unfolded proteins and previous studies on α-syn, we propose a simplified paradigm of possible α-syn structure-function relations as a working hypothesis for developing potential protective intrabodies against PD (Fig. 3). In this schematic, α-syn exists physiologically as a natively unfolded protein with the potential of transforming into the following three ordered structures. The available experimental evidence shows that the N terminal sequence of α-syn binds to acidic lipids and forms an α-helical structure. One hypothesis is that α-syn functions on cellular membranes once it acquires an ordered structure through binding to the membranes. In addition, like other natively unfolded proteins, α-syn may also bind to other multiple targets to play an important role as a modifier of various cellular pathways. While α-syn may utilize its unfolded structure to adopt several possible ordered structures to function, this possible biological benefit comes with the following potential downside: α-syn can form a stable β-sheet structure by self-binding under certain conditions. The latter include increased α-syn protein levels exceeding a threshold, genetic mutations which cause potency for self-oligomerization, changes in physiological conditions including oxidative stress, and post translational modifications including dopamine-α-syn adducts. As a result, when the protein quality control system of a cell becomes impaired, the physiological balance that favors the formation of beneficial α-syn structures may abnormally favor α-syn self aggregation. Such self-aggregation may in turn underlie the formation of α-syn aggregates, fibrils and LBs.
Figure 3.
A central molecular hypothesis for the intrabody therapy against the α-syn phathology. By blocking α-syn self-aggregation, intrabodies restore the balance between the natively unfolded structure of α-syn and its ordered philological structures, rendering therapeutic effects.
Based on the above model, it is tempting to develop intrabodies that can specifically block the self-binding of α-syn without affecting the binding of α-syn to its physiological partner molecules (Fig. 3). In the pioneer studies aimed at this goal, various scFvs against α-syn have been generated and we have listed the majority of these in Table 1. In the following, we will discuss several critical issues based on the available studies of anti-α-syn scFvs and intrabodies to shed light on future studies.
Table 1.
Published scFvs against α-syn
| Name | Source library |
Epitope (a.a.) |
Binding to α-syn aggregates |
Affinity (KD, M) |
In vitro effects | Intracellular effects |
|---|---|---|---|---|---|---|
| F8[49] | Phage | 101-111/27-37 | ? | 10-8 | Inhibit aggregation | Unstable expression |
| D10[25] | Phage | * | yes | 10-6 | ? | Inhibit α-syn aggregation and rescue impaired adhesion |
| 6E[50] | Phage | * | yes | ? | ? | ? |
| 3[52] | Phage | 71-85 | yes | ? | ? | ? |
| 14[52] | Phage | 106-120 | yes | ? | ? | ? |
| 15[52] | Phage | 117-131 | yes | ? | ? | ? |
| D5[51] | Phage | * | yes | ? | Inhibit aggregation | ? |
| NAC3[26] | Yeast | 61-78 | ? | ? | ? | No significant protection |
| NAC24[26] | Yeast | 61-78 | ? | ? | ? | Unstable expression and no significant protection |
| NAC32[26] | Yeast | 53-87 | ? | ? | ? | Inhibit α-syn aggregation and rescue cytoxicity |
not reported.
The scFvs D10, 6E, and D5 recognize α-syn conformational epitopes, although the amino acid sequences that constitute the epitopes are not identified.
Source of intrabodies
As potential therapeutics for human diseases, intrabodies are often generated by screening scFvs from human antibody libraries rather than by cloning scFv gene segments from mouse hybridomas. Current available anti-α-syn scFvs/intrabodies are selected from three phage libraries and one yeast library (Table 1). ScFvs F8 [54], D10 [25], 6E [55], D5 [56] are selected from two synthetic libraries that contain synthesized variable regions of scFvs. On the other hand, scFvs NAC3, NAC24, and NAC32 [26] are selected from a yeast library that is composed of scFv gene segments from the mRNA pool of 58 human adult spleens and lymph nodes. In addition, scFvs 3, 14 and 15 [57] are from a phage library generated using the mRNAs from human peripheral blood lymphocytes of a large (>100) pool of donors. Thus, all the scFv libraries currently used for generating anti-α-syn scFvs are naïve libraries in which scFv gene segments are from unimmunized individuals or are artificially synthesized. This suggests that, despite the unique unfolded structure of α-syn, it is feasible to screen anti-α-syn scFvs from current available large scFv libraries, no matter what display platform a library adopts, or what screening method a library employs. For example, phage libraries use a panning protocol in which immobilized antigens are coated on a solid surface, while yeast libraries use a flow cytometry sorting protocol with free antigens in solution (see [9] and [11] for detailed method descriptions). Meanwhile, it also indicates that the initial screened scFvs from these naïve antibody libraries may need to be engineered further to improve their affinities for the natively unfolded protein α-syn, or to increase their thermal stabilities and antigen-binding specificities.
Antigens used for screening scFv libraries
α-syn contains an N-terminal amphipathic region, a central hydrophobic region known as the nonamyloid component (NAC, aa 61-95), and a C-terminal acidic tail. It has been reported that the NAC region is the core sequence responsible for α-syn aggregation [58,59]. The N-terminal sequence has a propensity to bind with acidic lipids whereas the C-terminal region appears to be involved in α-syn aggregation [60,61]. Currently, the accurate molecular mechanisms of α-syn self-binding and the formation of stable oligomers and fibrils are unclear. Therefore, there are no clear strategies for predicting antigenic epitopes to produce anti-aggregation scFvs, but to explore all of the possibilities. That is to use α-syn peptides, α-syn monomers, and α-syn conformers (including α-syn oligomeric aggregates and fibrils) as antigens. With these antigens, it is hopeful that scFvs may be generated against two kinds of potentially useful epitopes to inhibit α-syn aggregation. One type of useful epitope is the linear α-syn sequences that are the self-binding sites of two or more α-syn monomers. Among scFvs that can bind to these sequences, it is reasonable to select those that can block the self-binding sites and thus inhibit α-syn oligomer formation. The identified sequences in the NAC region (aa 68-76 or aa 71-82), which are required for α-syn aggregation, are the most promising candidates in this aspect. Indeed, scFv NAC32 is selected against a peptide containing the amino acids 53-87 of α-syn. When tested in mammalian cells as an intrabody, it inhibited α-syn aggregation and reduced the cytotoxicity caused by mutant α-syn. However, we cannot exclude other N-terminal or C-terminal sequences as useful antigenic epitopes, since we do not know how these two regions play their roles in α-syn aggregation. Another type of useful epitope is the three-dimensional structures that are important for aggregation formation. ScFvs against this kind of epitope are also expected to block α-syn self-aggregation. For example, scFv D10 recognizes a conformational epitope, inhibits the formation of a high molecular weight α-syn species and rescues impaired cell adhesion caused by overexpressed α-syn. For these available anti-α-syn scFvs, a detailed description of the relationship between the epitopes and their scFvs’ capacity to inhibit α-syn aggregation can be found in Table 1.
In vitro versus in situ characterization of scFvs
As we mentioned before, antigen-binding efficiencies of an scFv between in vitro and intracellular conditions are often not consistent due to the reducing environment of the cytosol. For example, both scFv F8 and scFv D10 are selected against recombinant monomeric α-syn from a human synthetic antibody library. ScFv F8 shows high affinity (KD = ∼10-8 M) whereas scFv D10 has only low affinity (KD = ∼10-6 M). However, when expressed intracellularly, scFv F8 is very unstable whereas scFv D10 is quite stable. Surprisingly, scFv D10 is able to bind α-syn inside of mammalian cells and, further, to inhibit the formation of a high molecular weight α-syn species. The phenomenon of low-affinity scFvs being functional intrabodies might be addressed by two explanatory models of antigen-antibody interaction, the “lock-and-key” and “induced fit” models. In the “lock-and-key” model, the structures of the antigen-binding site and its corresponding antigen are highly complementary. Therefore, only minimum conformational changes are required when the antibody binds to its antigen, yielding high affinity. On the other hand, in the “induced fit” model, antigens, antibodies, or both, undergo conformational adjustments to facilitate antibody-antigen binding, thus resulting in low affinity [62]. It is likely that due to high intracellular stability, some low affinity scFvs, such as scFv D10, can accumulate to a certain amount in the cytosol and bind their antigens through an “induced fit” mechanism. Consistent with this hypothesis, other studies also revealed that the stability is much more critical than the in vitro affinity for an scFv to be a functional intrabody [29]. Therefore, while in vitro characterization is helpful for identifying scFv features, it is critical to assess the performance of scFvs inside of mammalian cells in order to generate functional intrabodies.
Intracellular characterization of intrabodies
From the above, it is apparent that intracellular stability and “intracellular affinity” are two critical aspects for evaluating the potential of an scFv to be a functional intrabody. To evaluate intracellular stability of scFvs, the pulse-chase method is a classic way to determine protein half-lives. In general, an scFv with a half-life of over 8 hours would likely have a high intracellular steady state level in order to be a functional intrabody. Meantime, it is also important that a stable scFv can fold correctly in the cytosol to maintain its antigen-binding efficiency in a reducing environment, i.e. “intracellular affinity”. Unfortunately, unlike in vitro affinity assays, such as BIAcore, a direct intracellular affinity assay is currently not available. Therefore, two intrabody binding assays have been established to test indirectly for binding efficacy between intrabodies and their antigens. One strategy is to express scFvs tagged with organelle-targeting sequences to gather scFvs at certain subcellular compartments, such as nuclei. If an scFv binds to its intracellular antigen efficiently, it can convey its antigen to the same organelle. As a result, both the scFv and its antigen can be found to be enriched and colocalized in the same organelle by immunocytochemistry. For example, co-overexpression of D10-NLS (nuclear localization signal) and α-syn causes the enrichment and co-localization of both proteins in the nuclei [25]. Of note, in scFv/antigen co-transfection experiments, the degree of accumulation of the scFv/antigen to a certain organelle varies from cell to cell. Thus, a strict quantification of the percentage and degree of the colocalization would be helpful. For this, one can use subcellular fractionation with Western blotting to assess the colocalization efficacy, which can be quantified with image quantification softwares, such as Scion Image and NIH Image. Another method to show the binding between intrabodies and their antigens is the co-immunoprecipitation assay. In this case, intrabodies and their antigens will be considered just as two proteins overexpressed intracellularly. Co-immunnoprecipitation is used to evaluate whether or not intrabodies and their antigens bind to each other inside of cells. For instance, the D10 intrabody shows dose-dependent co-immunoprecipition with α-syn [25]. In general, when the above two strategies are applied, they may reveal whether an scFv can bind its antigen effectively as an intrabody. For the future, a direct and real-time binding assay is needed to assess the “intracellular affinity” of intrabodies in a more accurate way so that an “intracellular KD” and a retention time can be given. Such elegant intracellular quantification assays will improve significantly the evaluation of intrabody characters.
Evaluation of anti-α-syn intrabody protective effects
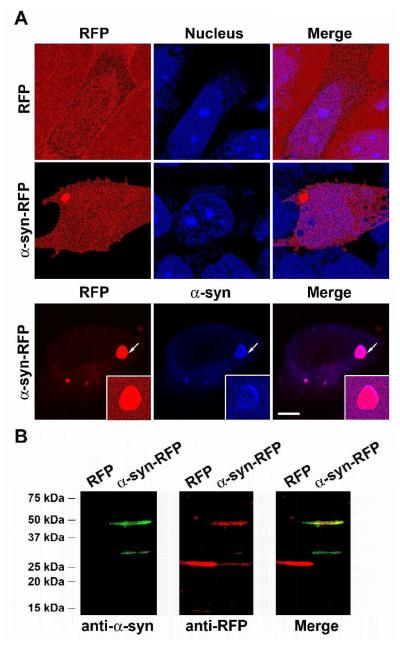
Once efficient anti-α-syn intrabodies have been identified, it is critical to evaluate whether or not these intrabodies are protective. Given that the physiological function of α-syn is still unclear, it is necessary to test the protective effect of intrabodies on both of the inhibition of α-syn aggregation and rescue from cytotoxicity caused by α-syn, which model the pathogenic hallmarks of PD --- Lewy body and neuron death. The thioflavin T aggregation assay and atomic force microscope (AFM) imaging are widely used for in vitro study of α-syn aggregation. Due to its aggregation-prone feature, α-syn forms self-aggregates in common solvents, such as phosphate buffers. By monitoring thioflavin T fluorescence intensities, the kinetics of α-syn aggregation can be established which allows one to evaluate whether or not an intrabody inhibits α-syn aggregation [49]. The AFM analysis, another in vitro tool, is often used to observe α-syn aggregates directly, allowing identification of various α-syn aggregation stages, from small aggregates to filaments [49, 52]. At the cellular level, except for analyzing α-syn by immunoblotting [25], fluorescence-tagged α-syn may be used to model α-syn aggregation. For instance, the α-syn-GFP fusion protein has been used to model α-syn aggregation and its abnormal effects in yeast [63]. Similarly, when a fusion protein of α-syn-red fluorescent protein (RFP) is overexpressed in human neuroblastoma SH-SY5Y cells, we observed that the majority of overexpressed α-syn-RFP formed perinuclear aggregates that are both α-syn and RFP positive (Fig. 4A). Furthermore, fluorescent immunoblotting shows that the majority of overexpressed α-syn-RFP is full-length protein (Fig. 4B), thus supporting α-syn-RFP overexpression as a model of α-syn aggregation in mammalian cells. Recently, cytotoxicity assays using ST14A neuronal cells have been utilized to test the protective effect of intrabodies against α-syn toxicity. Along this line, other established α-syn cellular models, such as inducible α-syn overexpression in PC12 cells [64] and overexpression of α-syn in primary cultured dopaminergic neurons [65], may also be useful for the evaluation of the key issue --- the protective effects of anti- α-syn intrabodies against neuronal death. Perhaps, the more challenging tests are whether anti-α-syn intrabodies can perform protective functions in vivo, especially in α-syn models in mice [66], Drosophila [67], Caenorhabditis elegans [68], etc.
Figure 4.
An α-syn aggregation model in human neuroblastoma SH-SY5Y cells. (A) While cells expressing RFP show homogenous distribution of RFP (red signals) without apparent protein aggregates, α-syn-RFP transfected cells have perinuclear RFP-containing protein aggregates (red signals). Nuclei are shown with DAPI labeling (blue signals). In addition, the RFP-positive protein aggregates (red signals) are also α-syn positive by the immunostaining with a mouse mononclonal anti- α-syn antibody (BD Biosciences, San Jose, CA) (blue signals). The protein aggregates indicated by arrows are shown at higher magnification in the insets. Scale bar, 5 μm. (B) The whole cell lysates extracted from RFP or α-syn-RFP transfected cells are subjected for fluorescent immunoblotting [82]. α-Syn is detected by the anti-α-syn antibody from BD Biosciences (green signals) and RFP is detected by a rabbit polyclonal anti-RFP antibody (Clontech Laboratories, Mountain View, CA) (red signals). The merged image shows that the ∼45 kDa protein species is both α-syn and RFP positive, thus indicating that the majority of α-syn-RFP is full-length (∼45 KD) and further supporting that the perinuclear protein aggregates, shown in (A), contain full-length α-syn-RFP.
Intrabody optimization
It is worth pointing out that after the initial identification of functional intrabodies that can bind to their antigens intracellularly, certain optimization strategies can be applied to obtain robust intracellular stability and “intracellular affinity” (Fig. 2). For example, the affinity of scFvs derived from naïve antibody libraries often can be further increased by random mutagenesis. Such mutagenesis can create sub-libraries with ∼106 mutated scFvs. If sub-libraries are screened against the same antigen, a set of scFvs with higher affinities can be obtained [69]. Recently, it has been shown that using yeast scFv display, scFvs with high thermal stability can also be achieved by mutagenesis [70]. With these further steps, intrabodies with optimized intracellular stability and affinity can be achieved eventually. Such robust intrabodies should be ready for further in vivo applications, including clinical trials.
Summary
As discussed above, anti-α-syn scFvs/intrabodies have been generated and tested for their potential protective effects against α-syn malfunction. While these intrabodies show promising protective effects, more extensive studies are needed in the following two areas. First, robust intrabodies with optimized intracellular stability and “intracellular affinity” are needed in order to make intrabody as an intracellular “magic bullet”. Currently, only two anti-α-syn scFvs, D10 and NAC32, have shown promising effects as intrabodies in inhibiting α-syn aggregation and in rescuing α-syn toxicity. While both scFvs show relative intracellular stability, they all have low affinity. Importantly, the production of anti-α-syn scFvs with high “intracellular affinity” is more likely to provide robust intrabodies as possible therapeutics for PD. To achieve this goal, as we proposed, it would be helpful to establish real-time analyses to evaluate intracellular intrabody-α-syn dynamic interaction. That way, the intracellular binding efficacy of anti-α-syn intrabodies can be quantified and further used to generate high “intracellular affinity” through mutagenesis. Currently, since such “intracellular affinity assays” are unavailable, it may be important to generate the scFvs that have both intracellular stability and in vitro high affinity, with the assumption that given the intracellular stability, the in vitro affinity may reflect a certain degree of “intracellular affinity”. A strategy for generating high affinity scFvs with high intracellular stability is given in Fig. 2. In addition, the specificity of anti-α-syn intrabodies for other synuclein family proteins, such as β-syn and γ-syn, need to be tested. For instance, the D10-NLS construct failed to convey β-syn to the nucleus, thus indicating its relative specificity for α-syn inside cells. This specificity issue also applies to other available anti-α-syn scFvs listed in Table 1. The highly specific scFvs against α-syn, but not its homologous proteins, are more likely to avoid disturbing other normal cellular functions. Second, the current studies of anti-α-syn intrabodies are pioneer work for demonstrating proof of principle. To generate valuable anti-α-syn intrabodies as therapeutics for PD, in-depth studies are needed to answer the following questions: how intrabodies inhibit α-syn aggregation; whether the binding of intrabodies with α-syn can change α-syn degradation; whether or not and how intrabodies can provide significant protection against dopaminergic neuron death; etc. Furthermore, to overcome the intrinsic limitation of studies using cellular models, in vivo intrabody effects on α-syn pathology could provide critical insight into the potential of intrabody therapy for PD.
LRRK2
In the following, we will discuss briefly a possible use of intrabodies as potential therapeutics against cellular toxicity caused by LRRK2. Recently, the importance of LRRK2 in PD has been highlighted by the fact that LRRK2 is currently the most frequent genetic cause of all PD cases.
LRRK2 is a large complex protein of 2,527 amino acids. It contains several predicted functional domains: Roc (Ras in complex proteins), COR domain (C terminal of ROC), a leucine-rich repeat (LRR), a protein kinase domain, a WD40 domain and an ankryin domain. The ROC domain has GTPase activity. Currently, the functions of some of these domains, such as Roc, LRRs, Ankyrin, and WD40 domains, are unknown.
Certain PD-linked LRRK2 mutations, such as G2019S and I2020T, increase LRRK2 kinase activity, thus leading to neuronal toxicity [71-73]. Accordingly, abolishing the kinase activity attenuates cytotoxicity caused by mutant LRRK2, confirming that the kinase activity is likely a prerequisite for the toxic effects of mutant LRRK2 [74]. Therefore, blocking LRRK2 kinase activity by intrabodies could be a potential therapeutic strategy against aberrant LRRK2 toxicity, especially when using kinase domain-specific intrabodies, which do not affect the function of other LRRK2 domains.
In addition, it has been shown that LRRK2 exists as a protein complex, including homodimers. LRRK2 homodimerization is dependent upon the interaction of the ROC GTPase domain and several other domains, such as the LRR and WD40 domains [75]. Interestingly, it has also been found that LRRK2 can form aggregates under certain circumstances. For example, overexpression of mutant LRRK2 has led to the formation of cytoplasmic aggregates or inclusions in various cell types, suggesting that LRRK2 mutations promote protein misfolding [74,76,77]. Therefore, intrabodies that can block LRRK2 misfolding may be protective.
Except for self-binding, LRRK2 has been shown to interact with other proteins, such as parkin [76] and Rab5b [78]. In addition, LRRK2 may function on cellular membranes [71,79,80] or through binding to microtubules [81]. With the function and toxicity of LRRK2 being further elucidated, intrabodies may be useful for blocking specific molecular events required for the gain of toxicity of LRRK2.
Prospectives
Clearly, intrabodies provide a useful tool to study protein function intracellularly and, more importantly, have the protential to serve as therapeutics for PD. Further development of intrabody technology, such as new approaches to generating robust intracellular stability and high “intracellular affinity”, novel intrabody-effector fusion proteins, organelle-specific intrabodies, and protein domain-specific intrabodies, will maximize intrabody therapeutic efficacy. In-depth studies of the intrabody characters and their anti-PD effects and mechanisms in various PD models, from in vitro to in vivo, will provide solid evidence for exploring the potential of intrabody therapeutic application. The development of gene delivery methods, which are the means of expressing intrabody in target cells, will increase the in vivo therapeutic efficiency of intrabodies. In addition, since the studies on the molecular mechanisms of PD pathology are advancing quickly, newly identified “toxic” molecules will provide additional targets for the intrabody therapy. Together, multi-field technological and knowledge advances related to the intrabody therapy hold one of the promises for the cure of PD.
Acknowledgements
We thank Drs. Anne Messer (Wadsworth Center, New York State Department of Health) and Howard Federoff (Georgetown University) for their insightful comments on the manuscript. The authors are supported by grants from the US National Institutes of Health (NS062180, NS11766-27A1, AG21617-01A1, NS38370-09), the US Department of Defense (DAMD 17-03-1-02), the Parkinson’s Disease Foundation, the Hartman Foundation, the Muscular Dystrophy Association/WOW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Lobato MN, Rabbitts TH. Intracellular antibodies as specific reagents for functional ablation: future therapeutic molecules. Curr. Mol. Med. 2004;4:519–528. doi: 10.2174/1566524043360384. [DOI] [PubMed] [Google Scholar]
- 2.Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 3.Braak H, Braak E, Yilmazer D, Schultz C, de Vos RA, Jansen EN. Nigral and extranigral pathology in Parkinson’s disease. J. Neural Transm. Suppl. 1995;46:15–31. [PubMed] [Google Scholar]
- 4.Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ, Roos J, Goedert M. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 5.Vila M, Przedborski S. Genetic clues to the pathogenesis of Parkinson’s disease. Nat. Med. 2004;10(Suppl):S58–S62. doi: 10.1038/nm1068. [DOI] [PubMed] [Google Scholar]
- 6.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 7.Miller RA, Maloney DG, Warnke R, Levy R. Treatment of B-cell lymphoma with monoclonal anti-idiotype antibody. N. Engl. J. Med. 1982;306:517–522. doi: 10.1056/NEJM198203043060906. [DOI] [PubMed] [Google Scholar]
- 8.von MM, Adams GP, Weiner LM. Monoclonal antibody therapy for cancer. Annu. Rev. Med. 2003;54:343–369. doi: 10.1146/annurev.med.54.101601.152442. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu. Rev. Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 11.Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JM, Yeung YA, Cochran JR, Heinzelman P, Colby D, Swers J, Graff C, Wiley HS, Wittrup KD. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat. Biotechnol. 2003;21:163–170. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 12.Hanes J, Pluckthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daugherty PS, Olsen MJ, Iverson BL, Georgiou G. Development of an optimized expression system for the screening of antibody libraries displayed on the Escherichia coli surface. Protein Eng. 1999;12:613–621. doi: 10.1093/protein/12.7.613. [DOI] [PubMed] [Google Scholar]
- 14.Chen G, Hayhurst A, Thomas JG, Harvey BR, Iverson BL, Georgiou G. Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS) Nat Biotechnol. 2001;19:537–542. doi: 10.1038/89281. [DOI] [PubMed] [Google Scholar]
- 15.Hudson PJ, Souriau C. Recombinant antibodies for cancer diagnosis and therapy. Expert. Opin. Biol. Ther. 2001;1:845–855. doi: 10.1517/14712598.1.5.845. [DOI] [PubMed] [Google Scholar]
- 16.Begent RH, Verhaar MJ, Chester KA, Casey JL, Green AJ, Napier MP, Hope-Stone LD, Cushen N, Keep PA, Johnson CJ, Hawkins RE, Hilson AJ, Robson L. Clinical evidence of efficient tumor targeting based on single-chain Fv antibody selected from a combinatorial library. Nat. Med. 1996;2:979–984. doi: 10.1038/nm0996-979. [DOI] [PubMed] [Google Scholar]
- 17.Yazaki PJ, Kassa T, Cheung CW, Crow DM, Sherman MA, Bading JR, Anderson AL, Colcher D, Raubitschek A. Biodistribution and tumor imaging of an anti-CEA single-chain antibody-albumin fusion protein. Nucl. Med. Biol. 2008;35:151–158. doi: 10.1016/j.nucmedbio.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olafsen T, Kenanova VE, Sundaresan G, Anderson AL, Crow D, Yazaki PJ, Li L, Press MF, Gambhir SS, Williams LE, Wong JY, Raubitschek AA, Shively JE, Wu AM. Optimizing radiolabeled engineered anti-p185HER2 antibody fragments for in vivo imaging. Cancer Res. 2005;65:5907–5916. doi: 10.1158/0008-5472.CAN-04-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R. Protein engineering of antibody binding sites: recovery of specific activity in an anti digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U S A. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 21.Adams GP, Schier R. Generating improved single-chain Fv molecules for tumor targeting. J. Immunol. Methods. 1999;231:249–260. doi: 10.1016/s0022-1759(99)00161-1. [DOI] [PubMed] [Google Scholar]
- 22.Wirtz P, Steipe B. Intrabody construction and expression III: engineering hyperstable V(H) domains. Protein Sci. 1999;8:2245–2250. doi: 10.1110/ps.8.11.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramm K, Gehrig P, Pluckthun A. Removal of the conserved disulfide bridges from the scFv fragment of an antibody: effects on folding kinetics and aggregation. J Mol Biol. 1999;290:535–546. doi: 10.1006/jmbi.1999.2854. [DOI] [PubMed] [Google Scholar]
- 24.Worn A, Pluckthun A. Stability engineering of antibody single-chain Fv fragments. J Mol Biol. 2001;305:989–1010. doi: 10.1006/jmbi.2000.4265. [DOI] [PubMed] [Google Scholar]
- 25.Zhou C, Emadi S, Sierks MR, Messer A. A human single-chain Fv intrabody blocks aberrant cellular effects of overexpressed alpha-synuclein. Mol. Ther. 2004;10:1023–1031. doi: 10.1016/j.ymthe.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Lynch SM, Zhou C, Messer A. An scFv intrabody against the nonamyloid component of alpha-synuclein reduces intracellular aggregation and toxicity. J Mol Biol. 2008;377:136–147. doi: 10.1016/j.jmb.2007.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visintin M, Tse E, Axelson H, Rabbitts TH, Cattaneo A. Selection of antibodies for intracellular function using a two-hybrid in vivo system. Proc Natl Acad Sci U S A. 1999;96:11723–11728. doi: 10.1073/pnas.96.21.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Portner-Taliana A, Russell M, Froning KJ, Budworth PR, Comiskey JD, Hoeffler JP. In vivo selection of single-chain antibodies using a yeast two-hybrid system. J Immunol Methods. 2000;238:161–172. doi: 10.1016/s0022-1759(00)00145-9. [DOI] [PubMed] [Google Scholar]
- 29.De JG, Fiers E, Eeckhout D, Depicker A. Analysis of the interaction between single-chain variable fragments and their antigen in a reducing intracellular environment using the two-hybrid system. FEBS Lett. 2000;467:316–320. doi: 10.1016/s0014-5793(00)01182-0. [DOI] [PubMed] [Google Scholar]
- 30.Worn A, uf der MA, Escher D, Honegger A, Barberis A, Pluckthun A. Correlation between in vitro stability and in vivo performance of anti-GCN4 intrabodies as cytoplasmic inhibitors. J Biol Chem. 2000;275:2795–2803. doi: 10.1074/jbc.275.4.2795. [DOI] [PubMed] [Google Scholar]
- 31.Carlson JR. A new means of inducibly inactivating a cellular protein. Mol Cell Biol. 1988;8:2638–2646. doi: 10.1128/mcb.8.6.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai J, Sui J, Zhu RY, Tallarico AS, Gennari F, Zhang D, Marasco WA. Inhibition of Tat-mediated transactivation and HIV-1 replication by human anti-hCyclinT1 intrabodies. J Biol Chem. 2003;278:1433–1442. doi: 10.1074/jbc.M208297200. [DOI] [PubMed] [Google Scholar]
- 33.Cattaneo A, Biocca S. The selection of intracellular antibodies. Trends Biotechnol. 1999;17:115–121. doi: 10.1016/s0167-7799(98)01268-2. [DOI] [PubMed] [Google Scholar]
- 34.Biocca S, Cattaneo A. Intracellular immunization: antibody targeting to subcellular compartments. Trends Cell Biol. 1995;5:248–252. doi: 10.1016/s0962-8924(00)89019-4. [DOI] [PubMed] [Google Scholar]
- 35.Marasco WA, Haseltine WA, Chen SY. Design, intracellular expression, and activity of a human anti-human immunodeficiency virus type 1 gp120 single-chain antibody. Proc Natl Acad Sci U S A. 1993;90:7889–7893. doi: 10.1073/pnas.90.16.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caron de FC, Gruel N, Venot C, Debussche L, Conseiller E, Dureuil C, Teillaud JL, Tocque B, Bracco L. Restoration of transcriptional activity of p53 mutants in human tumour cells by intracellular expression of anti-p53 single chain Fv fragments. Oncogene. 1999;18:551–557. doi: 10.1038/sj.onc.1202338. [DOI] [PubMed] [Google Scholar]
- 37.Cochet O, Kenigsberg M, Delumeau I, Virone-Oddos A, Multon MC, Fridman WH, Schweighoffer F, Teillaud JL, Tocque B. Intracellular expression of an antibody fragment-neutralizing p21 ras promotes tumor regression. Cancer Res. 1998;58:1170–1176. [PubMed] [Google Scholar]
- 38.Popkov M, Jendreyko N, McGavern DB, Rader C, Barbas CF., III Targeting tumor angiogenesis with adenovirus-delivered anti-Tie-2 intrabody. Cancer Res. 2005;65:972–981. [PubMed] [Google Scholar]
- 39.Wang CE, Zhou H, McGuire JR, Cerullo V, Lee B, Li SH, Li XJ. Suppression of neuropil aggregates and neurological symptoms by an intracellular antibody implicates the cytoplasmic toxicity of mutant huntingtin. J. Cell Biol. 2008;181:803–816. doi: 10.1083/jcb.200710158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deshane J, Siegal GP, Alvarez RD, Wang MH, Feng M, Cabrera G, Liu T, Kay M, Curiel DT. Targeted tumor killing via an intracellular antibody against erbB-2. J. Clin. Invest. 1995;96:2980–2989. doi: 10.1172/JCI118370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez RD, Barnes MN, Gomez-Navarro J, Wang M, Strong TV, Arafat W, Arani RB, Johnson MR, Roberts BL, Siegal GP, Curiel DT. A cancer gene therapy approach utilizing an anti-erbB-2 single-chain antibody-encoding adenovirus (AD21): a phase I trial. Clin. Cancer Res. 2000;6:3081–3087. [PubMed] [Google Scholar]
- 42.Lecerf JM, Shirley TL, Zhu Q, Kazantsev A, Amersdorfer P, Housman DE, Messer A, Huston JS. Human single-chain Fv intrabodies counteract in situ huntingtin aggregation in cellular models of Huntington’s disease. Proc Natl Acad Sci U S A. 2001;98:4764–4769. doi: 10.1073/pnas.071058398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colby DW, Chu Y, Cassady JP, Duennwald M, Zazulak H, Webster JM, Messer A, Lindquist S, Ingram VM, Wittrup KD. Potent inhibition of huntingtin aggregation and cytotoxicity by a disulfide bond-free single-domain intracellular antibody. Proc Natl Acad Sci U S A. 2004;101:17616–17621. doi: 10.1073/pnas.0408134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfgang WJ, Miller TW, Webster JM, Huston JS, Thompson LM, Marsh JL, Messer A. Suppression of Huntington’s disease pathology in Drosophila by human single-chain Fv antibodies. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11563–11568. doi: 10.1073/pnas.0505321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paganetti P, Calanca V, Galli C, Stefani M, Molinari M. beta-site specific intrabodies to decrease and prevent generation of Alzheimer’s Abeta peptide. J Cell Biol. 2005;168:863–868. doi: 10.1083/jcb.200410047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr. NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 47.Eliezer D, Kutluay E, Bussell R, Jr., Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 48.El-Agnaf OM, Jakes R, Curran MD, Middleton D, Ingenito R, Bianchi E, Pessi A, Neill D, Wallace A. Aggregates from mutant and wild-type alpha-synuclein proteins and NAC peptide induce apoptotic cell death in human neuroblastoma cells by formation of beta-sheet and amyloid-like filaments. FEBS Lett. 1998;440:71–75. doi: 10.1016/s0014-5793(98)01418-5. [DOI] [PubMed] [Google Scholar]
- 49.Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early onset Parkinson disease. Nat. Med. 1998;4:1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 50.Giasson BI, Uryu K, Trojanowski JQ, Lee VM. Mutant and wild type human alpha-synucleins assemble into elongated filaments with distinct morphologies in vitro. J. Biol. Chem. 1999;274:7619–7622. doi: 10.1074/jbc.274.12.7619. [DOI] [PubMed] [Google Scholar]
- 51.Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. α-synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc. Natl. Acad. Sci. USA. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 53.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emadi S, Liu R, Yuan B, Schulz P, McAllister C, Lyubchenko Y, Messer A, Sierks MR. Inhibiting aggregation of alpha-synuclein with human single chain antibody fragments. Biochemistry. 2004;43:2871–2878. doi: 10.1021/bi036281f. [DOI] [PubMed] [Google Scholar]
- 55.Barkhordarian H, Emadi S, Schulz P, Sierks MR. Isolating recombinant antibodies against specific protein morphologies using atomic force microscopy and phage display technologies. Protein Eng Des Sel. 2006;19:497–502. doi: 10.1093/protein/gzl036. [DOI] [PubMed] [Google Scholar]
- 56.Emadi S, Barkhordarian H, Wang MS, Schulz P, Sierks MR. Isolation of a human single chain antibody fragment against oligomeric alpha-synuclein that inhibits aggregation and prevents alpha-synuclein-induced toxicity. J Mol Biol. 2007;368:1132–1144. doi: 10.1016/j.jmb.2007.02.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maguire-Zeiss KA, Wang CI, Yehling E, Sullivan MA, Short DW, Su X, Gouzer G, Henricksen LA, Wuertzer CA, Federoff HJ. Identification of human alpha-synuclein specific single chain antibodies. Biochem Biophys Res Commun. 2006;349:1198–1205. doi: 10.1016/j.bbrc.2006.08.127. [DOI] [PubMed] [Google Scholar]
- 58.Giasson BI, Murray IV, Trojanowski JQ, Lee VM. A hydrophobic stretch of 12 amino acid residues in the middle of alpha-synuclein is essential for filament assembly. J Biol Chem. 2001;276:2380–2386. doi: 10.1074/jbc.M008919200. [DOI] [PubMed] [Google Scholar]
- 59.Bodles AM, Guthrie DJ, Greer B, Irvine GB. Identification of the region of non-Abeta component (NAC) of Alzheimer’s disease amyloid responsible for its aggregation and toxicity. J Neurochem. 2001;78:384–395. doi: 10.1046/j.1471-4159.2001.00408.x. [DOI] [PubMed] [Google Scholar]
- 60.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J. Biol. Chem. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 61.Kim TD, Paik SR, Yang CH. Structural and functional implications of C-terminal regions of alpha synuclein. Biochemistry. 2002;41:13782–13790. doi: 10.1021/bi026284c. [DOI] [PubMed] [Google Scholar]
- 62.Braden BC, Dall’Acqua W, Eisenstein E, Fields BA, Goldbaum FA, Malchiodi EL, Mariuzza RA, Schwarz FP, Ysern X, Poljak RJ. Protein motion and lock and key complementarity in antigen-antibody reactions. Pharm Acta Helv. 1995;69:225–230. doi: 10.1016/0031-6865(94)00046-x. [DOI] [PubMed] [Google Scholar]
- 63.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka Y, Engelender S, Igarashi S, Rao RK, Wanner T, Tanzi RE, Sawa A, Dawson L, Dawson TM, Ross CA. Inducible expression of mutant alpha-synuclein decreases proteasome activity and increases sensitivity to mitochondria-dependent apoptosis. Hum. Mol. Genet. 2001;10:919–926. doi: 10.1093/hmg/10.9.919. [DOI] [PubMed] [Google Scholar]
- 65.Zhou W, Hurlbert MS, Schaack J, Prasad KN, Freed CR. Overexpression of human alpha-synuclein causes dopamine neuron death in rat primary culture and immortalized mesencephalon-derived cells. Brain Res. 2000;866:33–43. doi: 10.1016/s0006-8993(00)02215-0. [DOI] [PubMed] [Google Scholar]
- 66.Giasson B, Duda JE, Quinn SM, Zhang B, Trojanowski JQ, Lee VM. Neuronal alpha-synucleinopathy with severe movement disorder in mice expressing A53T human alpha-synuclein. Neuron. 2002;34:521–533. doi: 10.1016/s0896-6273(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 67.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 68.Lakso M, Vartiainen S, Moilanen AM, Sirvio J, Thomas JH, Nass R, Blakely RD, Wong G. Dopaminergic neuronal loss and motor deficits in Caenorhabditis elegans overexpressing human alpha-synuclein. J. Neurochem. 2003;86:165–172. doi: 10.1046/j.1471-4159.2003.01809.x. [DOI] [PubMed] [Google Scholar]
- 69.Pavoni E, Flego M, Dupuis ML, Barca S, Petronzelli F, Anastasi AM, D’Alessio V, Pelliccia A, Vaccaro P, Monteriu G, Ascione A, De Santis R, Felici F, Cianfriglia M, Minenkova O. Selection, affinity maturation, and characterization of a human scFv antibody against CEA protein. BMC Cancer. 2006;6:41. doi: 10.1186/1471-2407-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weaver-Feldhaus JM, Miller KD, Feldhaus MJ, Siegel RW. Directed evolution for the development of conformation-specific affinity reagents using yeast display. Protein Eng Des Sel. 2005;18:527–536. doi: 10.1093/protein/gzi060. [DOI] [PubMed] [Google Scholar]
- 71.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson’s disease associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 73.West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM. Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 74.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 75.Greggio E, Zambrano I, Kaganovich A, Beilina A, Taymans JM, Daniels V, Lewis P, Jain S, Ding J, Syed A, Thomas KJ, Baekelandt V, Cookson MR. The Parkinson Disease-associated Leucine-rich Repeat Kinase 2 (LRRK2) Is a Dimer That Undergoes Intramolecular Autophosphorylation. J Biol Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, Dawson VL, Dawson TM, Ross CA. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc. Natl Acad. Sci. U. S. A. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 78.Shin N, Jeong H, Kwon J, Heo HY, Kwon JJ, Yun HJ, Kim CH, Han BS, Tong Y, Shen J, Hatano T, Hattori N, Kim KS, Chang S, Seol W. LRRK2 regulates synaptic vesicle endocytosis. Exp. Cell Res. 2008;314:2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 79.Biskup S, Moore DJ, Celsi F, Higashi S, West AB, Andrabi SA, Kurkinen K, Yu SW, Savitt JM, Waldvogel HJ, Faull RL, Emson PC, Torp R, Ottersen OP, Dawson TM, Dawson VL. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 80.Hatano T, Kubo S, Imai S, Maeda M, Ishikawa K, Mizuno Y, Hattori N. Leucine-rich repeat kinase 2 associates with lipid rafts. Hum Mol Genet. 2007;16:678–690. doi: 10.1093/hmg/ddm013. [DOI] [PubMed] [Google Scholar]
- 81.Gandhi PN, Wang X, Zhu X, Chen SG, Wilson-Delfosse AL. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J Neurosci Res. 2008;86:1711–1720. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]