Abstract
Psoriasis is a common immune mediated disorder that affects the skin, nails, and joints. To identify psoriasis susceptibility loci, we genotyped 438,670 SNPs in 1,409 European ancestry psoriasis cases and 1,436 controls. Twenty-one promising SNPs were followed-up in 5,048 psoriasis cases and 5,041 controls. Our results provide strong support for the association of at least seven genetic loci and psoriasis (each with p < 5×10−8 overall). Loci with confirmed association encode HLA-C, three genes involved in IL-23 signaling (IL23A, IL23R, IL12B), two genes that act downstream of TNF-α and regulate NF-κB signaling (TNIP1, TNFAIP3), and two genes involved in the modulation of Th2 immune responses (IL4, IL13). Although the proteins encoded in these loci are known to interact biologically, we found no evidence for epistasis between associated SNPs. Our results expand the catalog of genetic loci implicated in psoriasis susceptibility and suggest priority targets for study in other auto-immune disorders.
Psoriasis is a common inflammatory disease affecting ~1% of individuals. The most obvious cellular features of psoriasis are epidermal hyperplasia, altered keratinocyte differentiation, and inflammation1. Psoriasis susceptibility has a genetic component, partly explained by association between psoriasis and major histocompatibility complex (MHC) haplotypes bearing HLA-Cw62 and SNPs near IL12B and IL23R3. Patients with psoriasis show increased risk for other immune mediated disorders4 and some IL12B and IL23R polymorphisms are associated with Crohn’s disease and ulcerative colitis in addition to psoriasis (e.g.5).
To identify additional psoriasis susceptibility loci, we carried out a genome-wide association scan of 1,409 psoriasis cases and 1,436 controls in partnership with the Genetic Association Information Network (GAIN)6 (see Table 1 and Supplementary Table 1 for details of case and control collections). After samples were genotyped at Perlegen Sciences, we used a dataset that passed quality control filters and included 438,670 autosomal SNPs genotyped in 1,359 psoriasis cases and 1,400 controls to impute genotypes for 2.5M HapMap SNPs (see methods).
Table 1. Summary description of the samples used in this study.
All cases and controls were of white European ancestry.
| Cases |
Controls |
|||||||
|---|---|---|---|---|---|---|---|---|
| N | Age at Onset (Mean) | %Male | %Psoriatic Arthritis | N | Age at Exam (Mean) | %Male | Total | |
| Discovery Samples | ||||||||
| Collection of J.T. Elder | 480 | 23.0 | 52.1 | 25.2 | 702 | 40.5 | 49.3 | 1182 |
| Collection of G. Krueger | 476 | 28.4 | 42.9 | 30.0 | 473 | 29.7 | 42.7 | 949 |
| Collection of A. Bowcock | 453 | 27.2 | 49.9 | 26.5 | 261 | 57.4 | 36.0 | 714 |
| Discovery Sample Total | 1409 | 26.1 | 48.3 | 27.1 | 1436 | 40.0 | 44.7 | 2845 |
| Follow-up Samples | ||||||||
| Collection of J.T. Elder | 1642 | 30.8 | 46.3 | 16.4 | 1101 | 48.0 | 41.0 | 2743 |
| Collection of M. Weichenthal | 718 | 25.1 | 52.1 | 16.7 | 1464 | 40.4 | 51.0 | 2182 |
| Celera Follow-up Set 1, A. Begovich | 498 | 29.4 | 44.6 | 40.7a | 498 | 47.4 | 44.6 | 996 |
| Celera Follow-up Set 2, A. Begovich | 483 | 26.8b | 53.4 | 29.3a | 427 | 44.3b | 52.2 | 910 |
| Collection of D. Gladman | 691 | 29.4 | 59.9 | 71.6 | 217 | 41.8 | 47.7 | 908 |
| Collection of J. Fischer | 346 | 19.0 | 45.2 | 13.9 | 486 | n/ac | 47.2 | 832 |
| Collection of A. Bowcock | 302 | 28.0 | 49.0 | 34.1 | 500 | 59.0 | 48.0 | 802 |
| Collection of P. Rahman | 368 | 28.3 | 47.8 | 81.5 | 358 | 54.9 | 43.0 | 726 |
| Follow-up Sample Total | 5048 | 5051 | 10099 | |||||
In the Celera case samples, patients were only classified as psoriatic arthritis positive or negative 10 years after disease onset. In follow-up set 1, 98 of 241 patients followed-up for >10 years had psoriatic arthritis. In follow-up set 2, 63 of 215 patients met this criterion.
Information on age at disease onset and age at exam was available for 293 patients and 292 controls, respectively.
Age information for controls in this sample set was not tracked electronically in the sample database and is not readily accessible.
An initial comparison of case-control allele frequencies (genomic control λ=1.033) demonstrated association at established susceptibility loci HLA-C (rs12191877, p = 4×10−53), IL12B (rs2082412, p = 5×10−10), and IL23R (rs2201841, p = 3×10−7). Encouraged by these results, we selected 21 SNPs (representing 18 independent loci, see Methods) for genotyping in an additional 5,048 cases and 5,051 controls (see Table 1 and Supplementary Table 2). We found supporting evidence of association at 10 of these 18 loci (p < 0.05 in the follow-up sample, direction of effect matches discovery sample; Table 2). Evidence for association was particularly compelling at seven of these loci (p < 0.0005 in follow-up samples, combined p-value < 5×10−8). Due to the “winner’s curse”, odds ratios estimated in the discovery sample were larger than those estimated in the follow-up samples. To minimize this effect, we use follow-up sample odds ratios in the discussion that follows. Figure 1 summarizes the results of the association scan, with the seven regions of confirmed association detailed in Figure 2. Overall, our approach provides ~70% power to detect loci that are well tagged by genotyped SNPs, increase disease risk by >1.35-fold and have a frequency >20%.
Table 2. Loci with strongest evidence of association with psoriasis in the combined sample, including discovery and follow-up samples.
| Discovery Samples | Follow-up Samples | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alleles | (1359 cases, 1400 controls) |
(5048 cases, 5051 controls) |
Notable Nearby | ||||||||||
| SNP | Chr | Pos (Mb) |
risk/ non-risk |
Frequencya | OR | p-valued | Frequency | OR (meta) |
p-valued (meta) |
Combined p-valued |
Genes (relative position)c |
||
| Case | Control | Case | Control | ||||||||||
| rs12191877 | 6 | 31.36 | T/C | .313 | .141 | 2.79 | 4×10−53 | .301 | .147 | 2.64 | <10−100 | <10−100 | HLA-C(−13kb) |
| rs2082412 | 5 | 158.65 | G/A | .856 | .792 | 1.56 | 5×10−10 | .848 | .798 | 1.44 | 3×10−20 | 2×10−28 | IL12B(+24kb) |
| rs17728338 | 5 | 150.46 | A/G | .093 | .056 | 1.72 | 2×10−7 | .087 | .054 | 1.59 | 6×10−15 | 1×10−20 | TNIP1(−12kb) |
| rs20541 | 5 | 132.02 | G/A | .832 | .783 | 1.37 | 6×10−6 | .827 | .790 | 1.27 | 1×10−10 | 5×10−15 | IL13(non-synon) |
| rs610604 | 6 | 138.24 | G/T | .374 | .318 | 1.28 | 1×10−5 | .360 | .320 | 1.19 | 7×10−8 | 9×10−12 | TNFAIP3(intronic) |
| rs2066808b | 12 | 55.02 | A/G | .958 | .931 | 1.68 | 2×10−5 | .947 | .932 | 1.34 | 5×10−6 | 1×10−9 |
IL23A(+3.7kb) STAT2 (intronic) |
| rs2201841 | 1 | 67.47 | G/A | .350 | .286 | 1.35 | 3×10−7 | .325 | .295 | 1.13 | 4×10−4 | 3×10−8 | IL23R(intronic) |
| rs1076160 | 9 | 134.80 | T/C | .520 | .463 | 1.26 | 2×10−5 | .496 | .475 | 1.09 | 4×10−3 | 6×10−6 | TSC1 (intronic) |
| rs12983316 | 19 | 10.98 | G/A | .186 | .144 | 1.36 | 2×10−5 | .159 | .147 | 1.09 | 0.027 | 8×10−5 | SMARCA4 (intronic) |
| rs397211 | 2 | 113.6 | T/C | .718 | .677 | 1.21 | 1×10−3 | .709 | .696 | 1.08 | 0.025 | 4×10−4 | IL1RN (+0.5kb) |
Frequency of the risk allele.
Genotypes for rs2066808 were imputed using MaCH. The distribution of imputed posterior probabilities for each genotype was then compared between cases and controls. Similar evidence for association was observed at rs2066807 (combined p = 2×10−9), which maps nearby and was genotyped in discovery and follow-up samples.
Position of each SNP relative to notable nearby genes is given. +/− indicates whether the SNP is upstream (−) or downstream (+) of the transcription start site. SNPs that overlap the gene are labeled as “intronic”, “synonymous” or “non-synonymous”.
All p-values are two tailed.
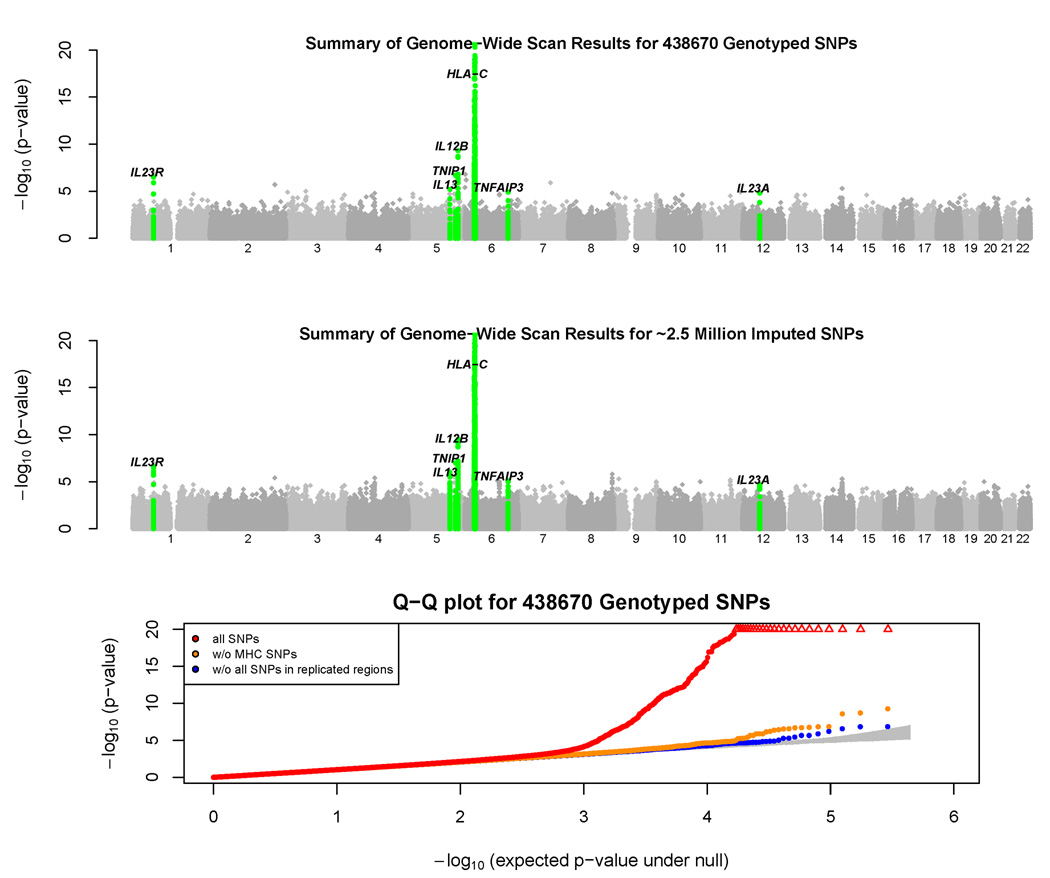
Figure 1. Bird’s Eye View of Association Scan Results.
The top panel summarizes the distribution of test statistics at genotyped SNPs across the genome. We used a simple chi-squared test to compare SNP allele frequencies in cases and controls and plotted the resulting -log p-values across the genome. Several p-values < 10−20 in the MHC region were truncated. Loci where we obtained confirmatory evidence of association in follow-up samples (see Table 2) are highlighted in green.
The middle panel summarizes the distribution of test statistics across the genome, after genotype imputation. We used a simple t-test to compare imputed allele counts in cases and controls and plotted the resulting –log p-values across the genome.
The bottom panel displays a Q-Q plot for our test statistics. Results are plotted including all SNPs (in red), after excluding SNPs in the MHC (in orange) and after excluding all SNPs in regions of replicated association (in blue). The shaded region represents a 90% confidence interval for the test statistics.
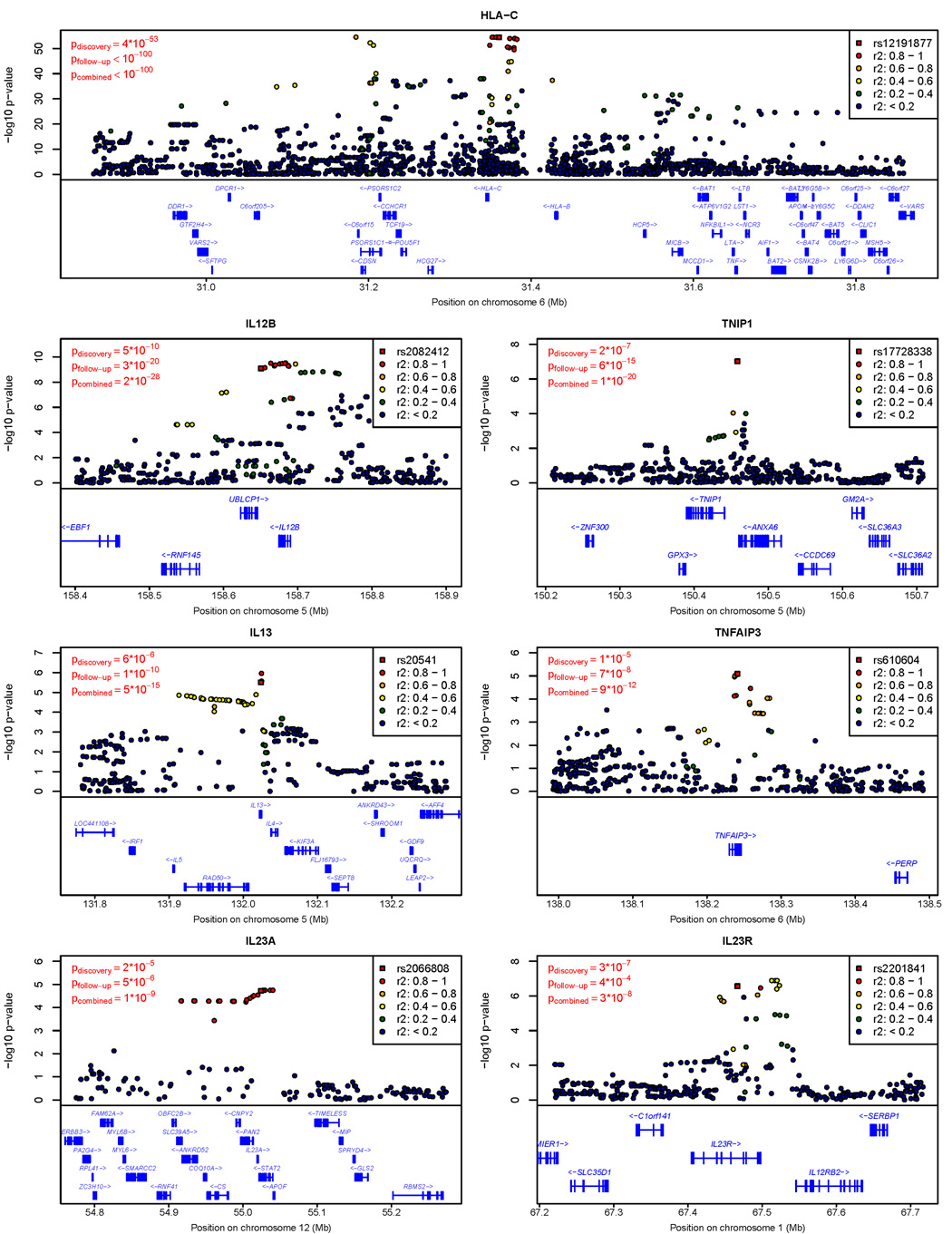
Figure 2. Evidence for Association in Confirmed Loci.
The figure summarizes evidence of association (in the discovery sample) in each region of confirmed association. Test statistics at the SNP selected for follow-up (typically, the genotyped SNP exhibiting strongest evidence for association in each locus) are highlighted with a square. Test statistics for other SNPs are drawn as circles and color coded according to the degree of linkage disequilibrium with the SNP selected for follow-up.
The results highlight the role of several key pathways in disease susceptibility. First, three SNPs exhibiting strong evidence of association map near IL12B (encoding the p40 subunit of IL-23 and IL-12), IL23A (encoding the p19 subunit of IL-23), and IL23R (encoding a subunit of the IL-23 receptor). The SNPs are rs2082412 (risk allele frequency in controls fcontrol = 0.80, odds ratio in follow-up samples ORfollow-up = 1.44, combined p-value pcombined = 2×10−28), rs2066808 (fcontrol = 0.93, ORfollow-up = 1.34, pcombined = 1×10−9), and rs2201841 (fcontrol = 0.29, ORfollow-up = 1.13, pcombined = 3×10−8), respectively. Genetic variants in the IL23A locus are implicated in psoriasis and auto-immune disease susceptibility for the first time by our study. IL-23 signaling promotes cellular immune responses by promoting the survival and expansion of a recently identified subset of T-cells expressing IL-17 that protects epithelia against microbial pathogens7. Dysregulated IL-23 signaling could predispose certain individuals to inappropriate, chronic immune responses that target epithelial cells and ultimately result in psoriasis.
Second, loci including TNFAIP3 (TNF-α induced protein 3) and TNIP1 (TNFAIP3 interacting protein 1), whose gene products work downstream of TNF-α to regulate NF-κB, show strong association with psoriasis. In these two regions, markers rs610604 (fcontrol = 0.32, ORfollow-up = 1.19, pcombined = 9×10−12) and rs17728338 (fcontrol = 0.05, ORfollow-up = 1.59, pcombined = 1×10−20) were sites of replicated association. TNFAIP3 encodes A20, a TNF-α-inducible zinc finger protein that temporally limits immune responses by inhibiting NF-κB activation and terminating NF-κB mediated responses8. Symptoms in a mouse model of psoriasis induced by administration of IL-23 are ameliorated by blocking of TNF-α9 and in a different mouse model, a region of mouse chromosome 10 encompassing Tnfaip3 promotes psoriasis in a TNF-α dependent manner10. Interestingly, this same region of the mouse genome has been also associated with atherosclerosis11, a major co-morbidity of psoriasis12. Note that both anti-IL-12/IL-23 p40 and anti-TNF-α monoclonal antibodies provide highly efficacious therapeutic regimens for many psoriasis patients13,14, and that five of the genes implicated here play key roles in pathways targeted by therapeutic interventions. Interestingly, common polymorphisms near TNFAIP3 have recently been associated with rheumatoid arthritis (e.g. rs6920220, rs10499194) (e.g.15) and systemic lupus erythematosus (e.g. rs5029939, rs13192841, rs2230926 and rs6922466)16,17. However, these polymorphisms show no association with psoriasis in our sample (all p > 0.30) and are not in linkage disequilibrium (LD, all r2 < 0.03) with the psoriasis associated alleles (e.g. rs610604).
Third, genes in the two other loci implicated here are also key modulators of immune response. One locus encodes the IL-4 and IL-13 cytokines that modulate humoral immune responses mediated by Th2 cells. In this locus, we replicated association at rs20541 (fcontrol = 0.79, ORfollow-up = 1.27, pcombined = 5×10−15). Dysregulation of IL-4 and IL-13 might polarize the immune response toward Th1-mediated cellular immune responses and support the marked expansion of IL-17-producing T cells observed in psoriatic lesions18. Our findings extend the promising results of a recent study19 to a genome-wide level of significance. Interestingly, our IL4/IL13 signal maps within ~200kb of the IBD5 Crohn’s disease susceptibility locus5. The two are not in LD (e.g., r2 between rs20541 and rs100777855 in HapMap CEU is < .01) but IBD5 does exhibit modest evidence for association with psoriasis (rs10077785, p = .03) suggesting it could be another locus that contributes to both diseases.
SNP rs12191877, the genotyped marker exhibiting strongest association with psoriasis (fcontrol = 0.15, fcase = 0.30, ORfollow-up = 2.64, pcombined ≪ 10−100), was in LD with HLA-Cw6 (r2 = 0.63). In a subset of cases and controls in which HLA-Cw6 genotypes were available, HLA-Cw6 was more strongly associated with psoriasis than any genotyped or imputed SNP, but could not fully account for all observed association signals (data not shown). To assess the evidence for multiple psoriasis susceptibility alleles within the MHC, we implemented a forward selection procedure to select a set of disease-associated variants in each locus (see Methods). This analysis resulted in a model with three imputed SNPs (Supplementary Table 3). The first two of these (rs12204500 and rs13191343, forward selection p-values of 8×10−57 and 2×10−10, respectively) are close to and in strong LD with HLA-Cw6 (r2 = 0.78 and 0.52, respectively), whereas the third one (rs2022544, p-value = 10−7) maps closer to HLA-DR and exhibits only weak LD with HLA-Cw6 (r2 = 0.01). These results endorse a search for additional psoriasis susceptibility loci within the MHC.
When we applied the same forward selection strategy to the other loci, two independent SNPs (r2 < 0.01) were selected in the IL12B and IL23R regions. Although only one SNP was selected in the four other regions (Supplementary Table 3), it is likely that independent disease-associated alleles exist in additional loci such as TNIP1 where rs884520 (a SNP only ~6 kb away from the peak of association at rs17728338) was suggestively associated with psoriasis (p = 9 × ×10−5 unadjusted, p = 0.051 after conservatively adjusting for 565 independent tests) in our conditional analyses. Fully characterizing the impact of these loci on psoriasis susceptibility will require characterization of the full spectrum of allelic variation at each locus in large case control samples.
Since all the loci implicated here are involved in regulation of immune responses, and several of the proteins they encode interact physically (e.g. IL-12B/p40, IL-23A/p19, and IL-23R and also TNIP1 and TNFAIP3), we assessed our data for evidence of epistasis. We considered all 21 possible pairings of the seven lead SNPs, testing for deviation from a log-additive risk model. Only the pairing involving rs12191877 near HLA-C and rs610604 near TNFAIP3 showed any evidence for epistasis under this model (p = 0.02 in combined sample). It is possible that tests of interaction will be more powerful once the causal variants at each loci have been identified, but it is striking that even when proteins encoded by the associated loci interact physically no evidence for epistasis was detected (a similar situation occurs for height20, among other traits).
To evaluate evidence for heterogeneity in the effect sizes at each of the seven replicated loci, we calculated I2 and Q statistics for a meta-analysis of follow-up samples (Supplementary Table 4). We observed no evidence for heterogeneity at non-MHC loci, and only modest evidence for heterogeneity at rs12191877 in the MHC (p = 0.007, Supplementary Table 4) – potentially reflecting sample differences in the proportion of familial cases and psoriatic arthritis. At several of the confirmed loci, we found modest differences in association signal strength for psoriatic arthritis compared to purely cutaneous psoriasis (Supplementary Table 5), supporting epidemiologic evidence for differences in genetic architecture of the two conditions21. In other stratified analyses, we found no evidence for heterogeneity between males and females (all p > .15) or between younger and older individuals (all p > .15, cases and controls stratified around median ages).
Psoriatic and uninvolved skin show significantly different expression for hundreds of genes, involved in both immune response and in the regulation of cellular differentiation and proliferation22. We reasoned that genes in the loci implicated by our study might also play the role of molecular triggers in disease progression. Therefore, we examined expression levels for the genes showing replicated evidence of association in skin biopsies from 64 GWAS controls and in biopsies of involved and uninvolved skin from 58 GWAS cases. The results are summarized in Supplementary Table 6 and in Supplementary Figure 1. Together these show that four of the genes investigated (HLA-C, IL12B, TNIP1 and IL23A) show highly significant differences in expression levels between involved and uninvolved skin (all with p < 10−9). Two of these (IL23A, TNIP1) also show differences in expression levels when we compared normal skin from controls and uninvolved skin from cases (p < 0.0003). The results are consistent with the hypothesis that the expression of particular HLA-C alleles and of IL23A and IL12B (encoding the two subunits of IL-23) in psoriatic skin, are key events in disease progression. However, the dosage of risk alleles at the seven psoriasis associated SNPs did not correlate with transcript levels for nearby genes in either involved, uninvolved or normal skin. It remains possible that association between these SNPs and gene expression patterns is stronger at specific time points during development, disease progression or in specific cell types.
Although this study represents a significant advance in our understanding of the genetic underpinnings of psoriasis, much work remains to be done. The association signals identified here account for a sibling recurrence risk (λs) of <1.35 (including ~1.25 due to HLA); consequently, much of the overall sibling recurrence risk for psoriasis, which has been estimated at ˜3 to 6-fold23, remains unexplained. Still, the rapid pace of advance in psoriasis genetics is encouraging. In the past 18 months, the number of independent genetic loci confidently associated with psoriasis has increased from one (HLA-Cw6 and other MHC variants) to at least ten, including the seven association signals reported in this paper, copy number variants in the beta-defensin24 and late cornified envelope (LCE) gene regions25, and a signal near ZNF313, a potential regulator of T cell activation26. The ZNF313 signal is supported by our data (see Supplementary Table 7 for analysis of previously reported GWAS26,27 signals in our data). Although we did not systematically characterize copy number variation, we note that rs4112788, a SNP proxy for the LCE deletion25,27, is associated with disease in our discovery sample (p = 0.001). In each of the loci identified here, fine-mapping and resequencing efforts together with further functional studies are required to pinpoint and characterize causal variants, confirm the identity of the implicated genes, and accurately quantify the contribution of the locus to disease susceptibility. In parallel, follow-up analyses with larger numbers of SNPs, execution of genomewide association scans in larger sample sets, meta-analyses of genomewide scan results, and large scale analyses of rarer variants should lead to additional susceptibility loci.
METHODS
Informed Consent
All participating subjects gave informed consent and protocols were reviewed and approved by local institutional review boards.
Genotyping
Perlegen Sciences (Mountain View, CA, USA) genotyped discovery samples using four proprietary, high-density oligonucleotide arrays. SNPs on the arrays were selected to tag common variation in European ancestry samples. Cases and controls from the same collection were genotyped together, and arranged to ensure similar proportions of cases and controls in each plate. Follow-up samples were genotyped using either Applied Biosystems Taqman assays, Sequenom single base extension assays, or allele-specific kinetic PCR. The 21 SNPs selected for follow-up included nineteen SNPs selected to represent loci exhibiting strongest evidence for association in our initial scan (including 2 SNPs per locus for hits near IL13, IL23A and PRKRIP1) and two SNPs in loci that included strong functional candidates (IL1RN and CNTN5) but more modest evidence of association (rs397211, p = 1×10−3, and rs12807920, p = 1×10−4).
Sample Quality Control
Eighteen samples failed genotyping for technical reasons. Among the remaining samples, we excluded those with call rates <95% (8 samples) and with outlier heterozygosities of <31% or >34% (24 samples; the average heterozygosity for all samples was 32.6% with standard deviation of 0.4%). We also excluded one individual from each pair of unexpected duplicates, 1st or 2nd degree relatives (36 individuals). This resulted in a dataset with 1,359 cases and 1,400 controls.
Quality Control of Genotype Data
Perlegen Sciences called >50% of genotypes for 556,383 SNPs. Before analysis, we excluded markers with <95% genotype call rates (99,963 SNPs), with minor allele frequency <1% in the combined dataset (6,106 SNPs), with HWE p-value < 10−6 (2,962 SNPs), with >2 mismatches among 48 duplicate pairs (62 SNPs) or with >2 Mendelian inconsistencies among 27 trios (41 SNPs). In total, 447,249 SNPs passed the quality control filters (average call rate of 99.2%). Here, we present analyses of 438,670 autosomal SNPs.
Genotype Imputation
As previously described28, we used information on patterns of haplotype variation in the HapMap CEU samples (release 21)29 to infer missing genotypes “in silico”. We only analyzed SNPs that were genotyped or could be imputed with relatively high confidence (estimated r2 between imputed SNP and true genotypes > 0.3; so that patterns of haplotype sharing between sampled individuals and HapMap samples consistently indicated a specific allele).
Assessment of Genotyping and Imputation Quality
A single plate containing 90 study samples was re-genotyped for 906,600 SNPs using the Affymetrix 6.0 chip. Comparison of 15,844,334 genotypes for 218,039 SNPs overlapping between the Perlegen and Affymetrix platforms resulted in an observed discrepancy rate of 0.25% per genotype (0.12% per allele). Comparison of 57,747,244 imputed and experimentally derived genotypes for 661,881 non-Perlegen SNPs present in both our imputed SNP set and the Affymetrix 6.0 array resulted in a discrepancy rate of 1.80% per genotype (0.91% per allele). Overall, the average r2 between imputed genotypes and their experimental counterparts, which provides an estimate of the relative power of analysis relying on imputation instead of direct genotyping, was 0.93. This r2 statistic exceeded 0.80 for >90% of SNPs suggesting excellent coverage of common variation in the genome.
Association Analyses
To evaluate the evidence for association between each genotyped or imputed SNP and psoriasis, we first calculated a single chi-squared statistic that contrasted observed or imputed allele counts between cases and controls. The 832 follow-up samples collected by Judith Fischer (see Table 1) and her colleagues were analyzed using a family-based approach30. To combine statistics across different samples, we first selected an arbitrary reference allele for each marker and then calculated a z-statistic characterizing the evidence for association in each study (summarizing both the p-value, in its magnitude, and the direction of effect, in its sign). We then calculated an overall z-statistic as a weighted average of the individual statistics and calculated the corresponding p-value. Weights were proportional to the square root of the number of individuals examined in each sample and were selected such that the squared weights sum to 1.0.
Conditional Analyses
We first selected the SNP exhibiting strongest association in each region. Then, conditioning on this SNP, we searched for the next most strongly associated SNP. If evidence for association at this second SNP was stronger than expected by chance (after adjusting for the number of SNPs tested), we sought a third strongly associated SNP and so forth.
Gene Expression
Six millimeter punch skin biopsies were obtained at the University of Michigan Department of Dermatology. One biopsy of normal skin was obtained from the buttock of 64 control individuals. Two biopsies (one involved, one uninvolved) were obtained from 58 psoriatic subjects. Involved skin biopsies were taken from psoriasis plaques, and uninvolved skin biopsies were taken from the buttocks, at least 10 cM away from the nearest plaque. RNA from each biopsy was isolated using the RNeasy kit (Qiagen). Samples were run on Affymetrix U133 Plus 2.0 arrays to evaluate expression of ~54,000 probes according to the manufacturer’s protocol. The raw data from 180 microarrays was processed using the Robust Multichip Average (RMA) method. Prior to analysis, we adjusted RMA expression values to account for batch and sex effects. To obtain a single expression value for each gene, we calculated the average of expression values of multiple probe sets on the microarray that were mapped to the same gene. Gene expression was contrasted between different groups of samples using two sample T-tests (for comparisons involving skin from normal controls and individuals with psoriasis) or paired T-tests (for comparisons involving involved and uninvolved skin from affected individuals). Comparisons of normal skin from controls and psoriatic skin from cases gave similar results (but slightly more significant p-values) to paired comparisons of involved and uninvolved skin from the same affected individual and are not shown. Re-analysis of a previously published dataset22 including paired biopsies of involved and uninvolved skin from 16 individuals gave results consistent with those reported here, suggesting that IL23A, IL12B and TNIP1 are over-expressed in involved skin. This independent dataset did not suggest differential expression of HLA-C.
Supplementary Material
Acknowledgements
We gratefully acknowledge funding from the National Institutes of Health, the Foundation for NIH’s Genetic Association Information Network and the National Psoriasis Foundation. Analysis and genotyping of follow-up samples was also supported by the German National Genome Research Network (BMFT 01GS 0171/BMBF NUW-S23T10) and POPGEN Biobank (BMBF 01GR0468), by the Canadian Institute of Health Research and the Arthritis Society of Canada, by the Centre National de Génotypage, Généthon and the Association Française contre les Myopathies (AFM), and by Celera Corporation.
Footnotes
Online Resources
The genotype and phenotype data described in this manuscript has been deposited in dbGap, at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000019.v1.p1
The microarray data have also been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE13355.
Conflict of Interest Statement.
ABB and SJS own stock and/or stock options in Celera Corporation.
REFERENCES
- 1.Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 2.Nair RP, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78:827–851. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cargill M, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yates VM, Watkinson G, Kelman A. Further evidence for an association between psoriasis, Crohn's disease and ulcerative colitis. Br J Dermatol. 1982;106:323–330. doi: 10.1111/j.1365-2133.1982.tb01731.x. [DOI] [PubMed] [Google Scholar]
- 5.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manolio TA, et al. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39:1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 7.Bettelli E, Oukka M, Kuchroo VK. TH-17 cells in the circle of immunity and autoimmunity. Nat Immunol. 2007;8:345–350. doi: 10.1038/ni0407-345. [DOI] [PubMed] [Google Scholar]
- 8.Lee EG, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan JR, et al. IL-23 stimulates epidermal hyperplasia via TNF and IL-20R2-dependent mechanisms with implications for psoriasis pathogenesis. J Exp Med. 2006;203:2577–2587. doi: 10.1084/jem.20060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, et al. A 9-centimorgan interval of chromosome 10 controls the T cell-dependent psoriasiform skin disease and arthritis in a murine psoriasis model. J Immunol. 2008;180:5520–5529. doi: 10.4049/jimmunol.180.8.5520. [DOI] [PubMed] [Google Scholar]
- 11.Idel S, Dansky HM, Breslow JL. A20, a regulator of NFkappaB, maps to an atherosclerosis locus and differs between parental sensitive C57BL/6J and resistant FVB/N strains. Proc Natl Acad Sci U S A. 2003;100:14235–14240. doi: 10.1073/pnas.1835672100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelfand JM, et al. Risk of myocardial infarction in patients with psoriasis. Jama. 2006;296:1735–1741. doi: 10.1001/jama.296.14.1735. [DOI] [PubMed] [Google Scholar]
- 13.Krueger GG, et al. A human interleukin-12/23 monoclonal antibody for the treatment of psoriasis. N Engl J Med. 2007;356:580–592. doi: 10.1056/NEJMoa062382. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhari U, et al. Efficacy and safety of infliximab monotherapy for plaque-type psoriasis: a randomised trial. Lancet. 2001;357:1842–1847. doi: 10.1016/s0140-6736(00)04954-0. [DOI] [PubMed] [Google Scholar]
- 15.Plenge RM, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham RR, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008 doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musone SL, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008 doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kryczek I, et al. Induction of memory IL-17+ T cell trafficking and expansion by IFN-gamma: Mechanism and pathological relevance. J Immunol. 2008;181:4733–4741. doi: 10.4049/jimmunol.181.7.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang M, et al. Variants in the 5q31 cytokine gene cluster are associated with psoriasis. Genes Immun. 2008;9:176–181. doi: 10.1038/sj.gene.6364451. [DOI] [PubMed] [Google Scholar]
- 20.Lettre G, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandran V, et al. Familial aggregation of psoriatic arthritis. Ann Rheum Dis. 2008 doi: 10.1136/ard.2008.089367. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, et al. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics. 2003;13:69–78. doi: 10.1152/physiolgenomics.00157.2002. [DOI] [PubMed] [Google Scholar]
- 23.Elder JT, et al. The genetics of psoriasis. Arch Dermatol. 1994;130:216–224. [PubMed] [Google Scholar]
- 24.Hollox EJ, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Cid R, et al. Deletion of the late cornified envelope (LCE) 3C and 3B genes as a susceptibility factor for psoriasis. Nat Genet. 2008;submitted doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capon F, et al. Identification of ZNF313/RNF114 as a novel psoriasis susceptibility gene. Hum Mol Genet. 2008;17:1938–1945. doi: 10.1093/hmg/ddn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease Loci. PLoS Genet. 2008;4:e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thornton T, McPeek MS. Case-control association testing with related individuals: a more powerful quasi-likelihood score test. Am J Hum Genet. 2007;81:321–337. doi: 10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.