Abstract
Ectopic expression of pluripotency gene sets provokes nuclear reprogramming in permissive somatic tissue environments, generating nduced pluripotent stem (iPS) cells. The evolutionary conserved function of sternness orthologs was tested here through interspecies transduction. A spectrum of human immunodeficiency virus (HIV)‐based lentiviral vectors was designed, and point mutations in the HIV‐1 capsid region were identified for efficient infectivity and expanded transspecies tropism. Human pluripotent gene sequences, OCT3/4, SOX2, KLF4, and c‐MYC, packaged into engineered lentiviral expression vectors achieved consistent expression in nonhuman fibroblasts. Despite variation in primary amino acid sequence between species, introduction of human pluripotent genes produced cell lines with embryonic stem cell‐like morphology. Transduced fibroblasts differentiated in vitro into all three germ layers according to gastrulation gene expression profiles, and formed in vivo teratoma with multilineage potential. Reprogrammed progeny incorporated into nonhuman morula to produce blastomeres capable of developing into chimeric embryos with competent organogenesis. This model system establishes a prototypic approach to examine consequences of human sternness factors‐induced reprogramming in the context of normal embryonic development by exploiting nonhuman early‐stage embryos. Thus, ectopic xenotransduction across species unmasks the promiscuous nature of sternness induction, suggesting evolutionary selection of core processes for somatic tissue reprogramming.
Keywords: chimera, iPS, HIV, lentiviral, ortholog, OCT3/4, SOX2, KLF4, c‐MYC
Introduction
Nuclear reprogramming of somatic cells into pluripotent progenitors was originally demonstrated through transfer of a nucleus into the cytoplasm of a competent host oocyte. 1 , 2 The resulting intraspecies nuclear‐to‐cytosol interaction catalyzed the epigenetic influence on phenotypic outcome, reversing differentiation to achieve genetic reprogramming of an adult source into an embryonic state. 3 , 4 , 5 The robustness of this process was further supported by somatic cell nuclear transfer applications in which xenogenic combinations of nucleus and cytoplasmic components were equally successful and revealed phylogenic conservation of the permissive oocyte environment. 6 A functionally conserved ecogenetic interface for dedifferentiation, although implied by the preserved responsiveness of somatic nucleus to multiple environments, has yet to be proven.
Beyond whole‐nucleus‐based transplantation, equivalent reprogramming of differentiated cytotypes has been achieved most recently by ectopic exposure to pluripotent components. 7 , 8 Delivery of sternness‐related genes Oct‐3/4, Sox2, Klf4, and c‐Myc reprogram adult somatic cells, promoting reacquisition of embryonic traits. 9 , 10 The resulting induced pluripotent stem (iPS) cells recapitulate the morphology and expression patterns of authentic embryonic stem cells. 11 _ 13 The alternative combination of Oct‐3/4, Sox2, Nanog, and Tin28 has also produced pluripotency induction from somatic tissues, suggesting mechanisms that can be reactivated by nonexclusive inductors. 14 iPS cells have been derived through intraspecies reprogramming with gene sets from murine, monkey, and human sequences, 15 , 16 , 17 , 18 , 19 , 20 however without evidence for interspecies pluripotent reprogramming.
Although orthologs are frequently assumed to be functionally conserved, 21 the differences in stem cell‐related genes between reproductively isolated species mandate an empiric examination of cross‐species gene–environment communication. Comparative studies of mammalian embryonic stem cells have uncovered both conserved and divergent factors required to maintain pluripotency. 22 Pathways requiring Oct3/4, Sox, Nanog as well as Stat3 signaling are essential for self‐renewal of both mouse and human embryonic stem cells and demonstrate highly conserved genetic structures across mammalian species. 23 In contrast, leukemia inhibitory factor (TIF) and basic fibroblast growth factor (FGF2) are exclusively required for self‐renewal in mouse or human embryonic stem cells, respectively. 24 , 25 Thus, the question arises whether sternness‐associated factors used to derive iPS cells are functionally conserved when expressed in distant mammalian species.
To examine the feasibility of interspecies induction of sternness traits requires platforms that enable consistent xenotransduction 26 of pluripotent transcription factors. Establishing functionality within an atypic environment necessitates readouts to capture innate pluripotent initiation, gastrulation, lineage specification, and ultimately organogenesis. 27 , 28 To this end, human sternness‐related factors were expressed here from a human immunodeficiency virus (HlV)‐based lentiviral vector system with enhanced cross‐species tropism and evaluated in distant mammalian systems. This enabled analysis of pluripotency characteristics according to escalating degrees of stringency that included the capacity for tissue chimerism during nonhuman embryonic organogenesis.
Utilizing the developed vector system and the validated hybrid progenitor cell analysis approach, this study demonstrates evolutionary a conserved function for pluripotent gene sets. Human sternness‐related factors induced morphological changes consistent with the embryonic stem cell phenotype, lineage‐specific gene expression, teratoma tissue formation, and contribution to organogenesis in nonhuman milieu. Together, these data indicate that genetic mechanisms responsible for reverting somatic tissue to pluripotent state represent a robust process of reprogramming that has been evolutionarily conserved to contribute to the overall fitness of developmental competency and regenerative capacity.
Methods
Fibroblasts
Mouse embryonic fibroblasts (MEFs) were obtained from embryos at 14.5 days post coitum (dpc). Internal organs and the head were removed prior to digestion with 0.25% trypsin‐EDTA (Invitrogen, Carlsbad, CA, USA). Digestion was performed three times. Obtained suspension was inactivated with equal volume of EmbryoMax Dulbecco's modified Eagle's medium (DMEM; Millipore, Billerica, MA, USA) supplemented with 10% fetal calf serum (FCS), 1% L‐glutamine (Invitrogen), and penicillin/ streptomycin (Invitrogen). Resulting fibroblasts were plated and grown to confluence in the same medium for two passages. Transduced MEFs were maintained in DMEM (Millipore) supplemented with pyruvate (Lonza, Basel, Switzerland) and L‐glutamine (Invitrogen), nonessential amino acids (Mediatech, Herndon, VA, USA), 2‐mercaptoethanol (Sigma‐Aldrich, St. Louis, MO, USA), 15% FCS (Invitrogen), and LIF (Millipore).
HIV packaging plasmid
The parental packaging plasmid pCMVR8.91 29 was used to engineer modifications in the HIV‐1 capsid region for increased vector transduction efficiency. 30 , 31 To generate HIV‐1 packaging constructs carrying the capsid mutations, the Apal, Bglll, and Spel sites in the uncoding region of pCMVR8.91 29 were deleted (p8.9Ex). Naturally occurring capsid amino acid substitutions, which affect the HIV cyclophilin A (Cyp A) dependency, were introduced into the capsid region of the gag gene, resulting in pEx‐HV, pEx‐QI, and pEx‐QV Vesicular stomatitis virus glycoprotein G (VSV‐G)‐expressing plasmid, pMD.G, 29 was used for pseudotyping HIV‐1 vector particles. Infectious HIV vectors were generated by packaging a green fluorescent protein (GFP)‐carrying HIV vector genome with the modified constructs and VSV‐G, and vector amounts were normalized by the levels of endogenous reverse transcriptase (RT) activity in vector particles. Human, simian, and murine cell lines were infected with various amounts of GFP‐expressing vectors, and GFP‐positive cell populations were analyzed using fluorescence‐activated cell sorting (FACScan, BD Biosciences, Franklin Lake, NJ, USA) and automated quantification (CELL QUEST software; Becton Dickinson, Franklin Lake, NJ, USA). Vector infectivity in each target cell line was determined by infectious units per nanogram RT activity. For MEF transduction, GFP‐carrying HIV vectors were generated with a conventional HIV packaging construct (p8.9Ex) or a packaging construct with the V83L, H87Q, and 191V capsid substitutions (pEx‐QV). To determine transduction efficiencies, 5 × 104 MEFs were infected with increasing amounts of unconcentrated vectors overnight. The number of infected MEFs was determined by GFP‐positive cells using FACScan.
HIV‐based transfer vectors
pSIN‐CSGWdlNotl was generated by deleting one of the two NotI sites in the GFP‐expressing HIV vector construct, pSIN‐SEW, 32 which allowed one‐step cloning of genes of interest by BamHI and NotI. Transfer vectors were generated with full‐length human Oct3/4, Sox2, Klf4, and c‐Myc cDNAs (Open Biosystems, Huntsville, AL, USA) amplified using the primer pairs Oct3/4 (5′‐ATAGGATCCGCCACCATGGCGGGACACCTGGCTTCG GAT‐3′ and 5′‐ATAGCGGCCGCTCAGTTTGAATGCATGGG AGAGCC‐3′, BamHI‐NotI), Sox2 (5′‐ATAGGATCCACCATGT ACAACATGATGGAGACGGAGC‐3′ and 5′‐ATAGCGGCCG CTCACATGTGTGAGAGGGGCAGTGT‐3′ BamHI‐NotI), Klf4 (5′‐GACGAATTCGGATCCACCATGAGGCAGCCACCTGGC GAGTCTG‐3′ and 5′‐GACCTCGAGCGGCCGCTTAAAAATG CCTCTTCATGTGTAAG‐3′, BamHI‐XhoI), and c‐Myc (5′‐GC CTGATCAAGGCTCTCCTTGCAGCTGCTTAGACG‐3′ and 5′‐ATAGCGGCCGCTTACGCACAAGAGTTCCGTAGCTG‐3′, BclI‐NotI) cloned into the pSIN‐CSGWdlNotl, resulting in pSIN‐Oct3/4, pSIN‐Sox2, pSIN‐Klf4, and pSIN‐c‐Myc. Human sternness‐related factors were driven by a spleen focus‐forming virus (SFFV) promoter. HIV vectors were produced by transient transfection of 293T cells using FuGene6 (Roche, Indianapolis, IN, USA) with a weight ratio of 2:1:1 of vector to packaging to VSV‐G plasmids. 30 Transfected cells were washed and grown for 48 hours, and supernatants were harvested and passed through a 0.45‐μm filter. Vector supernates (10 mL) were concentrated by ultracentrifugation (104 g, 2 hours at 4°C), resuspended in 500 μL of serum‐free media, aliquoted, and stored at −80°C For reprogramming, vector titers were determined in MEFs by FACS for GFP‐expressing vectors and by immunostaining for sternness factor‐encoding vectors.
Western blot
293T/17 cells (CRL‐11268; ATCC, Manassas, VA, USA) were maintained in DMEM (Invitrogen) supplemented with 10% FCS and antibiotics. Western blots were run on 12% SDS‐PAGE gels and transferred to P VDF membranes using the semi‐dry method. The membranes were then blocked overnight. Anti‐Oct4 (no. 2750S) and anti‐Sox2 (no. 2748S) antibodies (Cell Signaling, Boston, MA, USA), anti‐c‐Myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and anti‐KLF4 (ab26648–25) antibody (Abeam, Cambridge, MA, USA) were used to verify the expression of human sternness factors in vector‐infected cells.
Immunofluorescence
To determine the expression levels, native and transduced MEFs were labeled with anti‐Oct4, anti‐Sox2, anti‐c‐Myc, and anti‐KLF4 antibodies along with the FITC‐conjugated donkey anti‐mouse IgG secondary antibody for c‐Myc (Jackson Immuno Research, West Grove, PA, USA), and fiuorescein isothiocyanate (FITC)‐conjugated donkey anti‐rabbit IgG for Oct3/4, Sox2, and Klf4 (Jackson Immuno Research). To determine reactivation of pluripotent markers, isolated cell lines were stained with anti‐SSEAl antibody (MAB4301; dilution 1:50; Chemicon International) and anti‐Ki67 antibody (1:200; Neomarkers, Fremont, CA, USA) along with secondary antibodies, goat anti‐mouse Alexa Fluor 568 (1:250) and goat anti‐rabbit Alexa Fluor 488 (1:250; Invitrogen), to visualize markers of undifferentiation. The nuclei were labeled with 4,6′‐diamidino‐2‐phenylindole (DAPI; Invitrogen).
In vitro differentiation
Transduced cells were differentiated into three‐layer embryoid bodies (EBs) using the hanging‐drop method in differentiation media supplemented with 20% FCS without LIF. 33 , 34 , 35 Briefly, 25‐μL drops from a 25,000 cell/mL suspension were cultured on the lid of a plate for 48 hours. EBs were then flushed and kept in suspension for 2 days to allow spontaneous differentiation for a total of 5 days.
Detection of gastrulation markers
Expression of pluripotency and gastrulation markers was detected by RT‐PCR. 36 , 37 Total RNA was extracted with a combination of gDNA Eliminator and RNeasy columns (Qiagen, Valencia, CA, USA). cDNA was prepared from total RNA samples using Superscript III First Strand (Invitrogen). Mouse GAPDH (4352932E; Applied Biosystems, Foster City, CA, USA) was used as control. Analyzed genes included Fgf4 (Mm00438917_ml), Gsc (Mm00650681_gl), Soxl7 (Mm00488363_ml), Pou5fl (Mm00658129_gH), Zicl (Mm01239008_mH), and Sox2 (Mm00488369_sl; Applied Biosystems).
Teratoma formation
Native and transduced fibroblasts were injected subcutaneously into the flank skin of anesthetized athymic nude mice at a dose of 500,000/50 μL medium. Tumor growth was monitored daily until the tissue was harvested. Tumors were processed by rapid freezing and cut by cryosections at 7‐μm thickness to be stained with standard hematoxylin/ eosin procedures. 35
Chimeric blastocyst formation
In vivo contribution of transduced cells to embryonic development was assessed through diploid aggregation with preimplantation morula. 38 CD1 females at 3 weeks of age were superovulated using intraperitoneal injection of pregnant mare serum gonadotropin and human chorionic gonadotrophin, followed by pairing with adult CD1 males for timed pregnancy. Embryos at 2.5 days dpc were harvested, washed in EmbryoMax M2 medium (Millipore), and denuded from zona pelucida to produce morula competent for stem cell integration. After washing through M2 and EmbryoMax KSOM (Millipore) solutions, the embryos were plated as pairs in microweils to facilitate aggregation. Engineered stem cells were labeled for in vivo imaging by infection with a GFP‐carrying HIV vector generated with a conventional HIV packaging construct (p8.9Ex). Labeled cells cultured for at least two passages after thawing were partially digested using trypsin 0.25%‐EDTA (Invitrogen) and preplated for 45 minutes to allow attachment of feeders to the plate. Floating clumps (8–15 cells) were individually picked and washed in M2 medium and KSOM medium before being placed adjacent to the pair of embryos in microweils. The aggregation complex was incubated in a table‐top incubator (Thermofisher, Waltham, MA, USA) with continuous flow of a humidified gas mixture (5% C02/5% O2/90% N2) for 24 hours until cavitation of theblastocysts. 38
In utero organogenesis
CD1 females in estrus were identified and paired with vasectomized studs 2 days prior to aggregation to produce pseudopregnant mice. Surrogate mothers were anesthetized (2–3% inhaled isofiurane), their uteruses were dissected through a minimal flank incision, and blastocyst‐stage chimeric aggregates containing transduced cells were transferred into the distal portion of the uterus. 38 Pregnancy was supported by pseudopregnant females until 9.5 dpc when embryos were harvested and analyzed for transduced cell distribution using an LSM 510 laser scanning confocal microscope (Carl Zeiss, Oberkochen, Germany).
Results
Engineered HIV vector packaging constructs for improved transduction efficiency across species
Efficient HIV infection requires Cyp A in target human cells, 39 with sequence variations in the Cyp A‐binding loop of the capsid protein affecting viral infectivity. 30 , 31 , 40 Capsid mutations were used here to improve infectivity of HIV‐based vectors across species in order to test human stemness‐related factors in nonhuman cell types ( Figure 1A ). By generating GFP‐expressing HIV vectors containing specific mutations in the Cyp A‐binding loop region, the efficiency of infectivity was quantified in multiple cell lines. When the vector particle numbers were adjusted to the virion reverse transcriptase activities, the engineered packaging constructs pEx‐QV and pEx‐QI showed improved infectivity when screened in simian FrhK4 or CV1 cells as well as in murine P815 or EL4 cells compared with the parental p8.9‐Ex vector without aff ecting vector infectivity in human cells ( Figure 1A ). The absolute infectious vector yields were 2‐ to 3‐fold higher with pEx‐QV over p8.9‐Ex or modifi ed pEx‐QI. Furthermore, the pEx‐QVpackaged HIV vector transduced MEFs more efficiently than the parental p8.9‐Ex ( Figure 1B ). Th e validated packaging construct pEx‐QV was therefore selected to deliver stemness‐related factors.
Figure 1.
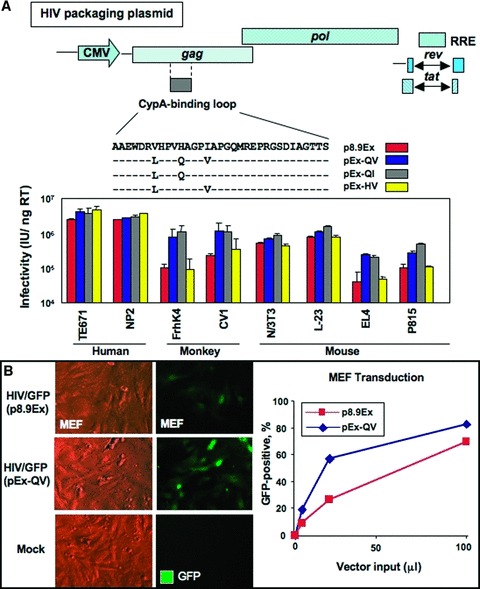
The H87Q capsid substitution in vector packaging constructs increases HIV vector infectivity across species. (A) Naturally occurring capsid substitutions were introduced into the cyclophilin A‐binding region of HIV‐1 gag gene of a vector packaging construct, p8.9Ex. Infectious HIV vectors were generated by packaging a GFP‐carrying HIV vector genome with the modified constructs, and the amounts of vectors were normalized by the levels of endogenous reverse transcriptase (RT) activity in vector particles. Human, simian, and murine cell lines were infected with various amounts of GFP‐expressing vectors, and GFP‐positive cell populations were analyzed by flow cytometry. Vector infectivity in each target cell line was determined by infectious units per nanogram RT activity. (B) GFP‐carrying HIV vectors were generated with a conventional HIV packaging construct (p8.9Ex) or a packaging construct with the H87Q capsid subsitution (pEx‐QV). MEFs (5 × 104) were infected with increasing amounts of unconcentrated vectors. The percentage of transfected cells was observed by comparing total cells to GFP‐positive cells under UV microscope 3 days after infection (left panels, with 20 μL of vector input) and analyzed by flow cytometry 5 days after vector infection (right panel).
Efficient expression of human stemness factors in nonhuman recipients
Human sequences were used to generate reprogramming vector sets to be tested in evolutionary distant somatic cell types. Gene sequences demonstrated a high degree of conservation, with the lowest percentage of homologies noted between Oct3/4 orthologs at 84%. This degree of homology is similar to the sequence for LIF, which does not conserve maintenance of pluripotency in human 24 as required for mouse stem cells ( Figure 2A ). Human cDNAs for stemness‐related factors were amplified by PCR, cloned into self‐inactivating vector plasmid, and packaged into the selected pEx‐QV HIV packaging construct to produce expression vectors encoding human OCT‐3/4, SOX2, KLF4, and c‐MYC ( Figure 2B ). Proper expression was verifi ed in human 293T cells with predicted molecular weight transgene products detected by immunoblotting with OCT‐3/4, SOX2, KLF4, and c‐MYC antibodies ( Figure 2C ). Robust transgene expression of the four human stemness‐related factors was also detected in >90% of MEFs ( Figure 2D ). Thus, the pEx‐QV HIV‐based lentiviral platform demonstrated cross‐species tropism and consistently delivered interspecies transduction of human pluripotent genes.
Figure 2.
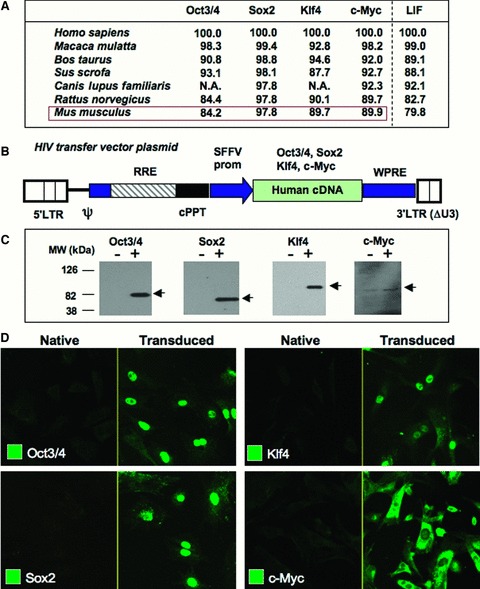
Efficient expression of sternness‐associated factors in human and murine cell types. (A) Percent amino acid homology among orthologous sternness‐related factors. Homology for LIF served as benchmark. Homology was determined by LALIGN program (EMBNet). NA = protein sequence not available. (B) Scheme of the HIV‐1 vector genome construct used to generate sternness factor‐expressing vectors. Ψ= packaging signal; LTR = long terminal repeat; RRE = Rev‐responsive element; cPPT = central polypurine tract; SFFV = spleen focus‐forming virus promoter; WPRE = Woodchuck hepatitis virus posttranscriptional regulatory element. OCT‐314, SOX2, KLF4, and c‐MYC cDNAs were driven by an internal SFFV promoter. The KLF4‐encoding vector lacks WPRE. (C) 29 3T cells (2 × 105) were infected with 50 μL of the sternness factor‐expressing vectors. Three days after infection, expression of full‐length sternness factors was verified by Western blotting with respective antibodies. (D) MEFs (5 × 104) were infected with 100 μL of unconcentrated vectors. Expression levels of transgene products were visualized by immunostaining 4 days following infection.
Virus‐transduced human stemness factors reprogram mouse fibroblasts
To determine whether human stemness‐related factors can reprogram mouse somatic cells, ectopic gene expression was achieved in MEFs. A GFP‐expressing vector was infected into MEFs at a multiplicity of infection (MOI) of 20 as the control to determine any spontaneous cellular changes ( Figure 3A ). Human pluripotent genes for OCT3/4, SOX2, c‐MYC, and KLF4 were transduced together in parallel experiments at an MOI of 5 each ( Figure 3B ). Two days post infection, transduced cells were passaged and monitored for the formation of embryonic stem cell‐like colonies, according to morphology, consisting of compact cell clusters. 12 In contrast to monomorphic single‐cell layered fibroblasts in GFP control groups, MEFs treated with the combination of four human factors produced numerous colonies after 7 days ( Figure 3B , inserts) and were of sufficient size to isolate individual clones after 10 days. Compared with native MEFs ( Figure 3C ), clonal expansion of isolated colonies produced rapidly dividing cell lines without contact inhibition and with maintained embryonic stem cell morphology through a minimum of 10 passages ( Figure 3D ). Transduced MEFs demonstrated ongoing cell divisions according to Ki67 expression ( Figure 3E and 3F ). In contrast to parental fibroblasts that lack pluripotent markers, transduced cells uniquely expressed the early stage‐specific mouse embryonic antigen (SSEA‐1) 41 as a characteristic of sternness ( Figure 3G and 3H ). Thus, the engineered platform based on human sternness factors induced efficient metamorphosis of murine fibroblasts into clonal populations, recapitulating growth kinetics and a cellular expression profile consistent with an embryonic stem cell phenotype.
Figure 3.
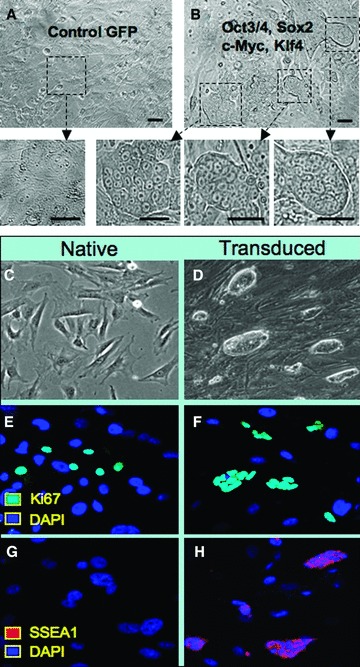
Transduced murine fibroblasts with human sternness factors reactivate stem cell phenotype. (A) Control virus expressing GFP demonstrated no discernible changes in MEF morphology. (B) Coinfection with human OCT‐314, SOX2, KLF4, and c‐MYC vectors produced multiple colonies with distinct stem cell‐like morphology that allowed isolation of individual clones. (C) Native MEFs continued to grow in monolayer and displayed contact inhibition at confluence. (D) Clonal expansion of transduced cells demonstrated morphology similar to embryonic stem cells. (E, F) Both native and transduced MEFs expressed markers of cell cycle activation indicated by Ki67 (cyan) in a subpopulation of progeny. (C, H) The stem cell marker SSEA1 (red) was uniquely expressed within transduced cells compared with native MEFs identified by nuclear staining with DAPI (blue).
In vitro differentiated transduced fibroblasts display a genetic pedigree characteristic of gastrulating tissues
To identify the diversity of lineage differentiation, gene expression analysis was performed according to protocols established for embryonic stem cells. 35 , 36 , 37 Parental MEFs provided the baseline for gene expression comparison. Following viral transduction, derived progenitors were differentiated in three‐dimensional cultures to allow spontaneous germ layer formation. 35 Sequential differentiation produced EBs at day 5 with dense compaction of cells within a sphere of tissue ( Figure 4A ). Gene expression analysis demonstrated a downregulation of pluripotent markers Oct4, Sox2, and Fgf4 by day 5 of differentiation ( Figure 4B ). Concomitantly, transduced progenitors gained the expression of mesoderm lineage marker Gsc, endoderm marker Soxl7, and ectoderm marker Zicl after 5 days of differentiation ( Figure 4C ). Thus, early lineage marker induction fulfilled in vitro criteria for xenogenic nuclear reprogramming with human sternness‐related genes.
Figure 4.
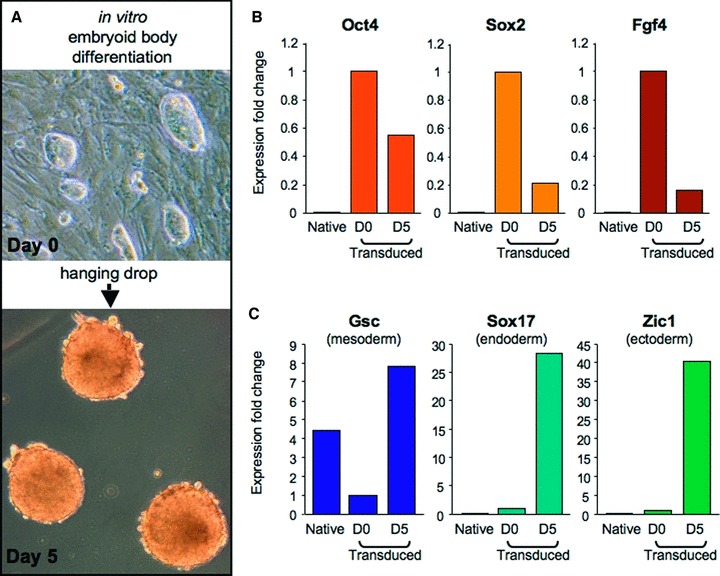
Gene expression following in vitro differentiation recapitulates gastrulation. (A) Differentiation of trans duced cells was facilitated by three‐dimensional clustering in a hanging drop to allow spontaneous maturation over a 5‐day time course. (B) Pluripotency markers, OCT4, SOX2, and FGF4, were highest in transduced cells at day 0 of differentiation, compared with either native MEFs or transduced counterparts at day 5 post initiation of differentiation. (C) The markers of mesoderm (Gsc), endoderm (Soxl7), and ectoderm (Zicl) were higher after 5 days of differentiation compared with day 0 of differentiation.
In vivo lineage differentiation of transduced fibroblasts
Pluripotent cells form spontaneous teratomas following transplantation into immunodeficient mice, an established assay to demonstrate multilineage developmental capacity. 42 , 43 , 44 Here, immunodeficient mice were subcutaneously injected with native MEFs or transduced counterparts. Only transduced cells gave rise to tumors, following injection at a dose of 500,000 cells, that enlarged to 1 cm in diameter within 4 weeks, in contrast to undetectable growth for native MEFs injected on the contralateral side ( Figure 5A ). Tumors derived from transduced MEFs were encapsulated and demonstrated a heterogenous appearance consisting of vascular networks and nonvascularized tissues on gross inspection ( Figure 5B ). Tissue histology revealed cellular architecture consisting of mesoderm lineages indicated by muscle ( Figure 5C ), ectoderm lineages denoted by keratinized tissues ( Figure 5D ), endoderm lineages composed of specialized epithelium ( Figure 5E ), and persistence of poorly differentiated cytotypes ( Figure 5F ). Together, these data documented the multiple tissues derived from in vivo differentiation and spontaneous formation of complex cytoarchitecture derived from mouse fibroblasts reprogrammed by human sternness‐related factors.
Figure 5.
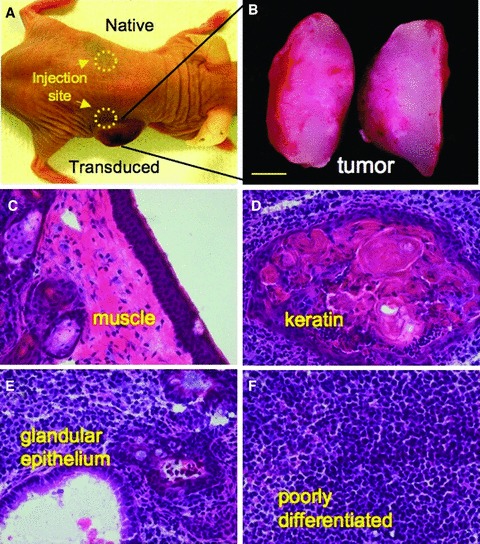
Multilineage in vivo differentation within tumors. (A) Spontaneous in vivo differentiation was monitored in immunodeficient mice following subcutaneous injection by comparing native and transduced MEFs. (B) Tumor growth was detected only from sites injected with transduced cells ater 1–2 weeks, followed by rapid expansion of tumor bulk, absent from native MEF injection sites. (C–F) Tissue was harvested at 4–6 weeks post injection. Cryosections and tissue staining demonstrated multiple lineages within the complex architecture of the nascent tumor and included muscle, keratin, glandular epithelium, and poorly differentiated tissues.
Contribution of transduced progeny into ex utero blastocysts
A hallmark characteristic of pluripotent stem cells is the ability to incorporate into 8‐cell embryos and form morula capable of developing into chimera blastocysts. 45 Primitive stem cell populations engraft within host 8‐cell embryos to form mosaic blastocysts, but are universally excluded upon the loss of functional pluripotency, 46 , 47 despite persistent expression of stem cell markers. 48 In order to determine the ability of reprogrammed MEFs to incorporate into early‐stage morula, the cells were lentivirally labeled with GFP, expanded in vitro, and prepared for diploid aggregation with unlabeled embryos ( Figure 6A and 6B ). GFP‐tagged transduced progeny retained the ability to engraft into 8‐cell embryos ( Figure 6C ) and contribute to chimeric embryos capable of spontaneous formation of blastocysts with appropriate cavitating morphology ( Figure 6D ). Diploid aggregation thus provided an efficient ex utero methodology to characterize functional properties of transduced MEFs within a permissive embryonic niche. Specifically, reprogrammed progenitors containing human sternness‐related factors revealed the primordial characteristic of pluripotency in embryos with morula integration and blastocyst development, expanding the stringency of pluripotency functional criteria.
Figure 6.
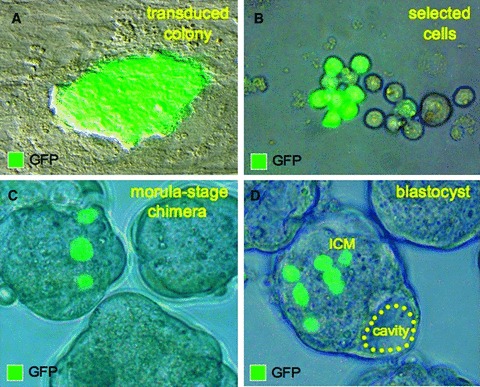
Transduced cells integrate into host morula. (A, B) Transduced MEFs were labeled with GFP tag for tracking and allowed selection for ex utero integration into early‐stage embryos. (C) Diploid aggregation between labeled transduced cells and normal morula produced chimeric early embryos. (D) Chimeric embryos developed into blastocysts, which displayed proper cavitation and formation of mosaic inner cell mass (ICM) with GFP‐labeled blastomeres.
High‐fidelity organogenesis from transduced progeny
Beyond ex vivo characterization, chimeric embryos establish in situ competency of transduced progeny during natural embryogenesis. Pluripotent stem cells contain the capacity to give rise to all lineages of the developing embryo upon blastocyst integration in a stochastic pattern, depending on the location of blastomere integration during early stage of preimplantation development. 49 Mosaic embryos produced by diploid aggregation using GFP‐labeled progenitors were transferred to the uterus of a pseudopregnant surrogate for in utero implantation and differentiation. Chimeric embryos were harvested at 9.5 dpc and analyzed for engraftment and differentiation of nonnative progeny. Embryos that demonstrated normal morphology and appropriate developmental stages of organogenesis were visualized for GFP expression. Transduced progenitors were identified throughout the embryo in multiple developing organs that included central nervous tissue ( Figure 7A and 7B ), pharyngeal arch ( Figure 7C ), the heart ( Figure 7D ), emerging limb buds ( Figure 7E ), and somites ( Figure 7F ). Thus, stochastic integration and widespread tissue contribution of reprogrammed cells demonstrated unrestricted differentiation potential and competitive fitness equal to that of native blastomeres, achieving a rigorous criterion to define pluripotency with competent in utero orchestrated organogenesis.
Figure 7.
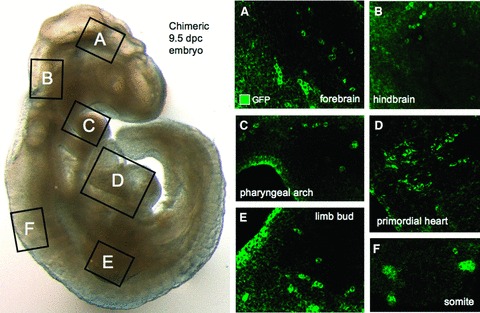
Organogenesis derived from transduced cells. (A‐F) Chimeric embryos were transferred into a surrogate mother for in utero differentiation and were harvested at 9.5 dpc for tissue analysis. Confocal microscopy revealed transduced progeny throughout the embryo including neuronal tissues of the forebrain (A) and hindbrain (B), along with the multilineage phaiyngeal arches that contained endoderm derivatives (C). Mesoderm‐derived lineages were present in the heart (D), limb bud (E), and somites (F).
Discussion
In the last decade, the advent of somatic cell nuclear transfer and reproductive cloning revealed the reversibility of stem cell pluripotency and a new paradigm for tissue dedifferentiation. 3 , 4 , 5 , 6 , 7 The atavistic environment of a fertilized egg cytoplasm demonstrated the ability to reprogram the genetic blueprint and reverse the differentiation state of the ectopic somatic nucleus. 1 , 2 With the more recent breakthrough of iPS cell technology, the scope of environmental regulation of reprogramming has now been extended to include the dynamic flux of nuclear transcription factors as a tool for bioengineering progenitor cells from somatic tissues. 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 The equilibrium of cytotype‐specific transcription factors can be titrated toward pluripotency and directed by ectopic intraspecies expression of gene sets for mouse, monkey, and human. 7 , 9 , 11 , 20 Although interspecies transfer of somatic cell nuclei demonstrates evolutionarily conserved reprogramming, 6 comparable xenotransduction of orthologous gene sets was untested prior to this study. The symmetry between sequence and function of orthologous genes cannot be assumed due to the genetic adaptation inherited through the process of evolution and thus requires empiric analysis. 21 Here, human sternness‐associated factors OCT‐3/4, SOX2, KLF4, and c‐MYC demonstrated functional equivalency between reproductively isolated species and produced pluripotent phenotype in nonhuman cytotypes, indicating evolutionary conservation of core process for nuclear reprogramming.
To enable interspecies cellular reprogramming from sternness‐related factors, an efficient delivery method was required that transduces across species and cytotypes. Typically, iPS derivation protocols employ retroviral or lentiviral vectors to deliver defined pluripotency‐related factors. 9 , 14 Unlike conventional retroviral vectors, which transduce only dividing cells, lentiviral vectors can deliver genes into both dividing and nondividing cells. 29 Lentiviral vectors also reach higher titers than retroviral vectors, making them particularly attractive vehicles to deliver multiple pluripotency‐associated factors. 32 Lentiviral vectors, however, display host range restriction and limited tropism. 26 For instance, HIV‐1‐based lentiviral vectors can transduce most human cell types efficiently, whereas their infectivity in monkey and mouse cells is relatively poor. 26 , 50 This is partly due to species‐specific host restriction factors in nonhuman cells. 51 , 52 , 53 For iPS generation, multiple genes need to be delivered simultaneously into a single cell. Thus, the present development of specialized lentiviral vectors with broader tropism offers a distinct advantage for iPS generation and provides a unique tool to examine interspecies activity of sternness‐related factors.
The engineered HIV‐based platform developed herein incorporates human OCT‐3/4, SOX2, KLF4, and c‐MYC genes, and efficiently reprogrammed mouse fibroblasts to establish cross‐species compatibility. Viral‐transduced cells demonstrated comprehensive reprogramming that fulfilled all essential postulates of pluripotent stem cells according to criteria spanning from in vitro expression profiles to in utero morphogenic potential. Transduced cells initially reactivated pluripotent markers such as SSEA‐1 expressed on multiple stem cell subpopulations. 54 Differentiating progeny, expanded in vitro as EBs and in vivo as subcutaneous tumors, spontaneously displayed features of pluripotent stem cells such as gastrulation markers along with meso derm, ectoderm, and endoderm lineages. These cellular traits are the hallmarks of pluripotency, previously thought to be unique to embryonic stem cells, and are now accepted features of bonafide induced pluripotency. 7 , 9 , 27
Beyond cell autonomous differentiation, embryonic development provides further stringency to discriminate the function of pluripotent progenitors based on the ability to integrate within the context of a host morula and produce an independent source of blastomeres that is equally competent to orchestrate complex organogenesis. 45 , 49 The development of functional dynamics between engineered progeny and native cytotypes in vivo is comprehensively examined during normal embryogenesis. 45 Thus, the overall morphogenic fitness of pluripotent progenitors programmed by human sternness‐related factors was validated by successful morula‐stage engraftment, efficient maturation of chimeric blastocysts, and synchronized organogenesis. Collectively, these developmental criterions indicate that human sternness‐related factors are able to effectively reprogram nonhuman cells and thus establish a prototypic model system to characterize gene–environmental interactions guided by human gene sets during native organogenesis.
In summary, nuclear reprogramming with ectopic expression of sternness‐inducing factors represents the next generation of bio engineering pluripotent stem cells. Xenotransduction of human gene sets into a nonhuman host environment triggered here interspecies nuclear reprogramming, providing direct evidence for a conserved process underlying cellular dedifferentiation and coordinated organogenesis. As such, the established model system provides a unique opportunity to further examine the consequences of human gene‐mediated nuclear reprogramming in the context of normal embryonic development by exploiting the permissive nonhuman early‐stage embryo. Promiscuous stemness induction underscores a primordial role for nuclear reprogramming in evolutionary survival and fitness.
Acknowledgments
We thank Dr. D. Trono and Dr. A. Thrasher for lentiviral vector plasmids, pCMVR8.91 and pMD‐G, and pSIN‐SEW, respectively. We also thank Lois A. Rowe for technical assistance in performing tissue and histological analysis, James E. Tarara for valuable expertise at the Mayo Clinic Optical Morphology and Flow Cytometry cores, and the personnel within the Mayo Clinic Advanced Genomics Technology Center. This work was supported by the National Institutes of Health (R56AI074363, T32HL007111, and R01HL083439), Marriott Individualized Medicine Program, Marriott Heart Disease Research Program, Mayo Foundation, National Hemophilia Foundation, La Caixa Foundation Graduate Program, and the American Society for Clinical Pharmacology and Therapeutics.
References
- 1. Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997; 385(6691): 810–813. [DOI] [PubMed] [Google Scholar]
- 2. Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996; 380(6569): 64–66. [DOI] [PubMed] [Google Scholar]
- 3. Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006; 441(7097): 1061–1067. [DOI] [PubMed] [Google Scholar]
- 4. Armstrong L, Lako M, Dean W, Stojkovic M. Epigenetic modification is central to genome reprogramming in somatic cell nuclear transfer. Stem Cells. 2006; 24(4): 805–814. [DOI] [PubMed] [Google Scholar]
- 5. Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Wolf DP, Mitalipov SM. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007; 450(7169): 497–502. [DOI] [PubMed] [Google Scholar]
- 6. Beyhan Z, Iager AE, Cibelli JB. Interspecies nuclear transfer: implications for embryonic stem cell biology. Cell Stem Cell. 2007; 1(5): 502–512. [DOI] [PubMed] [Google Scholar]
- 7. Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008; 132(4): 567–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henderson JT. Lazarus's gate: challenges and potential of epigenetic reprogramming of somatic cells. Clin Pharmacol Ther. 2008; 83(6): 889–893. [DOI] [PubMed] [Google Scholar]
- 9. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126(4): 663–676. [DOI] [PubMed] [Google Scholar]
- 10. Okita K, Ichisaka T, Yamanaka S. Generation of germline‐competent induced pluripotent stem cells. Nature. 2007; 448(7151): 313–317. [DOI] [PubMed] [Google Scholar]
- 11. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007; 131(5) 861–872. [DOI] [PubMed] [Google Scholar]
- 12. Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol 2007; 25(10): 1177–1181. [DOI] [PubMed] [Google Scholar]
- 13. Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007; 1(1): 55–70. [DOI] [PubMed] [Google Scholar]
- 14. Yu J, Vodyanik MA, Smuga‐Otto K, Antosiewicz‐Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007; 318(5858) 1917–1920. [DOI] [PubMed] [Google Scholar]
- 15. Yamanaka S. Pluripotency and nuclear reprogramming. Phil Trans R Soc B. 2008; 363(1500) 2079–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES‐cell‐like state. Nature. 2007; 448(7151): 318–324. [DOI] [PubMed] [Google Scholar]
- 17. Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008; 451(7151): 141–146. [DOI] [PubMed] [Google Scholar]
- 18. Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease‐specific induced pluripotent stem cells. Cell. 2008; 134(5): 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008; 322(5903): 945–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K, Qu X, Xiang T, Lu D, Chi X, Gao G, Ji W, Ding M, Deng H. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008; 3(6): 587–590. [DOI] [PubMed] [Google Scholar]
- 21. Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005; 39 309–338. [DOI] [PubMed] [Google Scholar]
- 22. Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell 2007; 128(4): 747–762. [DOI] [PubMed] [Google Scholar]
- 23. Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006; 38(4): 431–440. [DOI] [PubMed] [Google Scholar]
- 24. Daheron L, Opitz SL, Zaehres H, Lensch MW, Andrews PW, Itskovitz‐Eldor J, Daley GQ. LIF/ STAT3 signaling fails to maintain self‐renewal of human embryonic stem cells. Stem Cells. 2004; 22(5): 770–778. [DOI] [PubMed] [Google Scholar]
- 25. Xu C, Rosier E, Jiang J, Lebkowski JS, Gold JD, O'Sullivan C, Delavan‐Boorsma K, Mok M, Bronstein A, Carpenter MK. Basic fibroblast growth factor supports undifferentiated human embryonic stem cell growth without conditioned medium. Stem Cells. 2005; 23(3): 315–323. [DOI] [PubMed] [Google Scholar]
- 26. Ikeda Y, Collins MK, Radcliffe PA, Mitrophanous KA, Takeuchi Y. Gene transduction efficiency in cells of different species by HIV and EIAV vectors. Gene Ther. 2002; 9(14): 932–938. [DOI] [PubMed] [Google Scholar]
- 27. Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008; 2(3): 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mali P, Ye Z, Hommond HH, Yu X, Lin J, Chen G, Zou J, Cheng L. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells 2008; 26(8): 1998–2005. [DOI] [PubMed] [Google Scholar]
- 29. Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997; 15(9): 871–875. [DOI] [PubMed] [Google Scholar]
- 30. Ikeda Y, Ylinen LM, Kahar‐Bador M, Towers GJ. Influence of gag on human immunodeficiency virus type 1 species‐specific tropism. J Virol. 2004; 78(21): 11816–11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kootstra NA, Munk C, Tonnu N, Landau NR, Verma IM. Abrogation of postentry restriction of HIV‐1‐based lentiviral vector transduction in simian cells. Proc Natl Acad Sci USA. 2003; 100(3) 1298–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, Grez M, Thrasher AJ. High‐level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1‐based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002; 13(7): 803–813. [DOI] [PubMed] [Google Scholar]
- 33. Behfar A, Zingman LV, Hodgson DM, Rauzier JM, Kane GC, Terzic A, Puceat M. Stem cell differentiation requires a paracrine pathway in the heart. FASEBJ. 2002; 16(12): 1558–1566. [DOI] [PubMed] [Google Scholar]
- 34. Perez‐Terzic C, Behfar A, Mery A, Van Deursen JM, Terzic A, Puceat M. Structural adaptation of the nuclear pore complex in stem cell‐derived cardiomyocytes. Circ Res. 2003: 92(4): 444–452. [DOI] [PubMed] [Google Scholar]
- 35. Behfar A, Perez‐Terzic C, Faustino RS, Arrell DK, Hodgson DM, Yamada S, Puceat M, Niederlander N, Alekseev AE, Zingman LV, Terzic A. Cardiopoietic programming of embryonic stem cells for tumor‐free heart repair. J Exp Med. 2007; 204(2): 405–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Faustino RS, Behfar A, Perez‐Terzic C, Terzic A. Genomic chart guiding embryonic stem cell cardiopoiesis. Genome Biol. 2008: 9: R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nelson TJ, Faustino RS, Chiriac A, Crespo‐Diaz R, Behfar A, Terzic A. CXCR4+/FLK‐1+ biomarkers select a cardiopoietic lineage from embryonic stem cells. Stem Cells. 2008; 26(6): 1464–1473. [DOI] [PubMed] [Google Scholar]
- 38. Nelson TJ, Martinez‐Fernandez A, Terzic A. KCNJ11 knockout morula reengineered by stem cell diploid aggregation. Phil Trans R Soc B. 2008; 364(1514): 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Gottlinger HG. Functional association of cyclophilin A with HIV‐1 virions. Nature. 1994; 372(6504): 363–365. [DOI] [PubMed] [Google Scholar]
- 40. Chatterji U, Bobardt MD, Stanfield R, Ptak RG, Pallansch LA, Ward PA, Jones MJ, Stoddart CA, Scalfaro P, Dumont JM, Besseghir K, Rosenwirth B, Gallay PA. Naturally occurring capsid substitutions render HIV‐1 cyclophilin A independent in human cells and trim‐cyclophilin resistant in owl monkey cells. J Biol Chem. 2005; 280(48): 40293–40300. [DOI] [PubMed] [Google Scholar]
- 41. Solter D, Knowles BB. Monoclonal antibody defining a stage‐specific mouse embryonic antigen (SSEA‐l). Proc Natl Acad Sci USA. 1978; 75(11): 5565–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Damjanov I, Solter D. Animal model of human disease: teratoma and teratocarcinoma. Am J Pathol. 1976; 83(1): 241–244. [PMC free article] [PubMed] [Google Scholar]
- 43. Lensch MW, Schlaeger TM, Zon LI, Daley GQ. Teratoma formation assays with human embryonic stem cells: a rationale for one type of human‐animal chimera. Cell Stem Cell. 2007; 1(3): 253–258. [DOI] [PubMed] [Google Scholar]
- 44. Yamada S, Nelson TJ, Crespo‐Diaz RJ, Perez‐Terzic C, Liu XK, Miki T, Seino S, Behfar A, Terzic A. Embryonic stem cell therapy of heart failure in genetic cardiomyopathy. Stem Cells. 2008; 26(10): 2644–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wood SA, Allen ND, Rossant J, Auerbach A, NagyA . Non‐injection methods for the production of embryonic stem cell‐embryo chimaeras. Nature. 1993; 365(6441): 87–89. [DOI] [PubMed] [Google Scholar]
- 46. Stewart C. Aggregation between teratocarcinoma cells and preimplantation mouse embryos. J Embryo! Exp Morphol. 1980; 58: 289–302. [PubMed] [Google Scholar]
- 47. Fujii JT, Martin GR. Incorporation of teratocarcinoma stem cells into blastocysts by aggregation with cleavage‐stage embryos. DevBioi. 1980; 74(1): 239–244. [DOI] [PubMed] [Google Scholar]
- 48. Waters BK, Rossant J. The effect of retinoic acid pretreatment on the ability of murine embryonal carcinoma and inner cell mass cells to participate in chimaera development. J Embryol Exp Morphol. 1986; 98: 99–110. [PubMed] [Google Scholar]
- 49. Nagy A, Gocza E, Diaz EM, Prideaux VR, Ivanyi E, Markkula M, Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990; 110(3): 815–821. [DOI] [PubMed] [Google Scholar]
- 50. Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. Species‐specific, postentry barriers to primate immunodeficiency virus infection. J Virol. 1999; 73(12): 10020–10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV‐1 infection in Old World monkeys. Nature. 2004; 427(6977): 848–853. [DOI] [PubMed] [Google Scholar]
- 52. Zhang JX, Diehl GE, Littman DR. Relief of preintegration inhibition and characterization of additional blocks for HIV replication in primary mouse T cells. PLoS ONE. 2008; 3: e2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Noser JA, Towers GJ, Sakuma R, Dumont JM, Collins MK, Ikeda Y. Cyclosporine increases human immunodeficiency virus type 1 vector transduction of primary mouse cells. J Virol. 2006; 80(15): 7769–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Henderson JK, Draper JS, Baillie HS, Fishel S, Thomson JA, Moore H, Andrews PW. Preimplantation human embryos and embryonic stem cells show comparable expression of stage‐specific embryonic antigens. Stem Cells. 2002; 20(4): 329–337. [DOI] [PubMed] [Google Scholar]


