Abstract
BACKGROUND/AIMS
The genetic basis for ingestive behaviors is virtually unknown. Quantitative trait loci (QTLs) for carbohydrate and energy intake map to mouse chromosome 17 and were previously confirmed by a congenic strain bearing CAST/Ei (CAST) donor segment on the C57BL/6J (B6) background.
METHODS
We used microarray technology to facilitate gene identification. Gene expression was compared between the B6.CAST-17 (BC-17) congenic and B6 strains in two diets: 1) chow, and 2) carbohydrate/protein vs. fat/protein.
RESULTS
Within the QTL and unique to macronutrient selection, Agpat1 (acylglycerol-3-phosphate O-acyltransferase 1) was differentially expressed in hypothalamus. Irrespective of diet, the gene with the highest fold difference in congenic mice was trefoil factor 3 (Tff3) in liver. Several genes involved in fat metabolism were decreased in carbohydrate-preferring congenic mice, while genes associated with carbohydrate metabolism were increased. In particular, the glyoxalase pathway was enhanced including Glo1, Glo2, and dLDH. Higher expression of Glo1 mRNA in BC-17 congenic mice corresponded to increased protein expression revealed by Western blot, and to higher GLO1 activity in blood.
CONCLUSION
These genes represent new candidates for nutrient intake phenotypes. We propose that increased GLO1 in the BC-17 strain supports its need to protect against dietary oxidants resulting from high carbohydrate intake.
INTRODUCTION
Both humans (1–2, 3–7) and laboratory animal strains display highly variable intakes of high-fat and high-carbohydrate foods. In rodent models, a genetic basis for the observed variation in self-selected macronutrient intake was proposed as early as the 1940’s (8, 9) based on the observation of “similar appetites” among littermate rats. These observations were underscored by later evidence that intra-litter variation in fat and carbohydrate selection is smaller than inter-litter variation (10), as well as reports of within- and between-strain differences in rodents (11–15), which suggest a combination of genetic and environmental factors. In an evaluation of thirteen mouse inbred strains, we found continuous variation in macronutrient consumption across strains, thus providing clear evidence for a complex genetic trait (16). These results allowed for the identification of a suitable animal model, i.e., two strains displaying non-overlapping phenotypic distributions were chosen for genetic analysis: C57BL/6J (B6) mice self-select a higher proportion of energy from fat than CAST/Ei (CAST), a strain that consumes more carbohydrate (16). The CAST strain was also noted for its significantly higher calorie intake per body weight.
To begin identifying the complex trait genes underlying these phenotypic differences, we first established statistically significant, genome-wide linkage or association. Using a large F2 intercross population, the first mammalian QTL for macronutrient-specific intake (carbohydrate and fat) and for total energy intake were identified (17). A region on proximal Chr 17 revealed two significant QTL that co-localized for increased macronutrient intake-carbohydrate (Mnic1) and total kilocalorie intake (Kcal2), adjusted for body weight. Next, we developed a congenic strain to obtain independent evidence for linkage, to determine the locus more precisely, and to aid in identification of the underlying gene(s). The congenic line was constructed by introgressing CAST donor DNA from the Mnic1 and Kcal 2 QTL on Chr 17 in a genome identical to the background B6 strain (18). This B6.CAST-17 congenic strain confirmed the chromosome 17 locus by specifying the original linked traits; specifically, homozygous congenic mice consumed significantly more carbohydrate (27%) and total energy (17%) compared with wild-type B6 littermates (18). This complex QTL region has been localized to a 34.8-cM region, which corresponds to 60.3 Mb on mouse chromosome 17.
With the QTL captured in an interval-specific congenic strain that exhibits the original linked phenotypes, the molecular basis of the effects of this genetic locus on nutrient intake can be elucidated. Comprehensive microarray analyses of genes in the hypothalamus and liver were performed to reveal changes in expression between the wildtype B6 and B6.CAST-17 congenic mice (18). Transcriptional differences outside the target QTL were examined to identify regulatory pathways or downstream targets relevant to the control of food intake. To identify genes that were uniquely associated with macronutrient selection, and not with chow intake, two independent experiments were conducted with respect to diet, i.e., chow and 2-choice macronutrient diet. The results permit qualitative comparisons of the two sets of differentially expressed genes. The differential expression of genes was validated with quantitative RT-PCR using the same RNA samples that were subjected to microarrays. The results of this study provide the basis for further investigation of differentially expressed candidate genes within Mnic1 and Kcal2, with a potential role in preferential carbohydrate consumption and increased calorie intake. This is the first microarray study aimed at identifying candidate genes involved in the control of ingestive behaviors related to the selection of macronutrients.
METHODS
Animals
C57BL/6J (B6) and CAST/Ei (CAST) breeders were purchased from the Jackson laboratory (Bar Harbor, Maine). A full congenic strain, B6.CASTD17Mit19-D17Mit91 (B6.CAST-17) was developed on site at the Pennington Biomedical Research Center using the “speed congenics” method as previously described (18–19). Selection at each generation was based on the presence of desired genomic segment and the absence of contaminating donor DNA (19). The marker-assisted selection protocol produces congenic strains in less than half the number of generations required by the classic protocol (N12) and by S6, a “speed congenic” line has <0.25% contaminating donor genome (19). The B6.CAST-17 S6 (“speed” congenic generation 6) strain possesses a 34.8-cM CAST donor segment on the B6 background, extending at least from proximal marker D17Mit19 at 4.7Mb to distal marker D17Mit91 at 65.2Mb, containing 60.5 Mb. To generate experimental animals, heterozygous B6.CAST-17 mice were intercrossed to generate three (littermate) genotypes. All mice used as controls for the congenic animals were littermate B6. The use of littermate controls to test the phenotypic effect of a congenic segment is extremely important for overcoming the problems of background heterogeneity as well as the effects of litter, age, or other unidentified environmental factors.
Mice were bred and reared in polycarbonate cages with sterilized corn-cob bedding, and housed in temperature-controlled rooms at 23–24° C with a 14:10-h light-dark cycle. Mice were weaned onto standard rodent chow (no. 5001, LabDiet, Richmond, IN) which contained by energy 28% protein, 12% fat, and 60% carbohydrate. Animal studies were carried out with the approval of the PBRC Institutional Animal Care and Use Committee.
Phenotyping
Phenotyping methods have been previously described in detail, including diet composition (17). Briefly, 8 wk-old mice were adapted to single housing in hanging, wire-bottom mouse cages at 25–27° C until they regained positive weight balance. The light/dark cycle was set at 12:12-h. Mice were presented with a choice between two diets: fat/protein (F/P; vegetable shortening and casein) and carbohydrate/protein (C/P; corn starch, powdered sugar, and casein). The protein composition of the two diets was equivalent (22% of energy) and the balance of calories for each was contributed by fat or carbohydrate (78%). Both diets were supplemented with vitamins, minerals, and cellulose. Diet jars and all spillage were weighed daily.
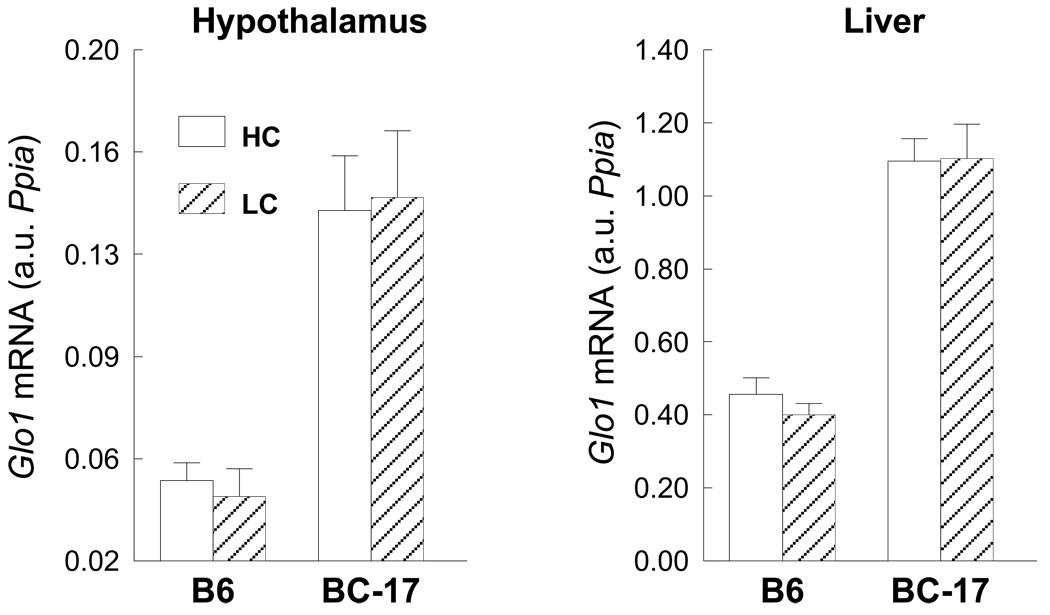
For the low- and high-carbohydrate study (Fig. 7), mice of each strain were fed a single nutritionally complete diet: D12331 or D12329, respectively, (Research Diets, Inc., New Brunswick, NJ) for two days. On the day of tissue harvest, mice were food-deprived at 0800 and euthanized at 1400 h.
Fig. 7.
qRT-PCR expression of Glo1 mRNA in wildtype B6 and homozygous B6.CAST-17 (BC-17) congenic mice fed a HC (high-carbohydrate; 73%) or LC (low-carbohydrate; 26%) diet for two days. Data are expressed as mean ± S.E. from 9 mice per strain, per diet, analyzed in duplicate.
Rationale for experimental design with respect to timecourse of diet exposure
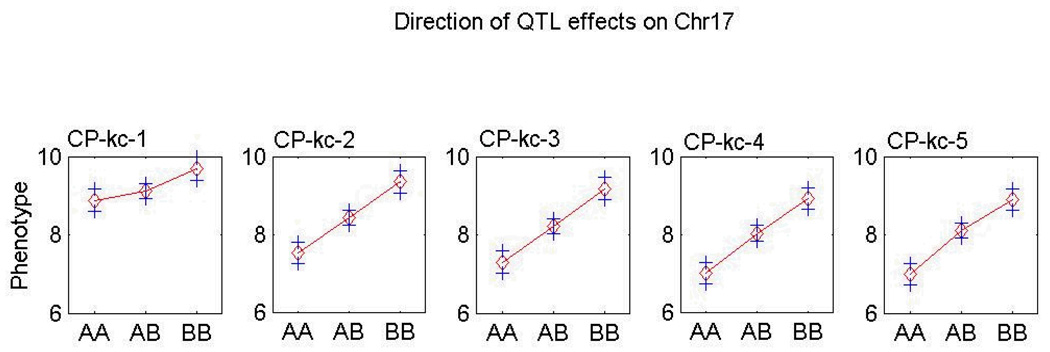
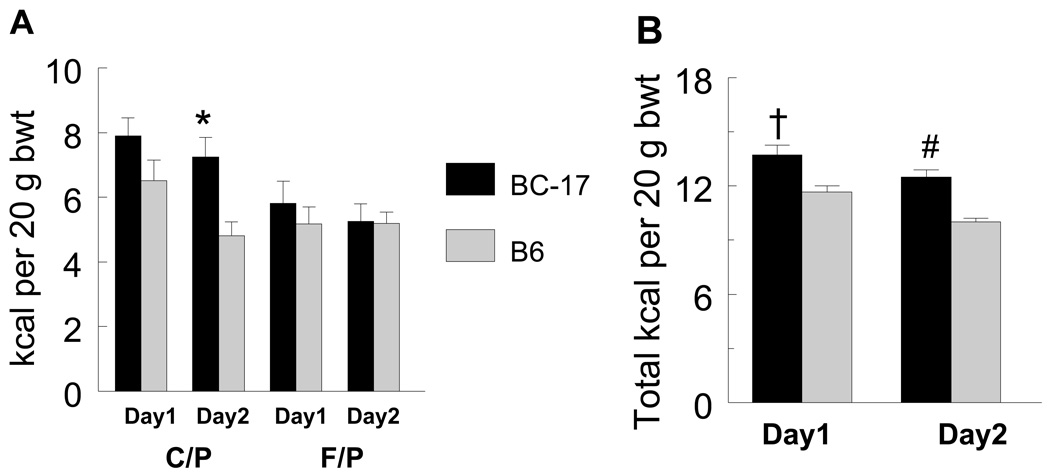
In the original QTL study, F2 mice showed an absence of genetic linkage on day 1 but not on days 2–10, for C/P kcal at the Mnic1 QTL locus (D17Mit100). This temporal variation was attributed to the B6 allele (17) (Fig. 1). This phenomenon also was observed in the congenic strain model used in the present study, i.e., there were no differences between day 1 and day 2 in C/P intake of the homozygous B6.CAST-17 mice (18). By contrast, the wild type B6 and heterozygous congenic mice ate more C/P calories on day 1, but then significantly reduced their carbohydrate intake beginning on day 2. Notably, there were no temporal variations in F/P intake (Fig. 2A). These results suggest that the choice of fat or carbohydrate may be affected by environmental factors during the first 24 h of exposure to the experimental diets but comes under a greater degree of genetic control with prolonged exposure. Based on the hypothesis that metabolic signals arising from ingestion of the diet on day 1 influenced the animals’ choice on day 2, tissues were collected shortly before onset of the dark feeding period on day 2.
Fig. 1.
Genetic linkage for carbohydrate/protein kcal (C/P kc) intake on days 1–5 in F2 mice derived from an intercross of C57BL/6J and CAST/EiJ (42). F2 mice showed an absence of genetic linkage on day 1, but not on days 2 and following, for C/P kcal at the Mnic1 QTL locus (D17Mit100), and the temporal variation in linkage was due to the B6 allele. AA = homozygous B6 allele; AB = heterozygous for B6 and CAST alleles; BB = homozygous CAST allele.
Fig. 2.
(A) Macronutrient diet selection data for carbohydrate/protein (C/P) and fat/protein (F/P) intake on days 1 and 2 of diet initiation in the 10 d phenotyping study. Wildtype B6 mice, but not homozygous B6.CAST-17 (BC-17) congenic mice, significantly reduced their carbohydrate intake from day 1 to day 2. *P < 0.001. By contrast, there was no difference in F/P intake between days 1 & 2 for either strain. (B) Total calorie (kcal) intake per 20 g body wt on days 1 and 2 of diet initiation. †P < 0.002, #P < 5.7 × 10−7.
Experimental design
Global gene expression, in both the hypothalamus and liver, was compared between the B6.CAST-17 homozygous congenic and B6strains in two diet conditions: Expt.1) chow (LabDiet #5001) and Expt. 2) carbohydrate/protein vs. fat/protein, 28 h after diet initiation, i.e., 4 h before lights out on day 2. For each experiment, RNA was pooled from six male B6.CAST-17 homozygous congenic mice and six male wild type B6 mice. In Expt. 2, animals were selected based on the absence of overlap in phenotypic values. To identify these individuals, mice of each strain (n = 20–25) were tested in the diet selection protocol for 10 d (data are presented in ref. 18). Six congenic and six B6 mice which displayed the most divergent phenotypic values for carbohydrate and total kcal intake (relative to body weight) were chosen for microarray analysis, returned to chow for 6 wks, and then exposed to the 2-choice diet for one day. With this design we aimed to study gene expression changes immediately prior to selection of the C/P or F/P diet, and to avoid the confounding effects of novelty on day 1, overnight fasting, or recent meals. On day 2, food was removed at 1300 during the light period; 4 h later mice were quickly anesthetized by isoflurane inhalation and decapitated, ~1 h before lights out.
Ingestive behavior is a highly integrated process involving a number of metabolic systems (20). Substantial evidence indicates that intracellular utilization of metabolic fuels generates signals that are used by the central nervous system (CNS) to control feeding (20) and several metabolic processes have been implicated, e.g., glucose utilization, fat oxidation, hepatic ATP content, and energy expenditure (22–26). We reasoned that genes encoding proteins involved in these processes are likely candidates for modifying the self-selected intake of macronutrients. The hypothalamus and liver were selected for analysis, based on their importance in the behavioral and metabolic control of food intake.
Genes with a 1.5 fold change or greater were chosen for further consideration. Results for selected candidates were validated using qRT-PCR to compare gene expression in B6.CAST-17 homozygous congenic and wild type B6 mice. Two independent experiments were conducted with respect to diet, i.e., chow and 2-choice macronutrient diet. The two datasets permitted qualitative comparisons of the sets of differentially expressed genes between experiments.
RNA isolation and cDNA synthesis
Total RNA was isolated from liver and hypothalamus of B6.CAST-17 congenics and wild type B6 mice using TRIzol (TRI reagent, Molecular Research center). RNA was purified using the RNeasy kit (Qiagen). The quantity and quality of RNA was checked using Agilent bioanalyzer (Agilent Technologies). The samples with RIN >8.0 were selected for microarray analysis. First-strand cDNA templates were synthesized by mixing 1 µg of RNA and 5 µM of the (dT)11- primer. The mixtures were heated for 5 min at 70° C and placed immediately on ice. Next, 1x RT buffer, 200 units Superscript II reverse transcriptase (Invitrogen), 25 µM dNTP, and 10 mM DTT were added and incubated for 90 min at 42°C in a final volume of 20 µl. After heat inactivation of enzyme for 5 min at 70° C, the cDNA was diluted to 1:5 with nuclease free water and stored at –20° C.
Microarray analysis
The Applied Biosystems (AB) Mouse Genome Survey Microarray was used to generate gene expression profiles. This microarray consists of approx. 34,000 features including a set of ~1,000 controls. Each microarray contains 32,996 probes targeted to 32,381 curated genes representing 44,498 transcripts. There are more than 1138 known genes in the Chr 17 QTL region (NCBI) and approx. 85% of these are represented in the AB mouse genome array. 1 µg of total RNA was used to synthesize Digoxigenin-UTP labeled cRNA using the AB chemiluminescent RT-IVT labeling Kit v2.0. Each microarray was first hybridized at 55°C for 1 h in hybridization buffer with blocking reagent. Next, 15 µg of labeled cRNA targets was fragmented into 100–400 bases by incubating with fragmentation buffer at 60°C for 30 min, mixed with internal control target and hybridized to each pre-hybridized microarray at 55° C for 16 hr. After hybridization, the arrays were washed and rinsed. Enhanced chemiluminescent signals were generated by first incubating arrays with anti-digoxigenin-alkaline phosphatase, enhanced with chemiluminescence enhancing solution and finally in chemiluminescence substrate. Images were collected using a 1700 analyzer (Applied Biosystems, Foster City, CA) equipped with high-resolution, large format CCD camera. Images were auto-girded and the chemiluminescent signals were quantified, corrected for background and spot, and spatially normalized. AB expression system software was used to extract assay signal and signal-to-noise ratio values from the microarray images. Bad spots (<0.5 % of genes) flagged by the software were removed. The assay signal was normalized across arrays using quantile-quantile normalization with R-script from Applied Biosystems (27). Signal-to-noise ratios of less than 3.0 and flagged genes with values greater than 5000 were eliminated from the analysis. The normalized, log-transformed, background-corrected data were then analyzed using Spotfire DecisionSite Software v16.0 (Spotfire, Somerville, MA). Fold changes were calculated for each gene using the normalized signal values. Hybridization data and parameter information were deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under the GEO series accession number GSE6507.
Bioinformatics approach to candidate gene identification
The Protein ANalysis THrough Evolutionary Relationships (PANTHER) gene ontology and classification data base (www.pantherdb.org) was used to identify perturbed signaling pathways and biological processes. This system classifies genes by their functions based on published reports, and uses evolutionary relationships to predict function in the absence of direct experimental evidence. Gene information and sequences were obtained from PANTHER and EMBL V31 (http://www.ensembl.org). A manual inspection of genes within the QTL region, to identify potential candidates not captured by PANTHER analysis, revealed that genes within the glyoxalase system were over-expressed in congenics. This observation led to an examination of genes upstream of the glyoxalase pathway and the discovery of several glycolytic genes with increased expression in congenic mice.
Selection of candidate genes for validation
The chromosomal location of genes was determined using AB annotation. Because genetic variation underlying the nutrient intake traits resides within the QTL, a search of the literature was carried out for all differentially expressed genes in this region with possible physiological relevance, e.g., those pertaining to carbohydrate or lipid metabolism, the regulation of food intake, peripheral sensing pathways, or energy homeostasis. QTL region genes with the highest fold differences, as well as any receptors or receptor signaling genes, were also selected.
The CAST/Ei and C57BL/6J strains are highly polymorphic. Because commercial microarray probes are usually designed for B6 DNA sequence, they may not be suitable for CAST/Ei. To overcome this problem, we validated the gene expression results with qPCR using SYBR green, with primers that amplified a region different from the probe binding site. In this way, false positives in the microarray results that could be due to a SNP(s) in the probe binding site were eliminated.
Quantitative real-time RT-PCR
Expression candidates were validated by RT-PCR by means of an AB Prism 7900HT Sequence Detection System using SYBR green and gene-specific primers designed using Primer Express software (AB). Briefly, each 30 µl reaction volume consisted of 1 µl of cDNA from 100 µl cDNA synthesized from 1 µg of total RNA, 30 pmoles of forward primer, and 30 pmoles of reverse primer in 2x SYBR green master mix (AB). Relative mRNA levels were determined in duplicate samples using a standard curve generated by a pool of RNA samples and normalized to values of the control gene Ppia. Ppia was chosen as the housekeeping gene because it does not show a constitutive difference in the strains and tissues of interest.
Western blots
Protein was extracted from the liver of B6.CAST-17 and WT mice. Approx. 500 mg of tissue was powdered in liquid nitrogen, homogenized and sonicated in 500 µl RIPA buffer containing 100 mM Tris-HCl (pH 8.0), 0.1 % SDS, 150mM NaCl, 50 mM EDTA, 0.1 % NP40, 0.1 % sodium deoxycholate, 100mM beta glycerophosphate and 50 µl of protease inhibitor cocktail (Sigma). ~200 µg of protein was separated in 12% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Millipore) for immunoblotting. The membrane was blocked in Odyssey buffer (LI-COR Bioscience, Lincoln, NE). The membrane was immunoblotted with antibody to Glo1 (kindly provided by Dr. Kenneth Tew) or mouse β-actin (Abcam, Cambridge, MA) overnight at 4 °C, followed by fluorescent-labeled secondary antibody (IRDye800™ or Cy5.5) for 1 h. Signal intensity was normalized with β-actin and quantified using the Odyssey imaging system (LI-COR Bioscience, Lincoln, NE).
Determination of glyoxalase 1 activity
Glyoxalase 1 activity was measured according to the method of Han et al. (28). An assay mixture containing 1 ml of glyoxalase buffer at pH 7.0 (0.028 M NaHPO4, 0.016 M KH2PO4, 0.1 M KCl, 0.01 M MgCl2-6H20), 20 µl of 0.316 M purified methylglyoxal (graciously provided by Dr. Dave Vander Jagt), and 20 µl of 0.1 M GSH (Sigma) was incubated for 2 min at room temperature to form hemithioacetal. Initial OD240 was measured. Next, 2 µl of blood sample was added and the enzymatic production of S-lactoylglutathione (ε3.37 mM−1 cm−1) was measured at 240 nm at 15 sec interval for 3 min. Specific activity was expressed as µmol S-lactoylglutathione formed/min/mg protein.
RESULTS
Macronutrient diet selection phenotypes
Consistent with our original linkage data (Fig. 2; ref. 17), the B6.CAST-17 congenic strain (n=20) consumed significantly more C/P kcal (34%) and total kcal (20%) per body weight on Day 2, but not on Day1 of the 10 d phenotyping period, when compared with wildtype B6 (n=25) (Fig. 2). By contrast, there was no strain difference in F/P intake. All mice were then placed back on chow for 6 wks. Next, a subset of mice with high and low phenotypic values were returned to the 2-choice diet for one day while the remaining animals were kept on chow (Expt. 1). The mice selected for microarray analysis in Expt. 2 (C/P vs. F/P diet) exhibited similar intakes in both the 10 d and 2 d phenotyping periods (Pearson r = 0.71, P < 0.01, df=12-2).
Expression of QTL transcripts on chow
Microarray analysis revealed expression differences in the hypothalamus and liver between BC-17 congenic and littermate wildtype B6 mice. The effects of diet on gene expression in these strains were studied by making qualitative comparisons of the two datasets obtained from separate sets of animals. In hypothalamus, 31 out of 375 expressed genes were differentially expressed within the QTL segment. Of these, 18 genes were upregulated and 13 were downregulated in congenic mice compared with WT (Table 3 & Table 4). In liver, 54 out of 346 expressed genes were differential between the two strains, of which 32 were downregulated and 22 were upregulated in congenic mice (Table 1 & Table 2).
Table 3.
QTL region genes that were differentially expressed in hypothalamus.
| Probe ID |
Gene name | UniGene ID | Chow | Macronutrient selection diet |
|---|---|---|---|---|
| 825725 | t-complex testis expressed 1 | Mm 948 | −3.58 | 2.60 |
| 797869 | Testis specific gene A2 | Mm 2743 | −3.24 | 4.82 |
| 667698 | Unassigned | Mm.195061 | −2.87 | 1.76 |
| 919579 | Cytochrome P450 family 4 subfamily f polypeptide 15 | Mm.26539 | −2.7 | 2.0 |
| 430779 | Unassigned | Mm.33042 | −2.48 | 1.61 |
| 595996 | Ribosomal protein L7-like 1 | Mm.218533 | −2.13 | 1.73 |
| 619090 | RAB26 member RAS oncogene family | Mm.266033 | −2.03 | 1.56 |
| 353516 | RIKEN cDNA B230354K17 gene | Mm.206588 | −1.93 | 1.99 |
| 905863 | Rab/GAB TBC related | Mm.301585 | −1.69 | 2.20 |
| 505490 | Zinc finger protein 40 | Mm.21025 | −1.56 | −3.95 |
| 880965 | Heat shock protein | Mm.372314 | −1.46 | 1.79 |
| 846025 | Unassigned | Mm.258491 | −1.38 | −1.74 |
| 904144 | Ribosomal protein S2 | Mm.362197 | −1.36 | 1.57 |
| 529509 | Zinc finger and BTB domain containing 12 | Mm 61119 | −1.3 | −1.76 |
| 919204 | RIKEN cDNA 1200007D18 gene | Mm.30496 | −1.29 | −2.90 |
| 896599 | Ribosomal protein S2 | Mm.328846 | −1.29 | 1.52 |
| 338441 | Methylmalonyl-Coenzyme A mutase | Mm.259884 | −1.23 | 1.61 |
| 841838 | Zinc finger protein 40 | Mm.21025 | −1.16 | −1.68 |
| 558496 | Unassigned | Mm.35016 | −1.16 | −1.85 |
| 919791 | Phosphodiesterase 10A | Mm.87161 | −1.03 | −1.50 |
| 432416 | Pre B-cell leukemia transcription factor 2 | Mm.7103 | 2.17 | 2.07 |
| 742093 | RIKEN cDNA 2400010G15 gene | Mm 2787 | 2.27 | −1.62 |
| 558559 | BTB (POZ) domain containing 9 | Mm.41220 | 2.38 | −1.51 |
| 418927 | N-acetylglucosamine-1-phosphotransferase gamma | Mm.38607 | 2.46 | −1.56 |
| 869343 | Radial spokehead-like 2 | Mm.359991 | 2.47 | −2.40 |
| 465717 | Mitochondrial carrier homolog 1 (C. elegans) | Mm.285322 | 2.66 | −2.27 |
| 915442 | Glyoxalase 1 | Mm.261984 | 2.71 | 1.59 |
| 741537 | Histocompatibility 2 K1 K region | Mm 6771 | 3.07 | −2.09 |
| 458119 | Phospholipase C-like 2 | Mm.217362 | 3.26 | −1.59 |
| 925126 | Trinucleotide repeat containing 5 | Mm.240325 | 3.48 | −1.94 |
| 768402 | cDNA sequence BC008155 | Mm.26783 | 3.5 | −1.87 |
| 742757 | Cullin 7 | Mm.329078 | 3.74 | −1.54 |
| 706669 | RIKEN cDNA 1500032D16 gene | Mm.28349 | 4.11 | 2.80 |
| 370857 | Ectonucleotide pyrophosphatase/phosphodiesterase 5 | Mm.30145 | 7.43 | −2.47 |
| 811338 | RAB5A member RAS oncogene family | Mm.329123 | 14.9 | −1.71 |
| 761588 | TATA box binding protein (TBP)-associated factor | Mm.267998 | 35.9 | −3.47 |
Table 4.
QTL region genes that were differentially expressed only in chow or macronutrient selection diet in hypothalamus.
| Probe ID |
Gene Name | UniGene ID | Fold Change |
|---|---|---|---|
| Chow | |||
| 548781 | Riken cDNA 1700110C19 | −2.23 | |
| 790322 | Unassigned | −1.57 | |
| 485013 | Nurim (nuclear envelope membrane protein) | Mm.279713 | 2.48 |
| Macronutrient selection diet | |||
| 853033 | Unassigned | −13.80 | |
| 871613 | Cytochrome P450, family 4, subfamily f, polypeptide 14 | Mm.426027 | −12.23 |
| 628220 | Histocompatibility 2, T region locus 24 | Mm.14573 | −8.83 |
| 795040 | CD2 associated protein | Mm.218637 | −3.53 |
| 906572 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | Mm.57043 | −2.65 |
| 350018 | Ubiquitin protein ligase E3 component n-recognin 2 | Mm.28234 | −2.56 |
| 731526 | RIKEN cDNA D130084M03 gene | Mm.130244 | −2.45 |
| 638437 | Hypothetical protein LOC638437 | −2.31 | |
| 300048 | Phospholipase A2, group VII | Mm.9277 | −2.27 |
| 561631 | Expressed sequence AI662250 | Mm.37290 | −2 60 |
| 932548 | 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) | Mm.8684 | −2.10 |
| 333538 | RIKEN cDNA 9030612M13 gene | Mm.38813 | −1.97 |
| 421019 | Carbonic anhydrase 15 | Mm.379878 | −1.94 |
| 647051 | Unassigned | −1.90 | |
| 764311 | DNA segment, Chr 17, Wayne State University 104, expressed | Mm.294870 | −1.88 |
| 928306 | Transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) | Mm.14814 | −1.86 |
| 734426 | CASK interacting protein 1 | Mm.70989 | −1.85 |
| 885645 | Cytochrome p450, subfamily 4F | −1.77 | |
| 571554 | Tumor necrosis factor (ligand) superfamily, member 9 | Mm.41171 | −1.76 |
| 834440 | cDNA sequence BC052046 | Mm.234640 | −1.74 |
| 581260 | Mannose-6-phosphate receptor binding protein 1 | Mm.311696 | −1.73 |
| 772704 | RIKEN cDNA 4932442K08 gene | Mm.35039 | −1.72 |
| 580017 | RIKEN cDNA 0610007P22 gene | Mm.180687 | −1.72 |
| 898923 | Gem (nuclear organelle) associated protein 6 | Mm.279985 | −1.68 |
| 439922 | Regulator of G-protein signaling 11 | Mm.109840 | −1.65 |
| 618358 | Tripartite motif protein 39 | Mm.40624 | −1.64 |
| 766610 | H2-K region expressed gene 2 | Mm.2948 | −1.61 |
| 835651 | Tumor necrosis factor receptor superfamily, member21 | Mm.200792 | −1.61 |
| 424542 | Alanyl-tRNA synthetase like | Mm.329063 | −1.58 |
| 639092 | RIKEN cDNA 9030612M13 gene | Mm.38813 | −1.54 |
| 454588 | AT rich interactive domain 1B | Mm.133401 | −1.53 |
| 672192 | Hypothetical gene supported by AK086736 | Mm.237134 | −1.52 |
| 766979 | Gene model 942 (NCBI) | Mm.329216 | −1.52 |
| 673839 | ATP-binding cassette, sub-family A, member 3 | Mm.250280 | −1.51 |
| 326220 | Mitochondrial ribosomal protein S10 | Mm.216250 | −1.51 |
| 795197 | Transducin (beta)-like 3 | Mm.100353 | −1.51 |
| 748793 | WD repeat domain 27 | Mm.379200 | 1.50 |
| 842988 | General transcription factor II H, polypeptide 4 | Mm.10182 | 1.51 |
| 915872 | Thrombospondin 2 | Mm.26688 | 1.52 |
| 850068 | Fibronectin type III domain containing 1 | Mm.379161 | 1.55 |
| 887858 | Protein phosphatase 1, regulatory subunit 10 | Mm.29385 | 1.55 |
| 414701 | Nuclear prelamin A recognition factor-like | Mm.24201 | 1.55 |
| 346435 | Lymphocyte antigen 6 complex, locus G5C | Mm.222643 | 1.56 |
| 490554 | Zinc finger protein 422, related sequence 1 | Mm.276296 | 1.56 |
| 344295 | SRY-box containing gene 8 | Mm.258220 | 1.57 |
| 656267 | RIKEN cDNA C230029D21 gene | Mm.373631 | 1.59 |
| 732848 | Histocompatibility 2, blastocyst | Mm.34289 | 1.60 |
| 801941 | Wilms’ tumour 1-associating protein | Mm.275521 | 1.72 |
| 750011 | Cytochrome P450, family 4, subfamily f, polypeptide 13 | Mm.254838 | 1.75 |
| 478237 | RIKEN cDNA A730055L17 gene | Mm.358998 | 1.81 |
| 586542 | Zinc finger protein 161 | Mm.29434 | 1.81 |
| 704678 | Lipin 2 | Mm.227924 | 1.88 |
| 773606 | Riken cDNA C530047H08 gene | Mm.124373 | 1.97 |
| 577870 | Mannosidase 2, alpha 1 | Mm.2433 | 2.37 |
| 298384 | Unassigned | Mm.268383 | 2.60 |
| 371577 | Transcription factor 19 | Mm.11434 | 3.03 |
| 602187 | t-complex-associated testis expressed 1 | Mm.1290 | 3.32 |
Table 1.
QTL region genes that were differentially expressed in liver (positive value in fold difference indicates an increase, and negative value indicates a decrease in congenic mice. Mm, mus musculus.
| Probe ID |
Gene Name | UniGene ID | Chow | Macronutrient selection diet |
|---|---|---|---|---|
| 716213 | 2-4-dienoyl-Coenzyme A reductase 2, peroxisomal | Mm.292869 | −6.29 | −4.93 |
| 761588 | TAF11 RNA polymerase II TATA box binding protein (TBP)-associated factor | Mm.267998 | −4.20 | −3.08 |
| 871613 | Cytochrome P450 family 4 subfamily f polypeptide 14 | Mm. 426027 | −3.42 | −2.95 |
| 925126 | Trinucleotide repeat containing 5 | Mm.240325 | −3.25 | −1.98 |
| 804439 | Apolipoprotein M | Mm.2161 | −3.09 | −2.92 |
| 460449 | Anaplastic lymphoma kinase | Mm.2536 | −2.66 | −1.59 |
| 885645 | Cytochrome P450 subfamily 4F | −2.51 | −2.00 | |
| 701966 | Lymphocyte antigen 6 complex locus G6C | Mm.215096 | −2.34 | −2.12 |
| 660078 | Unassigned | −2.25 | −2.09 | |
| 928591 | Histocompatibility 2 Q region locus 10;H2-Q10 | Mm.88795 | −2.21 | −2.26 |
| 467910 | Butyrophilin-like 1 | Mm.46284 | −2.18 | −1.84 |
| 333538 | RIKEN cDNA 9030612M13 gene | Mm.38813 | −2.12 | −3.82 |
| 707182 | Polycystic kidney disease 1 homolog | Mm.290442 | −2.07 | −1.65 |
| 356012 | Myeloid/lymphoid or mixed lineage-leukemia translocation to 1 homolog (Drosophila) | Mm.148748 | −1.97 | −1.56 |
| 747504 | Sema domain, transmembrane domain, and cytoplasmic domain (semaphorin) 6B | Mm.8029 | −1.92 | −1.71 |
| 749204 | Acetyl-CoA C-acyltransferase 2 | Mm.229342 | −1.91 | −2.02 |
| 634113 | Vitrin | Mm.292983 | −1.91 | −1.54 |
| 477069 | Fibroblast growth factor receptor substrate 3 | Mm.89912 | −1.87 | −1.60 |
| 561631 | Expressed sequence AI662250 | Mm.37290 | −1.82 | −1.72 |
| 640886 | Transmembrane protein 8 | Mm.304656 | −1.75 | 1.75 |
| 581260 | Mannose-6-phosphate receptor binding protein 1 | Mm.311696 | −1.75 | −2.45 |
| 914762 | G protein-coupled receptor 108 | Mm.28468 | −1.68 | −1.66 |
| 706669 | RIKEN cDNA 1500032D16 gene | Mm.28349 | −1.63 | −2.19 |
| 741537 | Histocompatibility 2 K1 K region | Mm.16771 | −1.61 | −1.79 |
| 619946 | FK506 binding protein-like | Mm.10025 | −1.59 | −1.87 |
| 742757 | Cullin 7 | Mm.329078 | −1.58 | −2.09 |
| 632987 | RIKEN cDNA 4930539E08 gene | Mm.248931 | −1.57 | −1.85 |
| 932548 | 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase alpha) | Mm.8684 | −1.52 | −1.75 |
| 592477 | Fibrillarin | Mm.379878 | −1.51 | −1.79 |
| 433605 | LEM domain containing 2 | Mm.29689 | −1.45 | −1.69 |
| 637912 | Histocompatibility 2 class II antigen A beta 1 | Mm.254067 | −1.44 | −2.22 |
| 807637 | Histocompatibility 2 M region locus 3 | Mm.14437 | −1.44 | −1.65 |
| 350018 | Ubiquitin protein ligase E3 component n-recognin 2 | Mm.28234 | −1.42 | −2.23 |
| 801941 | Wilms’ tumour 1-associating protein | Mm.275521 | −1.38 | −1.66 |
| 309261 | RAB11B member RAS oncogene family | Mm.35727 | −1.36 | −2.00 |
| 669667 | Unassigned | Mm.276405 | −1.33 | −1.65 |
| 425013 | RIKEN cDNA 1110018J12 gene | Mm.271817 | −1.31 | −1.65 |
| 493546 | F-box and leucine-rich repeat protein 17 | Mm.379894 | −1.22 | 1.81 |
| 764339 | RD RNA-binding protein | Mm.279907 | −1.20 | −1.69 |
| 904214 | Peptidyl-prolyl cis-trans isomerase A | −1.16 | −1.62 | |
| 479998 | EH-domain containing 3 | Mm.18526 | −1.13 | −1.56 |
| 864277 | Mitochondrial ribosomal protein L18 | Mm.290166 | −1.07 | −1.51 |
| 776724 | RIKEN cDNA 1700022C21 gene | Mm.26684 | −1.05 | −1.63 |
| 924756 | TEA domain family member 3 | Mm.6655 | −1.03 | 3.58 |
| 582898 | Unassigned | 1.03 | −1.80 | |
| 824604 | Ankyrin repeat and SAM domain containing 1 | Mm.32556 | 1.04 | 2.06 |
| 827226 | Insulin-like growth factor 2 receptor | Mm.213226 | 1.12 | 1.60 |
| 424542 | Alanyl-tRNA synthetase like | Mm.329063 | 1.13 | 1.99 |
| 672227 | ATP-binding cassette sub-family G member 5 | Mm.289590 | 1.20 | 1.86 |
| 816939 | Molybdenum cofactor synthesis 1 | Mm.22256 | 1.21 | −2.32 |
| 725803 | Multiple coagulation factor deficiency 2 | Mm.30251 | 1.24 | 1.65 |
| 710361 | High mobility group AT-hook 1 | Mm.4438 | 1.25 | 2.82 |
| 311119 | Cell division cycle 5-like (S. pombe) | Mm.28270 | 1.26 | 1.74 |
| 750011 | Cytochrome P450 family 4 subfamily f polypeptide 13 | Mm.254838 | 1.26 | 1.61 |
| 730857 | Serine/arginine repetitive matrix 2 | Mm.5222 | 1.30 | −1.53 |
| 915442 | Glyoxalase 1 | Mm.261984 | 1.32 | 1.66 |
| 631849 | Protein phosphatase 1 regulatory (inhibitor) subunit 11 | Mm.46176 | 1.50 | 1.63 |
| 735476 | HLA-B-associated transcript 1A | Mm.126043 | 1.51 | 1.64 |
| 585136 | KH-type splicing regulatory protein | 1.67 | 1.64 | |
| 926995 | Zinc finger protein 598 | Mm.219581 | 1.67 | 1.68 |
| 323513 | UDP-Gal:betaGalNAc beta 1 3-galactosyltransferase polypeptide 4 | Mm.11132 | 1.69 | 2.14 |
| 795197 | Transducin (beta)-like 3 | Mm.100353 | 1.73 | 1.87 |
| 353516 | RIKEN cDNA B230354K17 gene | Mm.206588 | 1.76 | 1.72 |
| 448315 | Expressed sequence AW049765 | Mm.204991 | 1.77 | 1.57 |
| 846025 | Differentially expressed in FDCP 6 | Mm.267359 | 1.80 | 1.65 |
| 621252 | Vascular endothelial growth factor A | Mm.282184 | 1.92 | 1.55 |
| 905863 | RAB/GAB related | Mm.301585 | 1.94 | 2.71 |
| 424409 | Cystathionine beta-synthase | Mm.206417 | 2.04 | 1.76 |
| 661148 | Mesothelin | Mm.17510 | 2.16 | 1.75 |
| 457113 | RIKEN cDNA 4930528F23 gene | Mm.52592 | 2.38 | 2.17 |
| 441362 | Sulfotransferase family cytosolic 1C member 2 | Mm.19320 | 4.71 | 2.67 |
| 856010 | Trefoil factor 3 | Mm.4641 | 79.74 | 9.61 |
Table 2.
QTL region genes that were differentially expressed only in chow or in macronutrient selection diet in liver.
| Probe ID |
Gene Name | UniGene ID | Fold Change |
|---|---|---|---|
| Chow | |||
| 431279 | Histocompatibility 2, class II antigen A, alpha | Mm.235338 | −3.54 |
| 848612 | Phosphatidylinositol glycan, Class W | −2.00 | |
| 542645 | Olfactory receptor 121 | Mm.223020 | −1.82 |
| 898280 | DNA segment, Chr 17, ERATO Doi 441, expressed | Mm.371601 | −1.53 |
| 841838 | Zinc finger protein 40 | Mm.21025 | 1.52 |
| 506800 | Triggering receptor expressed on myeloid cells-like 4 | Mm.131234 | 1.61 |
| 820967 | BING4 protein | Mm.2437 | 1.65 |
| 508816 | Unassigned | Mm.26553 | 1.98 |
| 697656 | Protein containing single MORN motif in testis | Mm.45208 | 2.03 |
| 586949 | Histocompatibility 2, O region alpha locus | Mm.116 | 2.29 |
| Macronutrient selection diet | |||
| 650378 | Histocompatibility 2, blastocyst | Mm.34289 | 5.06 |
| 618358 | Tripartite motif protein 39 | Mm.40624 | 3.60 |
| 729309 | Coiled-coil alpha-helical rod protein 1 | Mm.49544 | 2.38 |
| 897853 | PHD finger protein 20-related | 2.33 | |
| 706765 | Expressed sequence AW494914 | Mm.12412 | 1.88 |
| 436920 | Chloride intracellular channel 5 | Mm.37666 | 1.71 |
| 326220 | Mitochondrial ribosomal protein S10 | Mm.358901 | 1.69 |
| 502359 | cDNA sequence BC027072 | Mm.134151 | 1.65 |
| 740898 | cDNA sequence BC051226 | Mm.250393 | 1.64 |
| 452215 | DNA segment, Chr 17, Wayne State University 155, expressed | Mm.303441 | 1.62 |
| 324675 | Olfactory receptor 94 | Mm.377382 | 1.56 |
| 580896 | Ribosomal protein L10a | Mm.347060 | −1.58 |
| 599654 | Gametogenetin binding protein 1 | Mm.370478 | −1.59 |
| 682803 | Vomeronasal 2, receptor, 6 | Mm.246564 | −1.61 |
| 919265 | RIKEN cDNA 1600012H06 gene | Mm.28544 | −1.61 |
| 906020 | Phosphodiesterase 9A | Mm.10812 | −1.66 |
| 795040 | CD2 associated protein | Mm.218637 | −1.69 |
| 697457 | Chromosome transmission fidelity factor 18 homolog | Mm.102540 | −2.20 |
| 780705 | Unassigned | −2.89 | |
| 854091 | Triggering receptor expressed on myeloid cells-like 1 | Mm.90177 | −3.07 |
Expression of QTL transcripts on macronutrient selection diet
In hypothalamus, out of 426 expressed genes within the QTL interval, 38 were upregulated and 56 were downregulated in congenic mice compared with WT. In liver, 52 genes were downregulated and 39 genes were upregulated in congenic mice out of 364 expressed genes.
Expression of QTL transcripts on both chow and macronutrient selection diet
In the hypothalamus, only one metabolic gene, Glo1 (glyoxalase 1), was differentially expressed between strains in both diet experiments. In liver, all the metabolic genes that were altered in chow-fed animals were also regulated in mice on macronutrient selection diet. A number of genes involved in fat metabolism were downregulated in carbohydrate-preferring, congenic mice including: ApoM, (apolipoprotein M), Acat2 (acetyl-CoA C-acyltransferase 2), Agpat1 (acylglycerol-3-phosphate O-acyltransferase 1), and Decr2 (2–4-dienoyl-Coenzyme A reductase 2, peroxisomal). By contrast, several genes associated with carbohydrate metabolism were upregulated in the congenic strain including: Glo1 (glyoxalase 1), Glo2 (glyoxalase 2), Igf2r (insulin-like growth factor 2 receptor), and M6pbp4 (mannose-6-phosphate receptor binding protein 1) (downregulated). Other differentially expressed metabolic genes were: B3galt4 (UDP- beta 1,3-galactosyltransferase, polypeptide 4) (increased), and Cyp4f14 (cytochrome p450, family4; subfamily f) (decreased). Expression differences were also found for several transcription factors, i.e., Taf11 (TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor), Rnaset2 (ribonuclease T2), Rps6Ka2 (ribosomal protein S6 kinase, polypeptide 2), and Tead3 (transcription enhancer factor domain family member 3).
Glyoxalase pathway and Glo1 expression analyses
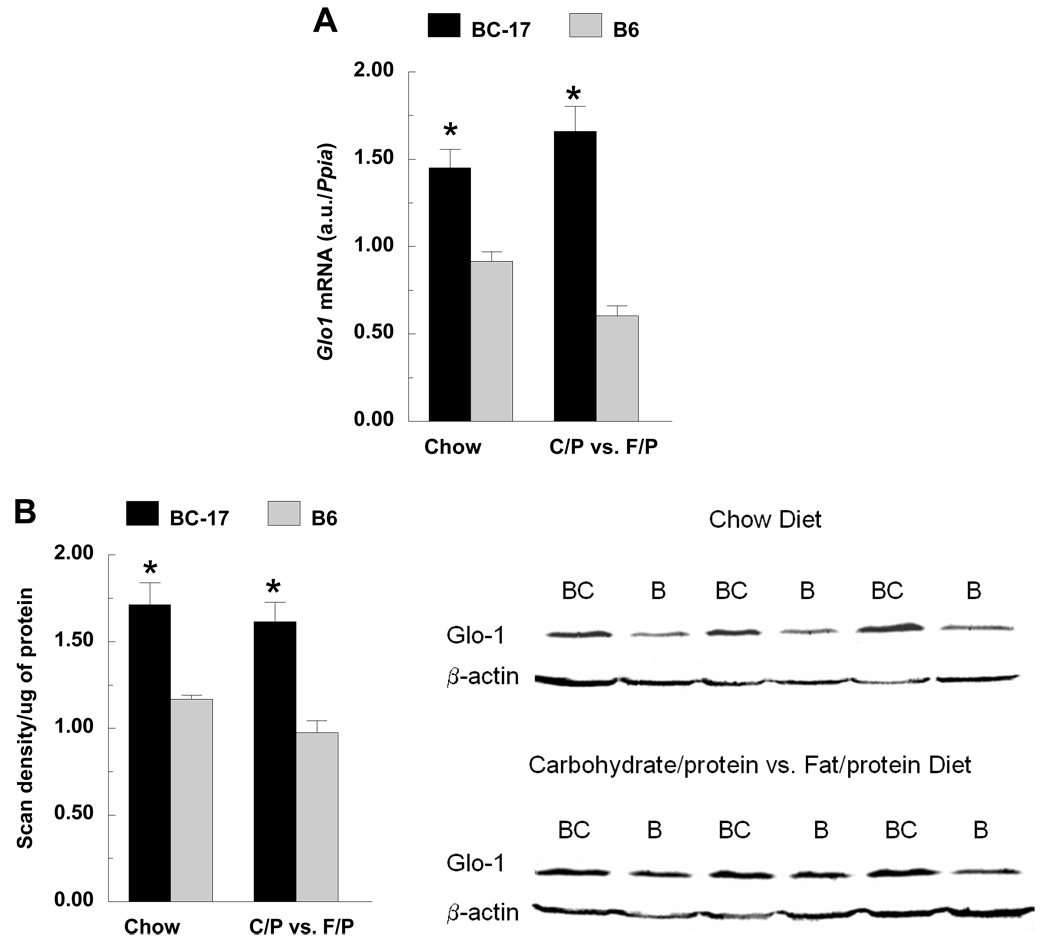
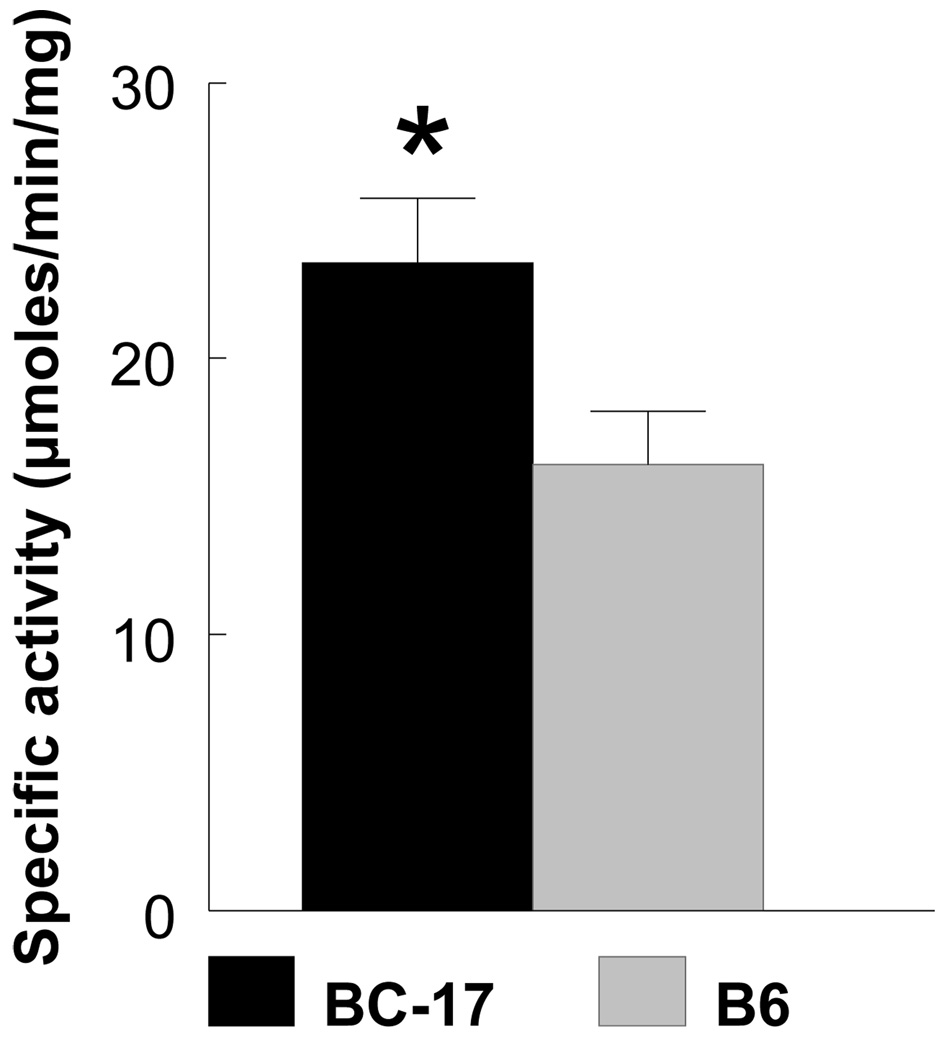
Transcriptional profiling revealed an upregulation of genes in the glyoxalase pathway including Glo1 and Glo2 in liver and hypothalamus, and dLDH (D-lactate dehydrogenase) in hypothalamus. Based on their biological function and location near the peak of the QTL, further analyses of Glo1 and Glo2 were conducted. Using qRT-PCR, Glo1 was upregulated by 1.6- (chow) and 3-fold (macronutrient selection) in B6.CAST-17 congenic mice compared with wildtype controls (Fig. 5A). Western blot analyses of Glo1 in liver yielded a single ~21 kDa band in both B6.CAST-17 congenic and wildtype B6 mice corresponding to the expected molecular weight of 20.7 kDa (Fig. 5B). Consistent with the observed strain difference in Glo1 mRNA and protein expression, congenic mice displayed higher enzymatic activity compared with WT (23.5 ± 2.3 vs. 16.1 ± 1.9 µmol/min/mg, P < 0.05) (Fig. 6). To investigate whether increased Glo1 expression in the congenic strain was primary or a result of high carbohydrate feeding, we fed the animals a high- (73%) or low-carbohydrate (26%) diet for two days and then examined Glo1 mRNA levels using qPCR. Within strain, we found no differences in Glo1 expression as a function of low- or high-carbohydrate feeding (Fig. 7). Morever, the strain difference observed on either diet was of similar magnitude both in hypothalamus (3–3.5 fold) and in liver (2.4–2.8 fold).
Fig. 5.
Expression pattern of Glo1 in mouse liver. A) qRT-PCR expression of Glo1 mRNA in wildtype B6 and homozygous B6.CAST-17 (BC-17) congenic mice. Data are expressed as mean ± S.E. from 7–11 mice per strain, per diet, analyzed in duplicate. B) Western blot analyses of liver yielded a single ~20 kDa band in wildtype B6 (B) and homozygous B6.CAST-17 (BC) corresponding to the known molecular weight of 21 kDa. Mouse β-actin antibody was applied as a loading control. *P < 0.01; #P < 0.005. Glo1, glyoxalase 1.
Fig. 6.
Glyoxalase 1 enzyme activity in the blood of wildtype B6 (n = 11) and homozygous B6.CAST-17 (BC-17) congenic (n = 7) mice. Activity was measured as the rate of production of S-lactoylglutathione (ε3.37 mM−1 cm−1) at 240 nm at 15 sec interval for 3 min and corrected for background by subtracting the OD240, measured before adding blood. Specific activity was expressed as µmol S-lactoylglutathione formed/min/mg protein. *P < 0.05.
Expression of QTL transcripts unique to chow
Out of ten genes that were differentially expressed in liver and unique to chow, none appeared to be promising candidates with respect to gene classification or known biological functions (Table 2). Of the three differential genes in hypothalamus that were unique to chow, two were unknown and one encodes for a nuclear envelope membrane protein (Nurim) of speculative function.
Expression of QTL transcripts unique to macronutrient selection diet
In the hypothalamus, 57 regulated genes were found to be unique to macronutrient selection diet, including a regulator of G-protein signaling (Rgs11; ref. 29) and several metabolic genes: Agpat1(1-acylglycerol-3-phosphate O-acyltransferase 1), Lpin2 (lipin 2), M6pbp4 (mannose-6-phosphate receptor binding protein 1), and Man2a1 (mannosidase 2, alpha 1) (Table 4). Recombinant lipin-2 has been shown to have phosphatidate phosphatase type-1 (PAP1) activity, which has a key role in glycerolipid synthesis (30). Also unique to the 2-choice diet was Probe ID #853033, an EST that was down-regulated by >13 fold in the hypothalamus of congenic mice (Table 4). In liver, 20 regulated genes were unique to the 2-choice diet including several ESTs and uncharacterized genes, but interestingly, none of these genes are involved in metabolism.
Gene expression differences outside the QTL
By comparing gene expression in the hypothalamus of congenic and WT mice while on chow diet, we identified 936 genes that were up-regulated and 996 genes that were down-regulated. In the liver, 775 genes were over-expressed and 812 genes were under-expressed in congenic mice. On macronutrient selection diet, 618 genes were over-expressed in the hypothalamus of congenic mice and 647 downregulated. In liver, 875 genes were over-expressed in congenic mice and 1019 downregulated.
Glycolysis pathway
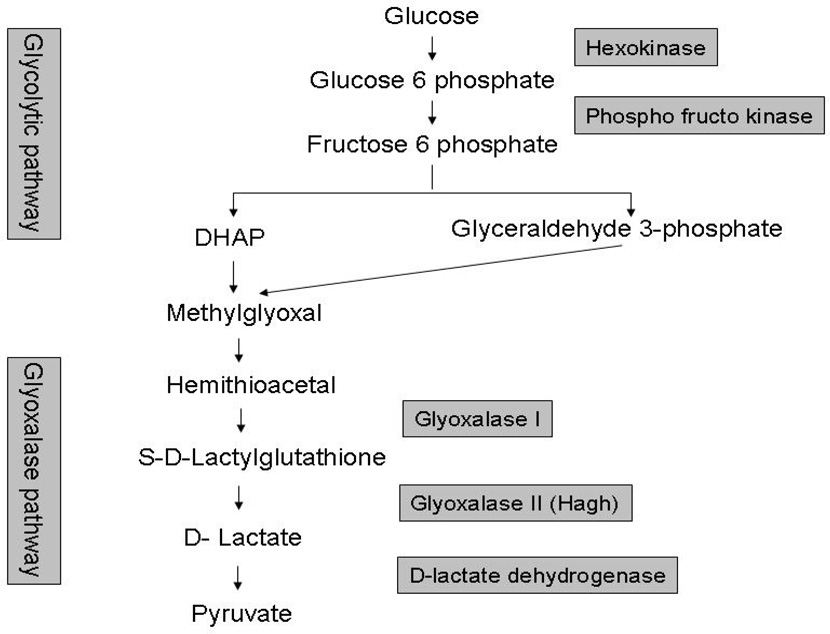
Our finding that the glyoxalase system was activated in B6.CAST-17 congenic mice led to an examination of genes upstream of this pathway, which revealed that several glycolytic genes outside the QTL were upregulated in this strain, in the macronutrient diet protocol, but not with chow diet (Table 6). Specifically, all genes that code for enzymes involved in the conversion of glucose to 3-carbon groups (glyceraldehyde 3 phosphate-G3P and di-hydroxy acetone phosphate-DHAP) were upregulated in the congenic strain (Fig. 4).
Table 6.
Differentially expressed glycolysis genes in liver, in the macronutrient selection diet protocol.
| Probe ID |
Gene Name | Fold change |
UniGene |
|---|---|---|---|
| 434034 | Hexokinase | 1.48 | Mm.196605 |
| 434336 | Phospho fructo kinase | 1.42 | Mm.132391 |
| 429576 | Aldolase 3C isoform | 1.48 | Mm.7729 |
| 486053 | Triose phosphate isomerase | −1.14 | Mm.391520 |
| 903599 | Glyceraldehyde 3 phosphate dehydrogenase | 1.39 | Mm.317779 |
| 336724 | Pyruvate kinase | 2.19 | Mm.8359 |
| 915442 | Glyoxalase 1 | 1.66 | Mm.261984 |
| 613660 | Glyoxalase 2 (Hagh) | 1.31 | Mm.43784 |
| 902722 | Glucose-6-phosphate dehydrogenase X-linked | 1.73 | Mm.27210 |
Fig. 4.
Pathways involved in glycolysis and the activation of the glyoxalase system. Methylglyoxal from glyceraldehydes-3-phosphate and dihydroxyacetone-phosphate (DHA-P) is converted to d-lactate by glyoxalase enzymes. D-lactate dehydrogenase converts d-lactate to pyruvate. Genes in shaded boxes were upregulated in the liver of B6.CAST-17 congenic mice.
Wnt signaling pathway
Several genes representing Wnt signaling and located outside the QTL, showed altered expression in liver, i.e., Hoxc4, Ccnd3, Fzd7, Wnt3a, Wnt10b, and Csnk1. The Wnt transcripts Wnt10b, Fzd8, Wnt3, and Wnt6 were down-regulated in congenic mice with macronutrient selection diet but not with chow. Wnt10b was shown to decrease oxygen consumption in FABP4-Wnt10b mice although an effect on food intake has not been demonstrated (30). The Wnt pathway has been linked to glycogen metabolism (32).
PDGF (platelet-derived growth factor) signaling
Expression differences were noted in only four PDGF signaling genes in chow-fed mice (Vav2, Arhgap1, Srf, and Nck), but after 24 h of macronutrient diet selection, sixteen were differentially regulated between strains in both liver and hypothalamus, i.e., Stat1, Pkn3, Srgap2, Vav2, Mapk1, Mapk15, Mapk7, Sos1, Rps6ka2, Myc, Stard13, Jun, Rasa2, Stat5b, Arghap1, and Rab11b. Most of these changes were due to over-expression in congenics. PDGF signaling is involved in glucose transport and insulin stimulation (33), thus these patterns of transcriptional activity suggest regulatory systems invoked to support the congenic strain’s high calorie intake in the form of carbohydrate.
Induction of inflammatory genes
A number of genes involved in inflammation were differentially regulated in congenic mice, which is not surprising considering that the MHC is located within the QTL interval (Table 2 & Table 4). The most markedly upregulated gene in the congenic strain was trefoil factor 3 (Tff3) located within the QTL region, which showed a high fold induction in liver that was validated by qPCR (>9 fold). Also in liver, histocompatibility 2 and CD2 associated protein were altered. In the hypothalamus, the activated inflammatory genes included histocompatibility 2-T region locus 24, CD2 associated protein, H2-K region expressed gene 2, histocompatibility 2, and t-complex-associated genes, tumor necrosis factor (ligand) superfamily, member 9 and tumor necrosis factor receptor superfamily, member21.
Validation of the microarray data by real-time qRT-PCR
Because the genetic variation underlying nutrient intake traits resides within the QTL, a literature search was performed for all differentially regulated transcripts in this region. Candidates were chosen for independent validation based on known function, e.g., in metabolism, signaling, or based on a large fold difference in the microarray. Three uncharacterized ESTs were selected which showed enriched expression in brain. Most of the genes subjected to qRT-PCR analysis were confirmed, with the exception of Rgs11 (Regulator of G-protein signaling), Igfals (Insulin like growth factor– acid labile subunit) and Mocs (Molybdenum cofactor synthesis 1) (Table 6), while the results obtained for Glo2 were marginal (P < 0.09). The novel Transmembrane protein 8 (Tmem8) drew interest because it was regulated in opposite direction with respect to diet, but when measured by qRT-PCR, was decreased in both diet protocols by 20–40 fold in congenic mice.
DISCUSSION
Increased carbohydrate and total calorie intake share overlapping linkage on mouse chromosome 17 (17). We have combined the use of microarrays with an interval-specific congenic strain in an unbiased approach to identify candidate genes for these complex traits. Previously we reported confirmation of these linked regions in a congenic strain, and an evaluation of positional candidates (18). QTL can be manifest by a polymorphism(s) that affects either the expression level of a gene or the amount or function of a protein/gene (34). Gene expression profiling has been used successfully to find genes and identify molecular mechanisms that underlie QTLs (35). Based on our analyses, a number of genes and pathways showed altered expression which may be involved in the preferential consumption of fat or carbohydrate.
Genomic responses unique to macronutrient diet selection, assayed in hypothalamus, include the more than 2-fold decrease in Agpat1 and several ESTs. Agpat1 was decreased in the hypothalamus after 24 h of diet self-selection but was unaltered on chow. Further studies will allow us to determine the effects of diet on gene expression of these candidates and thereby achieve a more complete understanding of these complex traits (36). Acyltransferases are involved in de novo biosynthesis of glycerolipids, such as phospholipids and triacylglycerol. AGPAT exists in at least five isoforms in humans, and all catalyze the same biochemical reaction (37). Agpat1 (1-acyl-sn-glycerol 3-phosphate acyltransferase; EC 2.3.1.51), also known as lysophosphatidic acid acyltransferase (LPAAT), catalyses the conversion of lysophosphatidic acid to phosphatidic acid (PA). For example, PA can be hydrolyzed to yield diacylglycerol, a second messenger that can activate the brain-specific gamma isotype of protein kinase C (PKC gamma) (38).
Irrespective of diet, a number of genes involved in lipid metabolism were decreased in carbohydrate-preferring, congenic mice, while several genes associated with carbohydrate metabolism were increased. With respect to carbohydrate metabolism, our analyses uncovered the clear transcriptional regulation of glyoxalase in the hypothalamus and liver of B6.CAST-17 congenic mice, in response to both the high-carbohydrate chow and macronutrient selection diets. In a separate experiment, we determined that this strain difference in Glo1 expression was present, in similar magnitude, with low- as well as high-carbohydrate feeding (Fig. 7). These results provide evidence for intrinsic strain variation in Glo1 expression which may constitute a basis for the QTL effect. We next sequenced the coding region of Glo1 and observed no amino acid changes in the open reading frame (18). The position of the QTL peak over Glo1 suggests that a regulatory variant interacting with Glo1 may be located near, or within the gene.
The two primary genes in this pathway, glyoxalase 1 (Glo1) and glyoxalase 2 (Glo2) are located within the confidence interval of the Chr 17 QTL which is responsible for increased carbohydrate consumption. Glyoxalase 1 catalyzes the conversion of methylglyoxal (MG) and glutathione to S-lactoylglutathione which is then coverted to D-lactate by glyoxalase 2. MG is a metabolic by-product of glycolysis that is also produced during food processing and can be toxic if excess levels are accumulated (39). If not catalyzed by the glyoxalase system, MG participates in the glycation of proteins and nucleotides, resulting in the formation of advanced glycation end products (AGEs) that can lead to mutagenesis, apoptosis, protein degradation and induction of pro-inflammatory cytokines (40). Given that the B6.CAST-17 congenic strain consumes more carbohydrate, we would expect that it also produces a higher flux of methylglyoxal. If MG flux is high, increased Glo1 expression may be required to protect the proteome and genome. In support of this proposition, we found a significant upregulation of glyoxalase 1 mRNA and protein expression in B6.CAST-17 congenic mice. Enzymatic activity paralleled the observed strain differences in expression. We propose that increased glyoxalase activity protects this congenic strain against dietary oxidants occurring as a result of high carbohydrate consumption.
Further evidence to suggest possible higher MG production in the B6.CAST-17 congenic line was found in that carbohydrate-preferring congenic mice showed enhanced expression of glycolytic genes upstream of di-hydroxy acetone phosphate (DHAP) formation, i.e., hexakinase and phosphofructo kinase (Fig. 4 & Table 6). Once fructose 1,6-diphosphate is cleaved into DHAP and glyceraldehyde-3 phosphate, these molecules act as precursors for the non-enzymatic formation of methylglyoxal. The end product of glyoxalase action is D-lactate which is converted to pyruvate by D-lactate dehydrogenase (ldhd), a mitochondrial enzyme (41). We observed that ldhd expression was >1.5 fold higher in the hypothalamus of congenic mice, providing evidence for the conversion of excess D-lactate to pyruvate (Fig. 4).
The available data from our study and others point to a possible link between Glo1 expression and ingestive behavior. We have shown previously that the 129/J and BALBcByJ prefer dietary carbohydrate over fat, whereas C57BL/6J and DBA/2J preferentially consume fat in a macronutrient diet selection paradigm (16). A recent study found that Glo1 was upregulated in the brain of the carbohydrate-preferring 129/J and BALBcByJ strains, and decreased in the fat-preferring C57BL/6J and DBA/2J (42). Together, these results suggest an association between increased Glo1 expression and carbohydrate preference in mouse inbred strains. An association of glyoxalase 1 with feeding behavior has not been identified but may yield new insight on the functional effects of this metabolic pathway. Glo1 has also been linked to anxiety (42), although this connection has been questioned (43), and the regulation of theta oscillations during sleep (44). The physiological function of Glo1 in the brain is mostly unknown. It has been proposed that neuronal damage by excess methylglyoxal could contribute to increased anxiety (43). Further studies are needed to examine mechanisms by which Glo1 expression and activity could alter neuronal function and ultimately, behavior.
Other QTL genes with key metabolic functions in lipid metabolism were decreased in the liver of carbohydrate-preferring mice, despite the equivalent fat intake between strains. The encoded protein for Decr2 increases the conversion of delta2-trans, delta4-trans-decadienoyl coenzyme A to delta3-enoyl coenzyme A and is involved in the peroxisomal oxidation of very long chain, polyunsaturated fatty acids (45). ApoM encodes an apolipoprotein found mainly in high-density lipoproteins (HDL), that appears to play a role in the prevention of atherosclerosis possibly through its anti-inflammatory effects (46). The ACAT2 enzyme is responsible for synthesis of plasma lipoprotein cholesteryl esters. A role for any of these candidates in pathways regulating food intake has not been established. Alternatively, these results may have relevance to other QTL on mouse Chr 17 for atherosclerosis (Ath26) (47) or for obesity (Obq4) (48).
A large number of genes associated with inflammatory processes were induced in congenic mice which showed no apparent clinical signs of inflammation. However, this result could reflect a genomic response to nutrient intake. For example, the increased expression of inflammatory genes in high carbohydrate-consuming congenic mice is consistent with reports of a pro-inflammatory effect of glucose intake in human subjects (49). Thus in our model, high carbohydrate intake may alter the expression of mRNAs related to inflammation.
The highest fold difference in gene expression and confirmed by qPCR, was for the hepatic expression of trefoil factor 3 (Tff3), located in the peak of the QTL (Table 5). TFF3 is an extracellular, secretory polypeptide that resists proteolytic degradation. TFF3 is expressed in several mammalian tissues but is found predominantly in the gastrointestinal tract where it protects and contributes to healing of the mucosal barrier (50). In liver, TFF3 contributes to epithelial cell migration and wound healing (51). Interestingly, higher Tff3 expression has been reported in the liver of B6 mice when compared with Tally Ho (TH) mice, a model of diabesity (52). Consistent with this observation, Tff3 maps to Obq4, an obesity QTL on mouse chromosome 17 (48). A possible role of Tff3 in dietary obesity remains to be established, along with its expression profile in relevant tissues such as muscle and adipose.
Table 5.
Validation by qRT-PCR of selected gene candidates identified by microarray analysis.
| Gene Name | Chow | Macronutrient selection (Fat vs. carbohydrate) |
||||
|---|---|---|---|---|---|---|
| Array Fold change |
Real-time qPCR | Array Fold change |
Real-time qPCR | |||
| Fold change |
P-value | Fold change |
P-value | |||
| Hypothalamus | ||||||
| 1-acylglycerol-3-phosphate O-acyltransferase 1 | −2.1 | −1.7 | 0.041 | |||
| Probe ID: 731526* | −2.45 | −2.19 | <0.001 | |||
| Probe ID: 853033* | −13.4 | −1.7 | <0.001 | |||
| Probe ID: 905863* | 2.71 | 1.45 | <0.001 | |||
| Glyoxalase 1 | 2.71 | 2.68 | <0.001 | 1.59 | 3.05 | <0.001 |
| Regulator of G-protein signaling 11 | −1.65 | −0.04 | 0.61 | |||
| Liver | ||||||
| Apolipoprotein M | −3.09 | −2.21 | <0.001 | −2.92 | −2.1 | <0.001 |
| Cytochrome P450 family 4 subfamily f polypeptide 14 | −3.42 | −2.9 | <0.001 | −2.95 | −2.2 | <0.001 |
| 2–4-dienoyl-Coenzyme A reductase 2, peroxisomal | −6.29 | −2.6 | 0.003 | −4.93 | −1.54 | 0.002 |
| Glyoxalase 1 | 1.32 | 1.59 | <0.001 | 1.66 | 2.72 | <0.001 |
| Glyoxalase 2 | 1.2 | 1.21 | 0.017 | 1.31 | 1.42 | 0.088 |
| Trefoil factor 3 | 79.74 | 31. 1 | <0.001 | 9.61 | 23.2 | <0.001 |
| Transmembrane protein 8 | −1.75 | −23.2 | <0.001 | 1.75 | −41.1 | <0.001 |
| Insulin like growth factor– acid labile subunit | 1.42 | 1.06 | 0.82 | 1.48 | 1.1 | 0.65 |
| Molybdenum cofactor synthesis 1 | 1.20 | 1.13 | 0.67 | 2.32 | 1.06 | 0.99 |
Independent validation, using real-time RT-PCR, of selected gene candidates identified by microarray analysis. Fold change refers to the ratio of expression values of B6.CAST-17 congenic mice relative to wild type B6. Statistical significance was calculated based on normalized threshold (CT) values.
The curated gene information is not currently available.
Because the mechanisms responsible for macronutrient diet selection are unknown, the key tissue(s) underlying the phenotypes can only be inferred at this stage of analysis. These results were obtained from an examination of gene expression in the hypothalamus and liver, yet genetic and genomic data from our laboratory (18) indicate that at least one molecular component of the QTL may reside in other tissues. Specifically, the Chr 17 QTL approximate the location of Glp1r (glucagon-like peptide 1 receptor), a compelling candidate gene for this QTL with respect to its expression in antral stomach (18). Elucidating the molecular basis of complex traits such as those involving ingestive behavior will likely require the characterization of gene expression in multiple tissues, the central nervous system notwithstanding.
In conclusion, our results reveal patterns of altered gene expression, resulting from a CAST segment of proximal Chr 17, for genes involved in metabolism, inflammation, and oxidative stress, any of which could be linked to phenotypic preferences for carbohydrate or fat. Genes in the Wnt signaling and PDGF signaling pathways were also up-regulated and thus may be implicated in carbohydrate metabolism. More specifically, these data support the hypothesis that mice genetically predisposed to eat more carbohydrate and total energy per body weight are protected against the stress of dietary oxidants through induction of the glyoxalase system. Sequence and expression analyses, as well as functional studies in our mice will establish the viability of candidates such as Agpat1 and Glo1, e.g., by overexpressing and/or silencing these genes by means of lentiviral transduction.
Supplementary Material
Fig. 3.
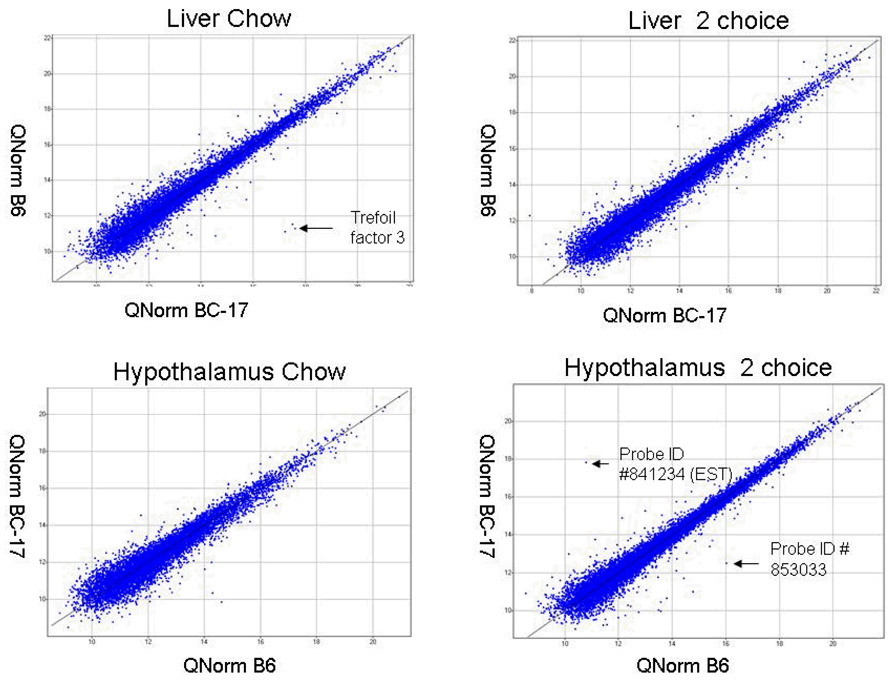
Expressed quantile normalized probes of all the microarray experiments. Quantile normalization of raw-signal intensity across arrays was done by R-Script. The normalized signals were visualized using SPOTFIRE decision site functional genomics. Probe #841234 is an EST located outside the QTL region and #853033 is within the QTL; little is known about these genes.
ACKNOWLEDGMENTS
We thank Dr. Barbara York for assistance with the tissue dissections and analyses of phenotypic data. This research was supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases to B.K.S.R. (DK-53113).
REFERENCES
- 1.Birch LL. Children’s preferences for high-fat foods. Nutr Rev. 1992;50:249–255. doi: 10.1111/j.1753-4887.1992.tb01341.x. [DOI] [PubMed] [Google Scholar]
- 2.Blundell JE, Cooling J. Routes to obesity: phenotypes, food choices and activity. Br J Nutr. 2000;83 Suppl 1:S33–S38. doi: 10.1017/s0007114500000933. [DOI] [PubMed] [Google Scholar]
- 3.de Castro JM. What are the major correlates of macronutrient selection in Western populations? Proc Nutr Soc. 1999;58:755–763. doi: 10.1017/s0029665199001032. [DOI] [PubMed] [Google Scholar]
- 4.Dreon DM, Frey-Hewitt B, Ellsworth N, Williams PT, Terry RB, Wood PD. Dietary fat:carbohydrate ratio and obesity in middle-aged men. Am J Clin Nutr. 1988;47:995–1000. doi: 10.1093/ajcn/47.6.995. [DOI] [PubMed] [Google Scholar]
- 5.Falciglia GA, Norton PA. Evidence for a genetic influence on preference for some foods. J Am Diet Assoc. 1994;94:154–158. doi: 10.1016/0002-8223(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 6.Geiselman PJ, Anderson AM, Dowdy ML, West DB, Redmann SM, Smith SR. Reliability and validity of a macronutrient self-selection paradigm and a food preference questionnaire. Physiol Behav. 1998;63:919–928. doi: 10.1016/s0031-9384(97)00542-8. [DOI] [PubMed] [Google Scholar]
- 7.Macdiarmid JI, Cade JE, Blundell JE. High and low fat consumers, their macronutrient intake and body mass index: further analysis of the national diet and nutrition survey of British adults. Eur J Clin Nutr. 1996;50:505–512. [PubMed] [Google Scholar]
- 8.Pilgrim J, Patton RA. Patterns of self-selection of purified dietary components by the rat. J Comp Physiol Psychol. 1947;40:343–348. doi: 10.1037/h0053575. [DOI] [PubMed] [Google Scholar]
- 9.Scott EM. Self selection of diet. J Nutr. 1946;31:397–406. doi: 10.1093/jn/31.4.397. [DOI] [PubMed] [Google Scholar]
- 10.York DA, Lin L, Smith BK, Chen J. Enterostatin as a regulator of fat intake. In: Berthoud HR, Seeley RJ, editors. Neural and Metabolic Control of Macronutrient Intake. Boca Raton, FL: CRC press; 2000. pp. 361–387. [Google Scholar]
- 11.Okada S, York DA, Bray GA, Mei J, Erlanson-Albertsson C. Differential inhibition of fat intake in two strains of rat by the peptide enterostatin. Am J Physiol. 1992;262:R1111–R1116. doi: 10.1152/ajpregu.1992.262.6.R1111. [DOI] [PubMed] [Google Scholar]
- 12.Overmann SR. Dietary self-selection by animals. Psychol Bull. 1976;83:218–235. [PubMed] [Google Scholar]
- 13.Smith BK, West DB, York DA. Carbohydrate vs. fat intake: differing patterns of macronutrient selection in two inbred mouse strains. Am J Physiol. 1997;272:R357–R362. doi: 10.1152/ajpregu.1997.272.1.R357. [DOI] [PubMed] [Google Scholar]
- 14.Smith BK, Berthoud H-R, York DA, Bray GA. Differential effects of baseline macronutrient preferences on macronutrient selection after galanin, NPY, and an overnight fast. Peptides. 1997;18:207–211. doi: 10.1016/s0196-9781(96)00318-x. [DOI] [PubMed] [Google Scholar]
- 15.Shor-Posner G, Ian C, Brennan G, Cohn T, Moy H, Ning A, Leibowitz SF. Self-selecting albino rats exhibit differential preferences for pure macronutrient diets: Characterization of three subpopulations. Physiol Behav. 1991;50:1187–1195. doi: 10.1016/0031-9384(91)90581-8. [DOI] [PubMed] [Google Scholar]
- 16.Smith BK, Andrews PK, West DB. Macronutrient diet selection in thirteen mouse strains. Am J Physiol. 2000;278:R797–R805. doi: 10.1152/ajpregu.2000.278.4.R797. [DOI] [PubMed] [Google Scholar]
- 17.Smith RichardsBK, Belton BN, Poole AC, Mancuso JJ, Churchill GA, Li R, Volaufova J, Zuberi A, York B. QTL analysis of self-selected macronutrient diet intake: fat, carbohydrate, and total kilocalories. Physiol Genomics. 2002;11:205–217. doi: 10.1152/physiolgenomics.00037.2002. Epub 2002 Dec 3. [DOI] [PubMed] [Google Scholar]
- 18.Kumar KG, Poole AC, York B, Volaufova J, Zuberi A, Smith Richards BK. Quantitative trait loci for carbohydrate and total energy intake on mouse Chromosome 17Congenic strain confirmation and candidate gene analyses (Glo1, Glp1r) Am J Physiol Regul Integr Comp Physiol. 2007;292:R207–R216. doi: 10.1152/ajpregu.00491.2006. Epub 2006 Aug 31. [DOI] [PubMed] [Google Scholar]
- 19.Wakeland E, Morel L, Achey K, Yui M, Longmate J. Speed congenics a classic technique in the fast lane (relatively speaking) Immunol Today. 1997;18:472–477. doi: 10.1016/s0167-5699(97)01126-2. [DOI] [PubMed] [Google Scholar]
- 20.Berthoud HR. An overview of neural pathways and networks involved in the control of food intake and selection. In: Berthoud HR, Seeley RJ, editors. Neural and Metabolic Control of Macronutrient Intake. Boca Raton, FL: CRC Press; 2000. pp. 361–387. [Google Scholar]
- 21.Cupples WA. Physiological regulation of food intake. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1438–R1443. doi: 10.1152/ajpregu.00195.2005. [DOI] [PubMed] [Google Scholar]
- 22.Friedman MI, Horn CC, Ji H. Peripheral signals in the control of feeding behavior. Chem Senses. 2005;30 Suppl 1:i182–i183. doi: 10.1093/chemse/bjh174. [DOI] [PubMed] [Google Scholar]
- 23.Horn CC, Addis A, Friedman MI. Neural substrate for an integrated metabolic control of feeding behavior. Am J Physiol. 1999;276:R113–R119. doi: 10.1152/ajpregu.1999.276.1.R113. [DOI] [PubMed] [Google Scholar]
- 24.Levin BE. Metabolic sensing neurons and the control of energy homeostasis. Physiol Behav. 2006;89:486–489. doi: 10.1016/j.physbeh.2006.07.003. Epub 2006 Aug 8. [DOI] [PubMed] [Google Scholar]
- 25.Smith Richards BK, Belton BN, York B, Volaufova J. Mice bearing Acads mutation display altered postingestive but not 5-s orosensory response to dietary fat. Am J Physiol Regul Integr Comp Physiol. 2004;286:R311–R319. doi: 10.1152/ajpregu.00488.2003. Epub 2003 Oct 30. [DOI] [PubMed] [Google Scholar]
- 26.Woods SC, Lutz TA, Geary N, Langhans W. Pancreatic signals controlling food intake; insulin, glucagon and amylin. Philos Trans R Soc Lond B Biol Sci. 2006;361:1219–1235. doi: 10.1098/rstb.2006.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 28.Han LP, Davison LM, Vander JagtDL. Purification and kinetic study of glyoxalase–I from rat liver, erythrocytes, brain and kidney. Biochim Biophys Acta. 1976;445:486–499. doi: 10.1016/0005-2744(76)90102-9. [DOI] [PubMed] [Google Scholar]
- 29.Dong MQ, Chase D, Patikoglou GA, Koelle MR. Multiple RGS proteins alter neural G protein signaling to allow C elegans to rapidly change behavior when fed. Genes Dev. 2000;14:2003–2014. [PMC free article] [PubMed] [Google Scholar]
- 30.Donkor J, Sariahmetoglu M, Dewald J, Brindley DN, Reue K. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J Biol Chem. 2007;282:3450–3457. doi: 10.1074/jbc.M610745200. Epub 2006 Dec 7. [DOI] [PubMed] [Google Scholar]
- 31.Longo KA, Wright WS, Kang S, Gerin I, Chiang SH, Lucas PC, Opp MR, MacDougald OA. Wnt10b inhibits development of white and brown adipose tissue. J Biol Chem. 2004;279:35503–35509. doi: 10.1074/jbc.M402937200. Epub 2004 Jun 9. [DOI] [PubMed] [Google Scholar]
- 32.Yamazaki H, Yanagawa S. Axin and the Axin/Arrow-binding protein DCAP mediate glucose-glycogen metabolism. Biochem Biophys Res Commun. 2003;304:229–235. doi: 10.1016/s0006-291x(03)00582-5. [DOI] [PubMed] [Google Scholar]
- 33.Whiteman EL, Chen JJ, Birnbaum MJ. Platelet-derived growth factor (PDGF) stimulates glucose transport in 3T3-L1 adipocytes overexpressing PDGF receptor by a pathway independent of insulin receptor substrates. Endocrinol. 2003;144:3811–3820. doi: 10.1210/en.2003-0480. [DOI] [PubMed] [Google Scholar]
- 34.Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- 35.Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, Al-Majali KM, Trembling PM, Mann CJ, Shoulders CC, Graf D, StLezin E, Kurtz TW, Kren V, Pravenec M, Ibrahimi A, Abumrad NA, Stanton LW, Scott J. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 36.Kaput J, Rodriquez RL. Nutritional genomicsthe next frontier in the postgenomic era. Physiol Genomics. 2004;16:166–177. doi: 10.1152/physiolgenomics.00107.2003. [DOI] [PubMed] [Google Scholar]
- 37.Lu B, Jiang YJ, Man MQ, Brown B, Elias PM, Feingold KR. Expression and regulation of 1-acyl-sn-glycerol- 3-phosphate acyltransferases in the epidermis. J Lipid Res. 2005;46:2448–2457. doi: 10.1194/jlr.M500258-JLR200. Epub 2005 Sep 8. [DOI] [PubMed] [Google Scholar]
- 38.Saito N, Shirai Y. Protein kinase C gamma (PKC gamma)function of neuron specific isotype. J Biochem (Tokyo) 2002;132:683–687. doi: 10.1093/oxfordjournals.jbchem.a003274. [DOI] [PubMed] [Google Scholar]
- 39.Ankrah NA, Appiah-Opong R. Toxicity of low levelsofmethylglyoxal: depletion of blood glutathione and adverse effect on glucose tolerance in mice. Toxicol Lett. 1999;109:61–67. doi: 10.1016/s0378-4274(99)00114-9. [DOI] [PubMed] [Google Scholar]
- 40.Thornalley PJ. Glutathione-dependent detoxification of a-oxoaldehydes by the glyoxalase system: involvement in disease mechanisms and antiproliferative activity of glyoxalase I inhibitors. Chem Biol Interact. 1998;111–112:137–151. doi: 10.1016/s0009-2797(97)00157-9. [DOI] [PubMed] [Google Scholar]
- 41.Bari L, Atlante A, Guarangnella N, Principata G, Passarella S. D-Lactate transport and metabolism in rat liver mitochondria. Biochem J. 2002;365:391–403. doi: 10.1042/BJ20020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hovatta I, Tennant RS, Helton R, Marr RA, Singer O, Redwine JM, Ellison JA, Schadt EE, Verma IM, Lockhart DJ, Barlow C. Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature. 2005;438:662–666. doi: 10.1038/nature04250. Epub 2005 Oct 23. [DOI] [PubMed] [Google Scholar]
- 43.Thornalley PJ. Unease on the role of glyoxalase 1 in high-anxiety-related behaviour. TRENDS in Molecular Medicine. 2006;12:195–199. doi: 10.1016/j.molmed.2006.03.004. Epub 2006 Apr 17. [DOI] [PubMed] [Google Scholar]
- 44.Tafti M, Petit B, Chollet D, Neidhart E, de Bilbao F, Kiss JZ, Wood PA, Franken P. Deficiency in short-chain fatty acid beta-oxidation affects theta oscillations during sleep. Nat Genet. 2003;34:320–325. doi: 10.1038/ng1174. [DOI] [PubMed] [Google Scholar]
- 45.Geisbrecht BV, Liang X, Morrell JC, Schulz H, Gould SJ. The mouse gene PDCR encodes a peroxisomal delta(2), delta(4)-dienoyl-CoA reductase. J Biol Chem. 1999;274:25814–25820. doi: 10.1074/jbc.274.36.25814. [DOI] [PubMed] [Google Scholar]
- 46.Huang XS, Zhao SP, Hu M, Luo YP. Apolipoprotein M likely extends its anti-atherogenesis via anti-inflammation. Med Hypotheses. 2007;69:136–140. doi: 10.1016/j.mehy.2006.10.052. Epub 2007 Jan 10. [DOI] [PubMed] [Google Scholar]
- 47.Smith JD, Bhasin JM, Baglione J, Settle M, Xu Y, Barnard J. Atherosclerosis susceptibility loci identified from a strain intercross of apolipoprotein E-deficient mice via a high-density genome scan. Aterioscler Thromb Vasc Biol. 2006;26:597–603. doi: 10.1161/01.ATV.0000201044.33220.5c. Epub 2005 Dec 22. [DOI] [PubMed] [Google Scholar]
- 48.Taylor BA, Phillips SJ. Obesity QTLs on mouse chromosomes 2 and 17. Genomics. 1997;43:249–257. doi: 10.1006/geno.1997.4835. [DOI] [PubMed] [Google Scholar]
- 49.Aljada A, Friedman J, Ghanim H, Mohanty P, Hofmeyer D, Chaudhuri A, Dandona P. Glucose ingestion induces an increase in intranuclear nuclear factor kappaB, a fall in cellular inhibitor kappaB, and an increase in tumor necrosis factor alpha messenger RNA by mononuclear cells in healthy human subjects. Metabolism. 2006;55:1177–1185. doi: 10.1016/j.metabol.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 50.Taupin D, Podolsky DK. Trefoil factors: initiators of mucosal healing. Nat Rev Mol Cell Biol. 2003;4:721–732. doi: 10.1038/nrm1203. [DOI] [PubMed] [Google Scholar]
- 51.Nozaki I, Lunz JG, 3rd, Specht S, Park JI, Giraud AS, Murase N, Demetris AJ. Regulation and function of trefoil factor family 3 expression in the biliary tree. Am J Pathol. 2004;165:1907–1920. doi: 10.1016/S0002-9440(10)63243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown AC, Olver WI, Donnelly CJ, May ME, Naggert JK, Shaffer DJ, Roopenian DC. Searching QTL by gene expression: analysis of diabesity. BMC Genet. 2005;6:12. doi: 10.1186/1471-2156-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.