To reach the mammalian gut, enteric bacteria must pass through the stomach. Many such organisms survive exposure to the harsh gastric environment (pH 1.5 – 4) by mounting extreme acid-resistance responses, one of which, the arginine-dependent system of E. coli, has been studied at levels of cellular physiology, molecular genetics, and protein biochemistry1–7. This multi-protein system keeps the cytoplasm above pH 5 during acid-challenge by continually pumping protons out of the cell using the free energy of arginine decarboxylation. At the heart of the process is a “virtual proton pump8” in the inner membrane - AdiC3,4 - that imports L-arginine (Arg) from the gastric juice and exports its decarboxylation product agmatine (Agm). AdiC belongs to the APC superfamily of membrane proteins6,7,9, which transport amino acids, polyamines, and organic cations in a multitude of biological roles, including delivery of arginine for nitric oxide synthesis10, facilitation of insulin release from pancreatic β-cells11, and, when inappropriately overexpressed, provisioning certain fast-growing neoplastic cells with amino acids12,13. High-resolution structures and detailed transport mechanisms of APC transporters are currently unknown. This report describes a crystal structure of AdiC at 3.2 Å resolution. The protein is captured in an outward-open, substrate-free conformation with transmembrane architecture remarkably similar to that seen in four other families of apparently unrelated transport proteins.
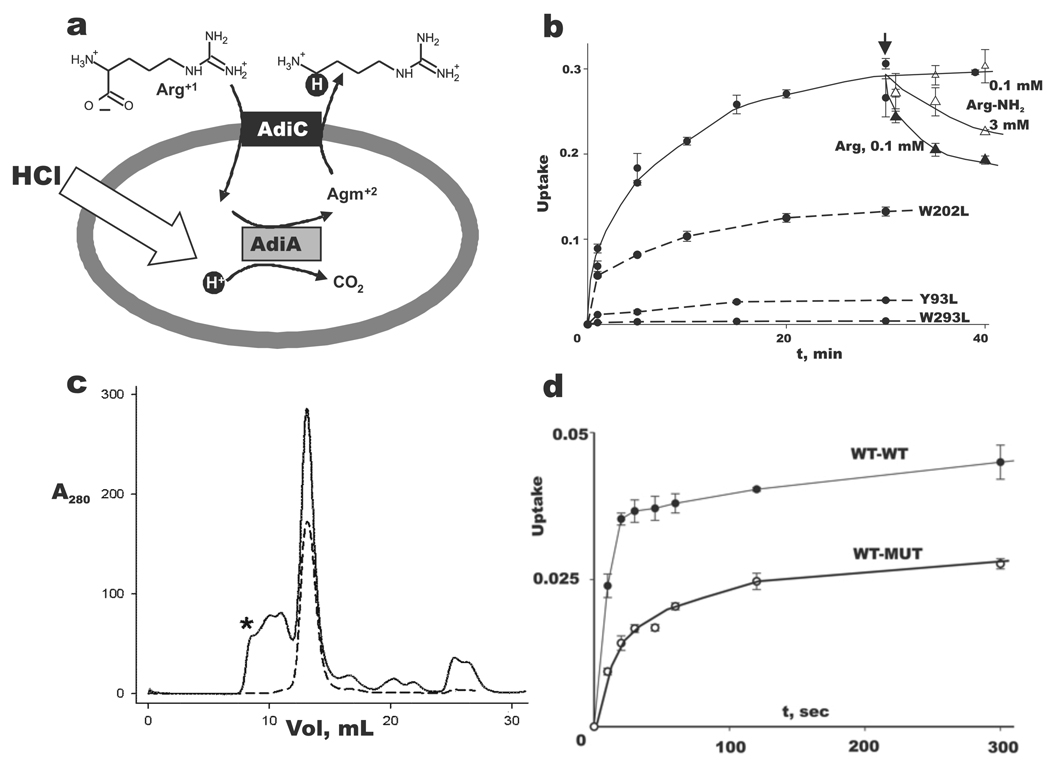
The proton-extrusion function of AdiC, formally an Arg-Agm antiporter, arises from linkage of transport to substrate decarboxylation (Fig 1a), a reaction carried out by a separate enzyme, AdiA, wherein an aqueous proton replaces the α-carboxyl group of Arg to form a C-H bond on Agm; export of this “virtual proton” counteracts cytoplasmic acidification that would otherwise occur at low extracellular pH. Physiological imperatives demand that AdiC specifically imports the deprotonated-carboxylate Arg+1 form, which below pH 2 represents a minor fraction of extracellular Arg2,6; transport of the predominant Arg+2 form would produce an futile cycle useless for acid-resistance, since a carboxyl proton would enter for each virtual proton pumped out. Transport of the Arg+2 form cannot be directly measured in our E. coli AdiC-reconstituted liposomes at pH 6, but argininamide+2 (Arg-NH2), an isosteric proxy for protonated Arg+2, is readily tested. In these experiments (Fig 1b), 14C-Arg is allowed to accumulate into liposomes pre-loaded with Arg. Radiolabel is then expelled from the liposomes by addition of a low concentration (0.1 mM) of unlabelled Arg to the external medium, according to competition for uptake between labeled and unlabeled substrate. A repeat of this assay with Arg-NH2 shows that this analogue is, as expected, a poor substrate for transport, since >30-fold higher concentration is required to mimic competition by Arg+1. A negatively charged carboxylate cannot be a general requirement for transport, however, since the physiological substrate Agm is an excellent substrate6. Figure 1b also presents negative transport-controls using mutations at certain conserved aromatic residues that inhibit activity without affecting protein assembly7. In contrast to many broad-spectrum amino acid transporters of the APC superfamily, AdiC and homologous virtual proton pumps exhibit substantial substrate specificity6,7,14, the structural determinants of which are not known.
Fig 1. AdiC physiology and function.
a. Virtual proton pumping in extreme acid resistance. Schematic of Arg-dependent acid resistance in E. coli, with AdiC-mediated Arg-Agm exchange across the inner membrane coupled to acid-activated Arg-decarboxylase AdiA. Virtual proton is shown in black circle.
b. Selection of α-carboxylate for transport. Uptake (fraction of total counts added) of 14C-Arg at 50 µM external concentration into AdiC-reconstituted liposomes (2 µg/mg lipid) was followed for 30 min, and then either Arg (filled triangles) or Arg-NH2 (open triangles) was added (arrow) externally to the indicated concentration. Additional Arg uptake experiments used the indicated AdiC mutants (dashed curves). Error bars indicate s.e.m. of triplicate experiments.
c. Proper assembly of tandem construct. Size-exclusion profiles of purified homodimeric AdiC (dashed trace) and WT-WT tandem (solid trace) in its final purification step immediately after elution from Co-affinity column. Material eluting between void volume of Superdex-200 column (asterisk) and main peak most likely represents improperly assembled, oligomeric tandems. Identical profile is obtained with WT-MUT tandem in which the second subunit contains the W293L mutation (data not shown). d. A half-dead heterodimer is functionally active. Arg uptake timecourses for WT-WT to WT-MUT tandems reconstituted at 0.2 µg/mg lipid, a low protein density where transporting liposomes carry only one copy to the reconstituted protein6,32. WT-WT transport rate is ~70% of normal homodimeric AdiC (data not shown).
AdiC is a homodimer in detergent micelles and phospholipid membranes6,7. Is subunit cooperation required for transport, or is each subunit a self-contained transporter? This fundamental question must be settled before details of mechanism can be sensibly examined. We therefore designed tandem constructs containing two AdiC subunits joined together by a short linker. The “WT-WT” tandem containing two wildtype subunits migrates identically to the WT homodimer on a size-exclusion column and shows similar transport activity (Fig 1c), a result etablishing that the tandem subunits fold and assemble normally. The transport-disruptive mutation W293L was then introduced into one of the subunits to form a “WT-MUT” tandem for comparison of its transport activity to WT-WT. The result is clear (Fig 1d): the WT-MUT tandem is functionally active, with an initial rate of Arg uptake roughly half of the WT-WT rate. Since the W293L substitution abolishes Arg binding7, this experiment eliminates any transport model that requires substrate binding to both subunits during a single transport cycle, as in cooperative, half-of-sites mechanisms. The result implies that each subunit is itself a transporter, and that the mechanistic underpinnings of substrate exchange are to be found within the subunit itself, as proposed previously on the basis of whole-cell studies for a homologous APC-superfamily protein14.
A Salmonella AdiC homologue, 95% identical to the E. coli sequence, produced crystals diffracting anisotropically to 3.5 Å. Crystals of AdiC complexed with a FAB fragment diffracted to 3.2 Å, and these, along with SeMet derivatives, were used for phasing (Supp Table 1). Serviceable crystals formed only in the absence of substrate. Experimental electron density maps of the FAB complex (Supp Fig 1a) were sufficient for model-building of the AdiC polypeptide chain, save for the first 10 and last 4 residues, and for two disordered loops (residues 174–182, 316–321). Poly-Ala was built into a 22-residue extracellular loop linking transmembrane (TM) helices 5 and 6 and into several poorly ordered regions of the FABs. The crystallographic arrangement of the complexes is somewhat unusual (Supp Fig 1b), as the asymmetric unit contains two AdiC homodimers but only two FABs, rather than four. This occurs because each FAB, by straddling the intracellular subunit interface near the homodimer’s twofold axis, occludes the symmetry-related epitope. Most of the crystal-contacts are mediated by FABs, and only a few by feeble head-to-head encounters at neighboring AdiC extracellular surfaces. A lower-resolution, molecular replacement structure without FAB closely recapitulates the TM helices in the complex (Supp Fig 1b), and thereby rules out problematic structural distortion by the FAB.
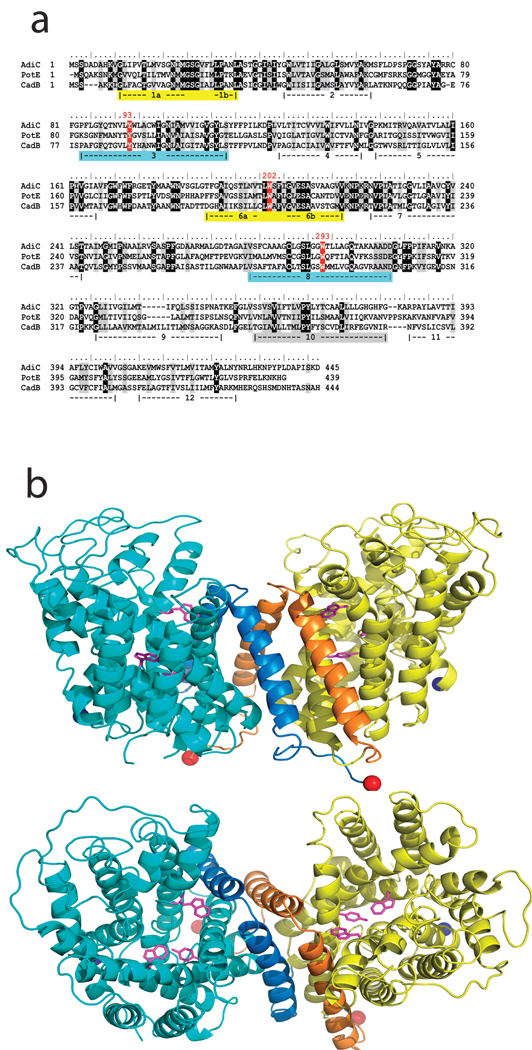
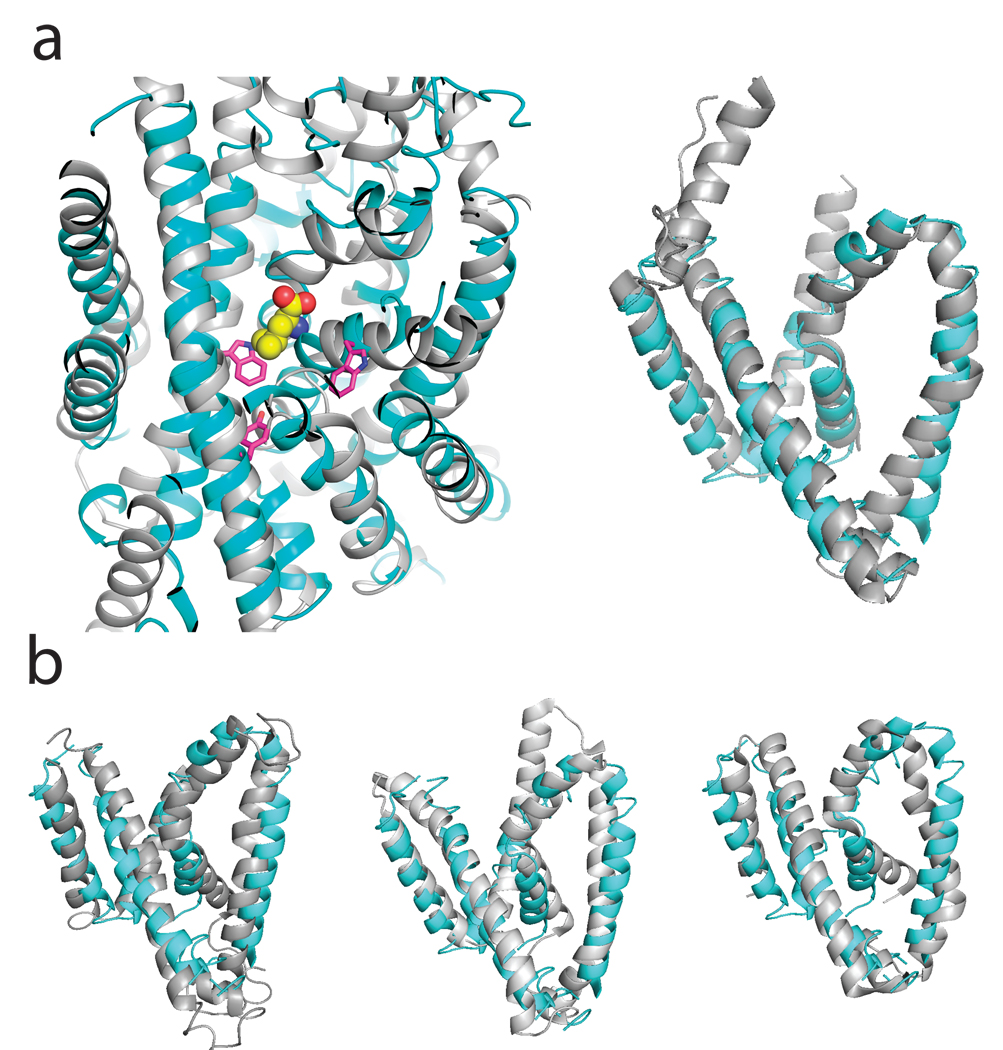
The roughly barrel-shaped AdiC subunit of ~45 Å diameter consists of 12 TM helices (Fig 2), TM1 and TM6 being interrupted by short non-helical stretches in the middle of their transmembrane spans. Biochemical analysis of homologues place the N- and C- termini on the intracellular side of the membrane15,16. TMs 1–10 surround a large cavity exposed to the extracellular solution. These ten helices reprise in AdiC the remarkable inverted structural repeats in membrane protein families as functionally disparate as water / glycerol channels, H+-coupled Cl− antiporters, and Na+-coupled symporters for a variety of bio-organic compounds17–22. TMs 1–5 of AdiC align well with TMs 6–10 turned “upside down” around a pseudo-twofold axis nearly parallel to the membrane plane (α-carbon rmsd ~ 3.4 Å, Supp Fig 2); thus, TM1 pairs with TM6, TM2 with TM7, etc, but no hint of this alignment appears in the primary sequence. Helices TM11 and TM12, non-participants in this repeat, provide most of the 2500 Å2 homodimeric interface. Moreover, AdiC mirrors the common fold observed unexpectedly in four phylogenetically unrelated families of Na+-coupled solute transporters19–23. This result, illustrated (Fig 3a) by a structural alignment with the amino acid transporter LeuT, dramatically confirms a recent prediction based on hydropathy analysis of APC-superfamily proteins24. Beyond the impressive match of the TM helices, the alignment also shows substrate bound in LeuT coincident with the region of AdiC where the functionally critical residues Y93 and W293 reside. As a Na+-independent antiporter joining the cluster of structurally similar families of Na+-coupled symporters (Fig 3b), AdiC further highlights emerging questions of convergent evolution vs deep links among ostensibly unrelated membrane proteins.
Fig 2. Structure of AdiC.
a. Sequence of AdiC aligned with two other E. coli virtual proton pump exchangers in the APC superfamily: PotE (ornithine/putrescine) and CadB (lysine/cadaverine). The five TMs lining the central cavity are highlighted, with matched colors representing helices paired by inverse repeat. Conserved aromatic residues discussed in the text are indicated in red. b. Ribbon diagrams of homodimer viewed from membrane, extracellular side up (upper panel) and from extracellular solution (lower panel), with Y93, W202, W293 side chains indicated (pink sticks). N- and C-termini indicated by blue, red spheres. TM 11,12 are indicated as darker hue in each subunit.
Fig 3. Structural alignment of AdiC with four transporter families.
Alignments of AdiC (cyan) with four transporters (grey) are shown. a. LeuT, a wide-spectrum amino acid transporter. Left panel: TMs 1–10, showing bound substrate (spacefilled) and conserved aromatics of AdiC (pink sticks); right panel: TM1-5 core. b. Alignments of AdiC TM 1–5 core with BetP (left panel), Mhp1 (center panel), and SGLT1 (right panel). Cα r.m.s.d. with AdiC: LeuT19 3.1 Å, SGLT120 4.0 Å, Mhp121 3.4 Å, BetP22 3.6 Å.
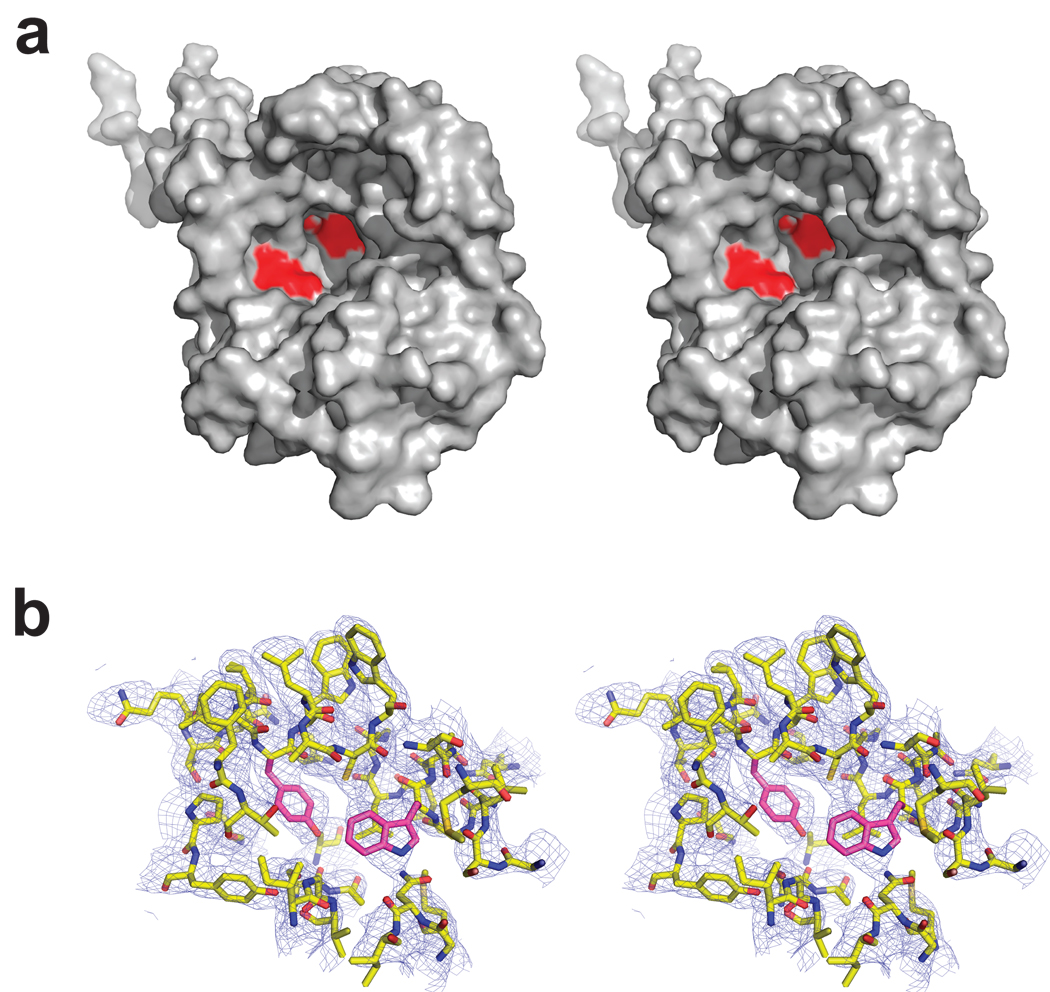
The central cavity stands out as a prominent feature of the structure (Fig 4). This extracellular-facing aqueous cavern, ~25 Å wide at the rim, tapers to a floor situated in the center of the protein about halfway through the membrane. The floor is formed by a pair of aromatic side chains, Y93 and W293, projecting inward from TM3 and TM8, the long, tilted helices paired by the inverted repeat-domains. An additional aromatic, W202, hangs 10–15 Å away on the cavity wall. These conserved residues have been variously proposed to contribute to substrate binding and transport in APC-type virtual proton pumps7,14,25, as indicated above for AdiC (Fig 1B). The wall, otherwise festooned with hydrophobic and polar moieties, is completely devoid of charged side chains. The cavity is unambiguously cut off from the cytoplasmic solution by a ~15 Å thickness of tightly packed protein.
Fig 4. AdiC cavity in outward-open conformation.
a. Oblique stereo view from extracellular side of single AdiC subunit, showing locations of aromatic sidechains (red).
b. Closeup view of proposed substrate-binding region, with 2Fo-Fc map contoured at 1.5 σ. Y93, W293 shown in pink sticks.
Discussion
The operation of any membrane transporter relies on a cycle of distinct protein conformations that expose substrate-binding sites alternately to the cytoplasmic and extracellular solutions and may additionally employ intermediate “occluded” states with substrates buried. Transport mechanisms are defined by rules linking substrate binding to transitions among these conformations. Since x-ray structures from the APC superfamily have not previously been described, the present view of AdiC in a single, substrate-free conformation is inadequate for laying out a mechanistic framework for this family of transporters. But the outward-facing structure here identifies a likely locale for substrate binding at the cavity’s floor. This suggestion is bolstered by three lines of argument. First, the narrowest part of a wide vestibule is a general expectation for the transport site in a coupled transporter26. Second, mutation of aromatic residues at this site inhibits transport activity in AdiC and homologues7,14,25. Finally, bound substrate in structurally aligned LeuT is found in this region of AdiC (Fig 3a).
Viewed as a binding site for extracellular Arg, the cavity raises questions of molecular recognition underlying the protein’s essential proton-extrusion function. Since Arg, Agm, and analogues with similarly disposed charged groups are transported by AdiC6,7, the protein might be expected to offer oppositely charged residues for salt-bridge stabilization of substrates. But AdiC’s physiological role in acid resistance quashes this expectation, since in a hydrated region exposed to pH 2, Glu/Asp side chains would be fully protonated and incapable of forming coulombic contacts. Indeed, charged residues are conspicuously absent from the cavity. The structure thus suggests - and we propose - that the outward-facing binding site neatly solves its electrostatic problem with aromatic sidechains, which stabilize substrates with cation-π interactions27,28. A similar “aromatic box” was recently observed at the binding site of a Na+/betaine symporter22. It is clear, though, that W202 is too far above the cavity’s floor for Arg α-amino and γ-guanidino groups to contact all of these aromatics simultaneously, and that the aromatics cannot by themselves mediate the nuanced substrate specificities of APC-type virtual proton pumps. We also note several backbone carbonyl oxygens on the repeat-related, non-helical stretches of TM1 and TM6, which might help stabilize substrates via any of the five H-bond donors of substrate guanidino groups. Resolution of the crucial biological issue - selectivity for the rare, negatively charged carboxylate of extracellular Arg+1 - will require crystals with bound substrate, which we have so far failed to obtain by co-crystallization or soaking.
An alternative possibility exists, however, which if true would nullify all the above ruminations on substrate selectivity. The structure here, while certainly outward-open, might not represent an Arg-binding form of the transport cycle at all, but rather an Agm-expelling form whose biological purpose is to efficiently rid the protein of substrate before beginning a new transport cycle. In other words, two different outward-open conformations, of high and low substrate affinities, might operate in transport; a similar situation might also apply to inward-facing forms. “Dual-open” mechanisms like this have not been seriously considered for antiporters, but they are not a priori implausible. Indeed, such a mechanism would be well-suited to the logic of virtual proton pumping, whereby Arg and Agm both move thermodynamiccaly downhill, binding at high concentration and dissociating at low, with gradients maintained by Arg decarboxylation. This structure provides no information about inward-open forms of AdiC, but a recent projection structure of AdiC in 2-dimensional membrane crystals7 could represent such a conformation, since it differs dramatically from the same projection of our x-ray structure (Supp Fig 3). Resolution of issues like this must await structures of AdiC under different conditions with substrates, and of other members of the APC superfamily.
Methods Summary
AdiC was expressed in E. coli, purified, and reconstituted in liposomes6. AdiC function was assessed using the E. coli homologue by 14C-Arg-Arg exchange (5 mM Arg inside, 50 µM 14C-Arg outside), at protein density 0.2–2 µg AdiC/mg lipid6. Two tandem constructs used a 6-residue linker (GSAGGT) connecting the C-terminus of the first AdiC subunit to the N-terminus of the second. Monoclonal antibodies were raised by inoculating mice with E. coli AdiC and screening ELISA-positive hybridomas for stable complexes by size-exclusion chromatography, crystallization, and diffraction quality. Approximately 25 monoclonals were tested for crystallization to obtain FAB fragment #21 used here, which was derived from a type-2a IgG.
Salmonella serovar typhimurium
AdiC complexed with FAB21 was purified on Superdex 200 in 100 mM NaCl, 5 mM decylmaltoside, 20 mM tris-HCl pH 8, concentrated to 8 mg/mL, mixed with an equal volume of 30–35%(w/v) PEG 400, 100–200 mM CaCl2, 100 mM glycine pH 9–9.5, and crystallized in hanging drops at 20°C. Crystals were frozen after 2–4 weeks, and datasets were collected at NSLS, APS, and ALS. SeMet derivatives were similarly treated. To increase redundancy of the anomalous signal in the P1 spacegroup, datasets were collected with 4–6 360° passes. Experimental phases determined to 3.5 Å resolution by anomalous dispersion from two SeMet crystals - one at one wavelength and the other at two wavelengths - were combined using Sigmaa29 and extended to 3.2 Å by noncrystallographic symmetry averaging (8-fold for AdiC in the two crystal forms and 2-fold for FAB21) and solvent flattening. Multi-domain, multi-crystal averaging using Dmmulti30 greatly improved the phases. Sharpening of the data by a temperature factor of −80 Å2 significantly enhanced the details of the electron density maps. Anomalous difference density maps identified 14 of the 15 Se atoms expected per subunit. Attempts to observe substrate density by soaking crystals in 5 mM substrates were unsuccessful.
Supplementary Material
Acknowledgments
YF was supported by NIH grant # T32 NS 07292. We appreciate the expert support of the beamline scientists at the Advanced Photon Source (GM-CAT, 23-ID), Advanced Light Source (8.2.1, 8.2.2), and National Synchrotron Light Source (X-25, X-29A). We are also grateful to J. Berry for help in hybridoma sequencing, to B. Bowman for crystallographic advice, to E. Gouaux for sharing information about an APC homologue, and to P. DeWeer, D.P. Krummel, H.H. Lim, K. Piasta, and J. Robertson for suggestions on the manuscript.
Footnotes
Coordinates and structure factors of the AdiC-FAB complex have been deposited in the PDB (Accession # 3HQK).
Reprints and permissions information are available at npg.nature.com/reprintsandpermissions
The authors have no competing financial interests
Contributor Information
Yiling Fang, Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts 02454.
Hariharan Jayaram, Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts 02454.
Tania Shane, Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts 02454.
Ludmila Kolmakova-Partensky, Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts 02454.
Fang Wu, Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts 02454.
Carole Williams, Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts 02454.
Yong Xiong, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520.
Christopher Miller, Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts 02454.
References
- 1.Audia JP, Webb CC, Foster JW. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int J Med Microbiol. 2001;291:97–106. doi: 10.1078/1438-4221-00106. [DOI] [PubMed] [Google Scholar]
- 2.Iyer R, Iverson TM, Accardi A, et al. A biological role for prokaryotic ClC chloride channels. Nature. 2002;419:715–718. doi: 10.1038/nature01000. [DOI] [PubMed] [Google Scholar]
- 3.Iyer R, Williams C, Miller C. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol. 2003;185:6556–6561. doi: 10.1128/JB.185.22.6556-6561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong S, Richard H, Foster JW. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J Bacteriol. 2003;185:4402–4409. doi: 10.1128/JB.185.15.4402-4409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 6.Fang Y, Kolmakova-Partensky L, Miller C. A bacterial arginine-agmatine exchange transporter involved in extreme Acid resistance. J Biol Chem. 2007;282:176–182. doi: 10.1074/jbc.M610075200. [DOI] [PubMed] [Google Scholar]
- 7.Casagrande F, Ratera M, Schenk AD, et al. Projection structure of a member of the amino acid/polyamine/organocation transporter superfamily. J Biol Chem. 2008;283:33240–33248. doi: 10.1074/jbc.M806917200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maloney PC. Bacterial transporters. Curr. Opin. Cell. Biol. 1994;6:571–582. doi: 10.1016/0955-0674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 9.Jack DL, Paulsen IT, Saier MH. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology (Reading, England) 2000;146:1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson B, Manner CK, Kleeman J, et al. Sustained nitric oxide production in macrophages requires the arginine transporter CAT2. J Biol Chem. 2001;276:15881–15885. doi: 10.1074/jbc.M010030200. [DOI] [PubMed] [Google Scholar]
- 11.Smith PA, Sakura H, Coles B, et al. Electrogenic arginine transport mediates stimulus-secretion coupling in mouse pancreatic beta-cells. J Physiol. 1997;499(Pt 3):625–635. doi: 10.1113/jphysiol.1997.sp021955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs BC, Bode BP. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin. Cancer Biol. 2005;15:254–266. doi: 10.1016/j.semcancer.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Kaira K, Oriuchi N, Imai H, et al. I-type amino acid transporter 1 and CD98 expression in primary and metastatic sites of human neoplasms. Cancer science. 2008 doi: 10.1111/j.1349-7006.2008.00969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashiwagi K, Kuraishi A, Tomitori H, et al. Identification of the putrescine recognition site on polyamine transport protein PotE. J Biol Chem. 2000;275:36007–36012. doi: 10.1074/jbc.M006083200. [DOI] [PubMed] [Google Scholar]
- 15.Kashiwagi K, Shibuya S, Tomitori H, et al. Excretion and uptake of putrescine by the PotE protein in Escherichia coli. J Biol Chem. 1997;272:6318–6323. doi: 10.1074/jbc.272.10.6318. [DOI] [PubMed] [Google Scholar]
- 16.Hu LA, King SC. Membrane topology of the Escherichia coli gamma-aminobutyrate transporter: implications on the topography and mechanism of prokaryotic and eukaryotic transporters from the APC superfamily. Biochem. J. 1998;336:69–76. doi: 10.1042/bj3360069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu D, Libson A, Miercke LJ, et al. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 18.Dutzler R, Campbell EB, Cadene M, et al. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita A, Singh SK, Kawate T, et al. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 20.Faham S, Watanabe A, Besserer GM, et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321:810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weyand S, Shimamura T, Yajima S, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322:709–713. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, et al. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature. 2009;458:47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]
- 23.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459:347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lolkema J, Slotboom D-J. The major amino acid transporter superfamily has a similar core structure as Na+-galactose and Na+-leucine transporters. Molec. Membr. Biol. 2008;25:567–570. doi: 10.1080/09687680802541177. [DOI] [PubMed] [Google Scholar]
- 25.Soksawatmaekhin W, Uemura T, Fukiwake N, et al. Identification of the cadaverine recognition site on the cadaverine-lysine antiporter CadB. J Biol Chem. 2006;281:29213–29220. doi: 10.1074/jbc.M600754200. [DOI] [PubMed] [Google Scholar]
- 26.Guan L, Kaback HR. Lessons from lactose permease. Ann. Rev. Biophys. Biomol. Structure. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burley SK, Petsko GA. Amino-aromatic interactions in proteins. FEBS Lett. 1986;203:139–143. doi: 10.1016/0014-5793(86)80730-x. [DOI] [PubMed] [Google Scholar]
- 28.Zacharias N, Dougherty DA. Cation-pi interactions in ligand recognition and catalysis. Trends Pharm. Sci. 2002;23:281–287. doi: 10.1016/s0165-6147(02)02027-8. [DOI] [PubMed] [Google Scholar]
- 29.Read R. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Cryst. A. 1986;42 [Google Scholar]
- 30.Cowtan KD, Main P. Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr D Biol Crystallogr. 1996;52:43–48. doi: 10.1107/S090744499500761X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.