Abstract
The rough coat (rc), an autosomal-recessive mutation, arose spontaneously in C57BL/6J mice. Homozygous rc mice develop severe skin and hair abnormalities, including cyclic and progressive hair loss and sebaceous gland hypertrophy. The rc locus was previously mapped to Chromosome 9. To elucidate the genetic basis underlying the rc phenotype development, we carried out positional cloning, and mapped the rc locus to a 246-kb interval. We identified a missense mutation within a novel open reading frame in the rc/rc mice, which is predicted to encode a cell adhesion molecule with the highest homology to myelin protein zero (MPZ) and myelin protein zero-like 2 (MPZL2, also called epithelial V-like antigen). We therefore named this gene Mpzl3 (myelin protein zero-like 3). The mutation in the rc/rc mice occurred at a highly conserved residue within the conserved Ig-like V-type domain, thus likely altering the MPZL3 protein function. Reverse transcriptase-PCR and Western blot analyses revealed expression of the Mpzl3 gene in various adult organs, including the skin. Using indirect immunofluorescence, we detected MPZL3 protein in the keratinocytes and sebocytes in the skin. Results from this study identified a novel gene encoding a predicted adhesion protein whose mutation in the rc/rc mice likely caused the rc phenotype.
INTRODUCTION
The rough coat (rc) mutation arose spontaneously in the C57BL/6J (B6/J) inbred mouse strain at the Jackson Laboratory in 1966 (Dickie, 1966). The rc mutation is autosomal recessive. Homozygous rc mice are born with no apparent abnormalities, but display unkempt looking hair coats by weaning age, and develop cyclic and progressive hair loss thereafter (Figure 1) (Hayashi et al., 2004). In addition, histological analysis of skin sections revealed sebaceous gland hypertrophy in the rc/rc mice (Figure 2) (Ruvinsky et al., 2002; Hayashi et al., 2004). Both male and female homozygous rc mice are fertile (Dickie, 1966), although only about a quarter of the pups born to rc/rc female mice survive (Hayashi et al., 2004). Linkage analysis with DNA markers assigned the rc locus to 32.0 cM on Chromosome 9, close to the Mpi1 gene (two recombinants among 107 backcross offspring) (Eicher et al., 1977) at 57.57 megabase (Mb) (Ensembl Mouse Genome Database v38, released in April 2006, www.ensembl.org/Mus_musculus), but the gene mutation remained unknown.
Figure 1. Gross phenotype of the rc/rc mice.
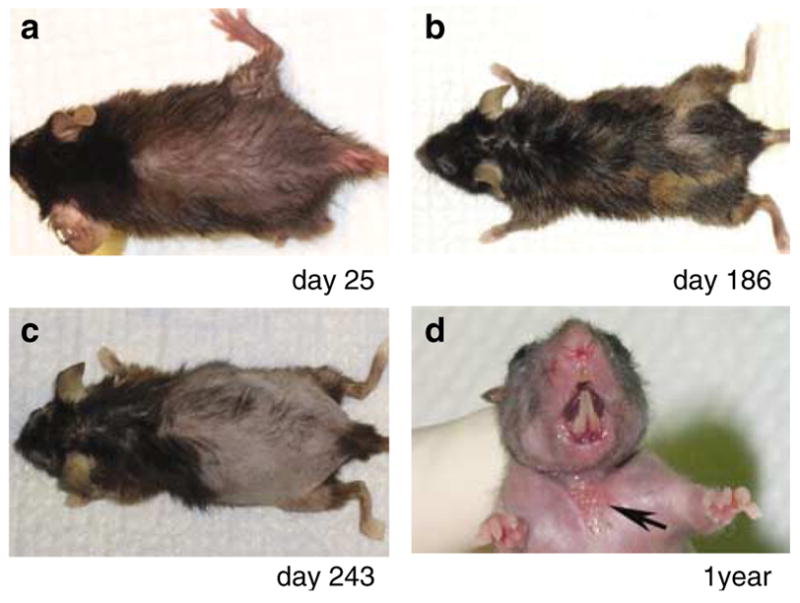
(a) Hair loss is apparent on the dorsal trunk of an rc/rc mouse on day 25 after birth. (b) New hair coat retains the rough coat phenotype, sometimes with reduced pigmentation. (c) Hair loss is progressive in older rc/rc mice. (d) In more than 50% of rc/rc mice older than 1 year, ulcerated wounds develop spontaneously in the ventral neck region (arrow).
Figure 2. Sebaceous gland hypertrophy in the rc/rc mice.
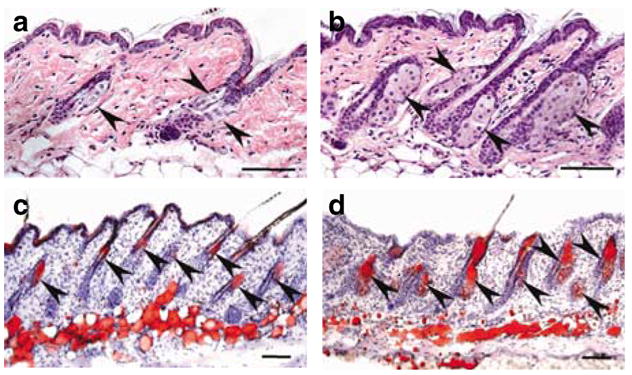
(a, b) Hematoxylin and eosin staining and (c, d) oil red O staining of lipids in back skin sections from (a, c) a +/rc and (b, d) an rc/rc mouse. Arrowheads point to the sebaceous glands. (b, d) Notice the hypertrophic sebaceous glands in the rc/rc mouse. The sebocytes (d) in the rc/rc mouse are functional lipid-secreting cells as (c) in the normal mouse. Bar = 100 μm.
The appearance of rough hair coats in rc/rc mice is similar to matted (ma) mice, another strain with a spontaneous mutation (Searle and Spearman, 1957). However, tests for allelism through breeding experiments with ma and several other mutant strains such as ichthyosis (ic), plucked (pk), fuzzy (fz), and rough (ro) have all been negative (Dickie, 1966). In addition, a recent study excluded rc as allelic to abnormal feet and tail (aft) (Ruvinsky et al., 2002). Interestingly, mice homozygous for rough fur (ruf), another spontaneous mutation mapped to Chromosome 9, demonstrated similar skin abnormalities with the rc/rc mice, such as the “unkempt” and “wet” appearance and sebaceous gland hypertrophy (Sweet et al., 1990; Park et al., 2001). However, allelism has not been tested for rc and ruf, and the mutation in ruf mice has not been identified.
In our previous studies, we showed linkage of the rc locus with two microsatellite markers, D9Mit162 at 49.954 Mb (one recombinant among 129 backcross offspring) and D9Mit104 at 65.953 Mb (three recombinants among 129 backcross offspring) (Hayashi et al., 2004). We also excluded the loxl gene (at 58.317–58.342 Mb) located between these two markers as allelic to rc (Hayashi et al., 2004). In this study, to elucidate the genetic basis and better understand the molecular mechanisms of rc phenotype development, we carried out positional cloning in backcross mice.
RESULTS
Cyclic and progressive hair loss and sebaceous hypertrophy in the rc/rc mice
Homozygous rc pups were born with no apparent abnormalities and developed normal hair coats that became plush 7 days after birth, thus indicating that the initial hair growth (follicular neogenesis) was normal. However, by day 14, the rc/rc pups started to show unkempt-looking hair coats (“rough” coat) and a loss of glossiness. By day 18, hair loss became apparent on the dorsal, ventral, and lateral trunk. At the next anagen, the dorsal skin became dark and thickened, and the newly grown hair coats retained the “rough coat” phenotype. Thereafter, the rc/rc mice underwent cyclic hair loss and hair growth, sometimes with reduced pigmentation in the new hair coat (Figure 1). Hair loss was progressive, however, with some older rc/rc mice becoming nearly bald. No breakage of the hair shaft was observed in the rc/rc mice by scanning electron microscopy (data not shown). In addition to hair loss, we observed a high incidence of spontaneous and persistent ulcerated lesions on the ventral skin of the neck in rc/rc mice over 1 year of age (17 of 27, 63%) (Figure 1d). Histological analysis of such lesions revealed typical features of chronic wounds, including inflammation in the wound bed and hyperplasia of the epidermal keratinocytes at the wound edge (data not shown).
We observed severe hypertrophy of the sebaceous glands in skin sections from rc/rc mice at day 16 (Figure 2a and b). This observation was confirmed by oil red O staining of lipids in differentiated sebocytes (Figure 2c and d). Similar observations were made in rc/rc skin at days 24, 34, and 76 (data not shown). Sebaceous gland hypertrophy was a result of sebocyte hyperplasia: there were twice as many sebocytes per sebaceous gland section in rc/rc mice as in +/rc mice (14.9±2.8 vs 6.2±2.5 in the day 76 samples examined, P ≪ 0.01).
High-resolution linkage analysis
Before this study, no mapping interval was defined for the rc locus, even though it had been mapped close to the Mpi1 gene (Eicher et al., 1977) and two microsatellite markers, D9Mit162 and D9Mit104 (Hayashi et al., 2004). To define a mapping interval for the rc locus, we outcrossed B6/J-rc/rc mice with both CAST/Ei mice and BALB/cJ mice to obtain F1 hybrids (+/rc) on two mixed strain backgrounds to avoid a potential low rate of recombination within the rc region (Fernandez-Gonzalez et al., 2002). Female F1 hybrids were backcrossed with male B6/J-rc/rc mice to obtain F2 hybrids.
We monitored F2 hybrids daily from birth for the rc phenotype development. F2 hybrids that showed unkempt hair coats by day 16, hair loss by day 24, ensuing hair growth, and subsequent hair loss were considered phenotypic and homozygous for the rc mutation (rc/rc). F2 hybrids that never showed hair abnormality at these stages were considered non-phenotypic and heterozygous for the rc mutation (+/rc). Penetrance of the rc phenotype was 100% in both F2 hybrid backgrounds.
We analyzed linkage between the rc locus and five microsatellite markers in 200 B6/J-BALB/cJ F2 hybrids (Table 1). Linkage was shown between the rc locus and all five microsatellite markers (P ≪ 0.0001). All four recombinants with D9Mit191 also showed recombination with D9Mit162 and D9Mit73, and none of them showed recombination with D9Mit67 or D9Mit328. All six recombinants with D9Mit328 also showed recombination with D9Mit67, and none of them showed recombination with D9Mit73, D9Mit162, or D9Mit191. These recombinations defined the rc interval between D9Mit328 and D9Mit191.
Table 1.
Summary of linkage analysis of five microsatellite markers and the rc locus in 200 C57BL/6J-BALB/cJ F2 offspring
| Marker | Position (Mb) | Number of recombination | Number of mice analyzed | Distance (cM) | LOD score |
|---|---|---|---|---|---|
| D9Mit67 | 36.961 | 11 | 200 | 5.5 | 41.7 |
| D9Mit328 | 41.822 | 6 | 200 | 3.0 | 48.5 |
| rc | |||||
| D9Mit191 | 46.647 | 4 | 200 | 2.0 | 51.7 |
| D9Mit162 | 49.954 | 6 | 200 | 3.0 | 48.5 |
| D9Mit73 | 71.609 | 25 | 200 | 12.5 | 27.5 |
cM, centimorgan; LOD, logarithm of the odds; Mb, megabase.
P ≪ 0.0001 for all markers. The Mb positions are according to the Ensembl Mouse Genome Database (www.ensembl.org/Mus_musculus), v38 (released in April 2006), based on the NCBI m35 assembly (released in December 2005).
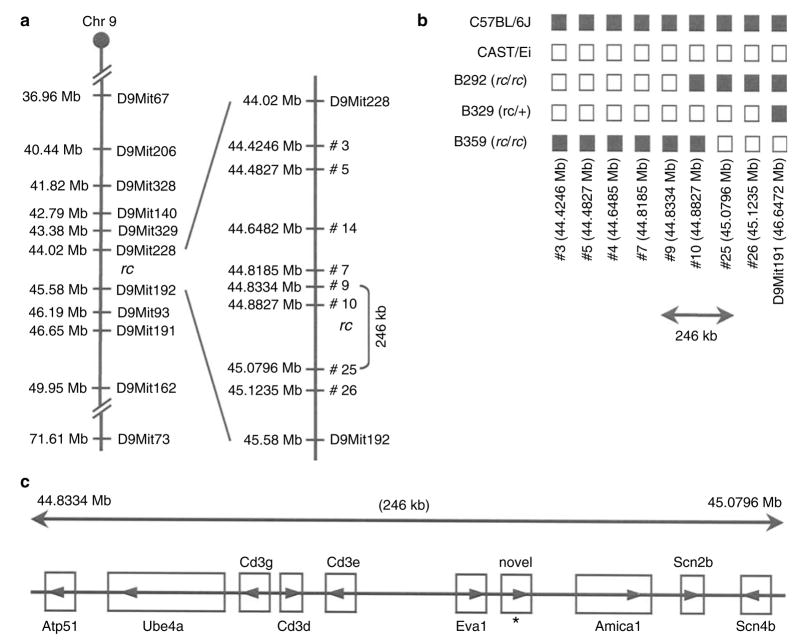
Using a similar approach, we analyzed linkage between the rc locus and the published polymorphic microsatellites (shown in Figure 3a) in 361 B6/J-CAST/Ei F2 hybrids. We were able to reduce the rc interval to 1.560 Mb, between D9Mit228 and D9Mit192. We then identified eight novel microsatellite polymorphisms within the D9Mit228–D9Mit192 interval between B6/J and CAST/Ei strains (Figure 3a). The chromosomal locations and primer sequences of these novel polymorphisms are shown in Table 2. Haplotype analysis of F2 hybrids B292, B329, and B359 revealed that the rc locus lay within a 246-kb interval, between 44.8334 Mb (microsatellite #9) and 45.0796 Mb (microsatellite #25) (Figure 3b).
Figure 3. High-resolution genetic map of the rc interval and the candidate genes.
(a) High-resolution genetic map of the rc interval based on linkage analysis in 200 C57BL/6J-BALB/cJ and 361 C57BL/6J-CAST/Ei F2 hybrid offspring. Microsatellite markers in the Ensembl Mouse Genome Database (www.ensembl.org/Mus_musculus) are shown on the left with their chromosomal locations in megabases (Mb). Novel microsatellite polymorphisms between the C57BL/6 and CAST/Ei strains identified in this study are shown on the right. Marker #10 was concordant with rc in all the F2 hybrids we analyzed, and its location relative to the rc locus could not be determined in our linkage analysis. (b) Haplotype analysis of the three recombinants with D9Mit191 or marker #3 among the 361 C57BL/6J-CAST/Ei F2 hybrid offspring. These results show that the rc locus lies within a 246-kb interval. (c) Candidate genes in the 246 kb mapping interval for the rc locus (Ensembl Mouse Genome Database, www.ensembl.org/Mus_musculus).
Table 2.
Summary of linkage analysis of novel microsatellite polymorphisms identified between B6/J and CAST/Ei strains and the rc locus in 361 C57BL/6J-CAST/Ei F2 hybrids
| Marker | Chromosomal position (Mb) | Number of recombinants | Primer sequences |
|---|---|---|---|
| #3 | 44.4246 | 1 | F: CTGGCTCTACAGGCGTGTAC |
| R: TGCTAAGTAAGGAGAGAGGG | |||
| #5 | 44.4827 | 1 | F: TGATCTCCTGGTCCCATGAG |
| R: GGATCCACTTGTAAATGTGC | |||
| #14 | 44.6485 | 1 | F: CTCTGCTCTCCACACTCATC |
| R: TGCACACGCTTGTGACCATG | |||
| #7 | 44.8185 | 1 | F: TCAATGGGAGAGTGCTTGCC |
| R: GAATCTCTCTCAGTGCCTCC | |||
| #9 | 44.8334 | 1 | F: GGAGAGTGAGAAAGCAGGAC |
| R: CCCTAGCCTATGAGATCTCC | |||
| #10 | 44.8827 | 0 | F: AGGTTTTGTAGAATCCAGGC |
| R: CTGACCTCCACACTACACTC | |||
| #25 | 45.0796 | 1 | F: TCACAGACATGGCAGGAGTC |
| R: CCTCTGCTTCTGGTTGCTAC | |||
| #26 | 45.1235 | 1 | F: GGCTAATTCAGCGTACACAC |
| R: CTCAGGTCATTTGGCACCAG |
cM, centimorgan; F, forward; Mb, megabase; R, reverse.
The Mb positions are according to the Ensembl Mouse Genome Database (www.ensembl.org/Mus_musculus), v38 (released in April 2006), based on the NCBI m35 assembly (released in December 2005).
Mutation detection
Within this 246 kb interval, there are 10 candidate genes, including nine known genes and one novel gene (Figure 3c). By reverse transcriptase (RT)-PCR analysis, we detected expression of nine of the 10 candidate genes in normal as well as rc/rc skin (data not shown). As we could not pinpoint a most likely “functional” candidate gene, such as one whose expression in the rc/rc skin was drastically down-regulated, we carried out sequence analysis of all the coding sequences and flanking splice sites of all 10 candidate genes. There are a total of 71 exons among these genes, 68 of which contain coding sequences, according to the Ensembl Mouse Genome Database (www.ensembl.org/Mus_musculus). These numbers included all exons predicted from different transcripts in the Ensembl database (www.ensembl.org/Mus_musculus, NCBI m35 assembly released in December 2005, with reference to NCBI m36 assembly released in April 2006). We did not identify any mutation in the known genes in the rc/rc DNA.
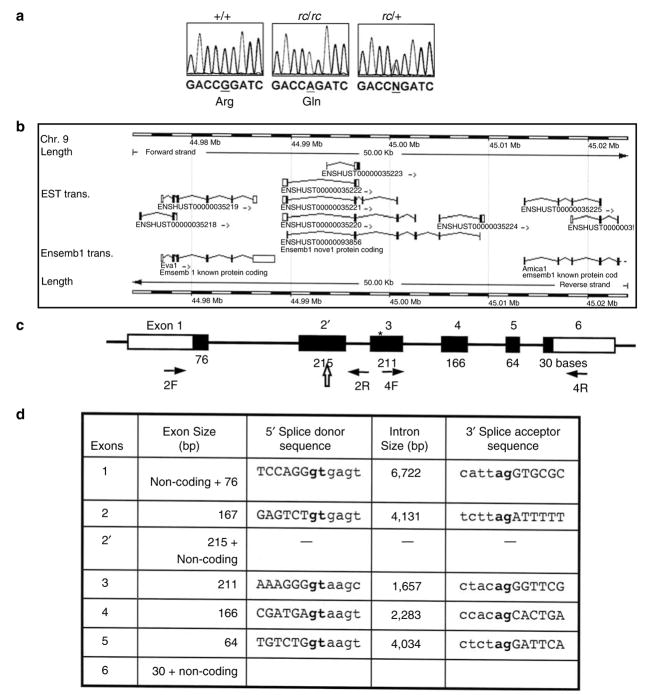
However, we have identified a point mutation in the open reading frame within the novel gene ENSMUSG00000070305, located at 44.989–45.009 Mb (Figure 4, Table 3). This novel gene consists of six exons, and encodes a polypeptide, ENSMUSP00000091378 (from transcript ENSMUST00000093856), of 230 amino acids. A closer examination of this entry, although, revealed that the coding sequence neither started with an ATG nor ended with a STOP codon. We therefore considered it incomplete. This novel gene has a human homolog ENSG00000160588 at 117,602,619– 117,628,245 bp on Chromosome 11, and a rat homolog ENSRNOESTG00000015598 at 48,004,303–48,024,405 bp on Chromosome 8. The human homolog and rat homolog encode polypeptides of 235 (ENSP00000278949) and 236 (ENSRNOESTP00000021062) amino acids, respectively. By comparison with the human and rat homolog and analysis of the mouse genomic sequence and expressed sequence tags (ESTs), we predicted additional amino acids at the amino and carboxyl termini of the encoded mouse polypeptide. There are an additional four amino acids at the amino terminus, starting with a methionine encoded by ATG. We also identified an in-frame STOP codon 36 bp upstream from this ATG within the 5′ flanking genomic sequences. Furthermore, we predicted an additional three amino acids at the carboxyl terminus of the mouse polypeptide, followed by a STOP codon. Thus, we predicted a polypeptide of 237 amino acids encoded by the mouse gene ENSMUSG00000070305. We have submitted this novel gene/mRNA sequence to GenBank (GenBank accession number EF102773).
Figure 4. Gene structure and mutation analysis of a novel gene in which a mutation was identified in the rc/rc DNA.
(a) Sequence analysis identified a point mutation in a predicted open reading frame in the rc/rc genomic DNA, which was confirmed in rc/+ genomic DNA and rc/rc cDNA. We have named this gene “Mpzl3” based on the similarity of the predicted domain structure of its encoded protein to MPZ and MPZL2 (also called EVA1). (b) The Mpzl3 gene (ENSMUSG00000070305), predicted on the basis of multiple Ensembl transcripts based on ESTs, in its genomic context. Eva1 (Mpzl2) gene is upstream, and Amica1 gene is downstream. (c) Our prediction of the murine Mpzl3 gene that is homologous to the human gene ENSG00000160588 and rat gene ENSRNOESTG00000015598. Exon and EST information was based on the Ensembl Genome Database (www.ensembl.org) and our prediction by sequence comparison between species. Numbers underneath each exon indicate the length of the coding sequence in that exon (filled portion). Exon 2 has 167 bp of coding sequence, and exon 2′ has 215 bp of coding sequence (48 bp longer than exon 2) as well as a 3′ UTR. Block arrow points to the splice site in exon 2′ used to generate the six-exon transcript. The arrows denote primers 2F, 2R, 4F, and 4R used for RT-PCR analysis in Figure 6. *The position of the mutation detected in rc/rc DNA. (d) Exon–intron information of the predicted murine Mpzl3 gene.
Table 3.
Sequences of primers used to PCR amplify the six exons of the novel mouse gene ENSMUSG00000070305 (later named Mpzl3) for sequence analysis
| Exons | Forward primer | Reverse primer | Amplicon (bp) |
|---|---|---|---|
| Exon 1 | ATCAGATCCTCCTGAGAGTC | TCAAGTCTCACAAGGTGGTC | 518 |
| Exon 2/2′ | TGAAGCATCTCTCATGTTCAC | AAACTTGCACAGCAGGTGAC | 433 |
| Exon 3 | ACAGCCAAGGGAAGAGAAGC | ACCTTGACACAGTGATCCTC | 422 |
| Exon 4 | CTTTTACGAACATGCGTCCTG | TCAGCAGTGGACCAAACGTC | 441 |
| Exon 5 | GCTCCGATATGTGCTTCACG | GTTCATACGTTCCTGTGCTG | 307 |
| Exon 6 | GAGCATAGGTGTGCTCTCAG | GATCTTCTGTCACTGCTGTC | 267 |
cDNA, complementary DNA.
The reverse primer for exon 2/2′ was also used to PCR amplify the two-exon Mpzl3 cDNA (primer 2R in Figure 4c).
The mutation we have identified in the rc/rc DNA is a G → A transition in exon 3 of this gene, resulting in an Arg100 → Gln substitution in the predicted 237-amino-acid polypeptide (Figure 4). This mutation was confirmed in DNA samples from multiple rc/rc mice of different parents, rc/+ mice, as well as C57BL/6J-rc genomic DNA purchased from the Jackson Laboratory (Bar Harbor, ME).
To determine whether this mutation could be a polymorphism, we carried out analysis of the coding sequence of this gene in multiple mouse strains. This mutation was absent in wild-type Balb/c, CAST/Ei, B6C3F1 (F1 hybrid of C57BL/6NCr and C3H/HeN MTV-), B6D2F1 (F1 hybrid of C57BL/6NCr and DBA/2NCr), CD1, and SwissGP mice.
The protein encoded by the mutated gene
According to our prediction based on mouse genomic sequence and homology to human and rat counterparts, the full-length polypeptide encoded by the murine ENSMUSG00000070305 gene consists of 237 amino-acid residues, and has a predicted molecular weight of 26,058 Da and isoelectric point of 7.73. It is predicted to be a type I transmembrane protein, with a signal peptide at the amino terminus (amino acids 1–32), an Ig-like V-type domain at amino acids 33–149, and a transmembrane domain at amino acids 160–182 (Figure 5). The Ig-like V-type domain is predicted to be extracellular upon cleavage of the signal peptide, and the carboxyl terminus is predicted to be cytoplasmic. The conserved cysteines in the Ig-like domain are at amino acids 53 and 129, and there is a putative N-glycosylation site (NXS/T) at amino acid 124. Arg100 is a highly conserved residue within the Ig domain.
Figure 5. Sequence comparison of the human, murine, and rat MPZL3 protein and murine MPZ and MPZL2 (EVA1).
The signal peptides and transmembrane domains are underlined, flanking the Ig-like V-type domains (in box), with conserved residues highlighted bold. N-linked glycosylation sites and the Arg100 → Gln substitution caused by the G → A mutation in the rc/rc mice are shown in small boxes. Conserved “signature” cysteines for disulfide bond formation in the Ig-like domains are marked by arrowheads. The GenBank accession number for murine Mpzl3 gene/mRNA is EF102773.
Among known murine proteins, the highest sequence homology to this 237-amino-acid protein was identified in myelin protein zero (MPZ) and myelin protein zero-like 2 (MPZL2, also called epithelial V-like antigen (EVA1)) (Figure 5). We therefore registered this novel gene as Mpzl3 (myelin protein zero-like 3) with the International Committee on Standardized Genetic Nomenclature for Mice and the Mouse Genomic Nomenclature Committee (MGNC) through the Mouse Genome Informatics (MGI) Resource (The Jackson Laboratory, Bar Harbor, ME). Both MPZ and MPZL2 (EVA1) proteins have been implicated in cell–cell adhesion (Guttinger et al., 1998), and the Ig-like domains in a number of other proteins have been shown to mediate homophilic cell–cell adhesion. Hence, it is likely that the MPZL3 protein is also involved in cell adhesion through its Ig V-type domain, and that substitution of the highly conserved Arg100 within this domain alters its function.
The coding sequence of the six-exon murine Mpzl3 transcript shares an 84.5% nucleotide identity with its human homolog, and the murine and human MPZL3 proteins share an 86.8% identity and 93.2% similarity. However, within the conserved Ig-like V-type domain, the murine and human MPZL3 proteins share a 93.3% identity and 96.6% similarity. The coding sequence of the six-exon murine Mpzl3 transcript shares a 93.3% nucleotide identity with its rat homolog, and the murine and rat MPZL3 proteins share a 96.6% identity and 97.9% similarity. Interestingly, EST evidence also suggests a two-exon Mpzl3 transcript in mice (ENSMUST00000035222) and rats (ENSRNOT00000032837), encoding a polypeptide of 96 amino acids. The 96-amino acid mouse and rat polypeptides are highly homologous, with 90.6% identity and 95.8% similarity.
A search of the available genome databases showed MPZL3 homolog in human, chimpanzee, rhesus monkey, rat, mouse, bovine, dog, opossum, and chicken. In all of these organisms, Arg100 is highly conserved. However, the function of the MPZL3 protein is not known in any of these organisms.
The murine Mpzl3 gene
The murine Mpzl3 gene consists of six exons spanning over 19 kb on mouse Chromosome 9 (44.989–45.010 Mb) (Ensembl v38; Vega release 18, 8 May 2006) (Figure 4b). According to our prediction based on homology to human and rat counterparts, the coding sequences in the murine Mpzl3 exons varied from 30 bp (exon 6) to 215 bp (exon 2′), and the introns varied from 1,657 bp (intron 3) to 6,722 bp (intron 1) (Figure 4c and d). EST analysis provided evidence for at least two transcripts through alternative splicing, both containing coding sequences flanked by a 5′ UTR that included an in-frame STOP codon and a 3′ UTR. One transcript consisted of two exons encoding a polypeptide of 96 amino acids. EST evidence suggested a 5′ UTR of at least 53 bp and a 3′ UTR of at least 3,160 bp for this transcript. This transcript, however, does not contain a mutation. Interestingly, there is a consensus splice donor site within the coding sequence of exon 2′ (Figure 4d). When this site is used, a transcript containing six exons would result from the Mpzl3 gene, encoding a protein of 237 amino acids. EST evidence suggested a 5′ UTR of at least 57 bp and a 3′ UTR of at least 261 bp for this transcript. In the rc/rc mice, the G → A missense mutation we identified in exon 3 would result in Arg100 → Gln substitution.
Expression of the Mpzl3 gene
We carried out RT-PCR analysis to determine the tissue distribution of the Mpzl3 transcripts in normal adult mice. Primers were designed to amplify the complete coding sequences from both the six-exon (845 bp amplicon using primers 2F and 4R) (Figure 4c) and two-exon (457 bp amplicon using primers 2F and 2R) (Figure 4c) transcripts based on EST predictions (Figure 6). As shown in Figure 6a, the two Mpzl3 transcripts were detected in a variety of organs examined, with high levels of expression in the brain, heart, liver, and skin. The tissue distribution patterns of the two-exon and six-exon transcripts were similar.
Figure 6. RT-PCR analysis of Mpzl3 gene expression.
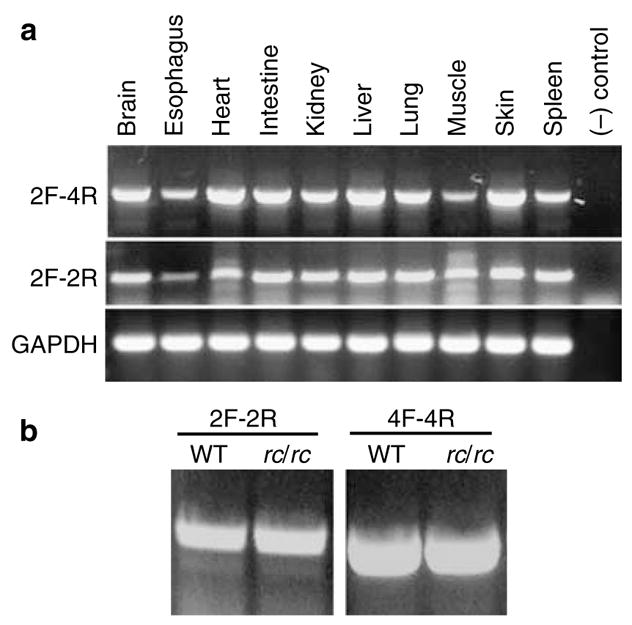
(a) Expression of the six-exon Mpzl3 transcripts (2F-4R) and the two-exon Mpzl3 transcripts (2F-2R) in normal adult mouse organs. The positions of the primers are shown in Figure 4c. The two transcripts had similar tissue distribution. (b) Expression of the six-exon Mpzl3 transcripts (4F-4R to amplify exons 3–6) and the two-exon Mpzl3 transcripts (2F-2R) in wild-type (WT) and rc/rc mouse skin.
RT-PCR analysis also showed expression of both Mpzl3 transcripts in the rc/rc mouse skin (Figure 6b). We sequenced the RT-PCR products from both normal and rc/rc skin complementary DNA (cDNA), and confirmed the G → A mutation in exon 3 of Mpzl3 in the rc/rc cDNA derived from the six-exon transcripts.
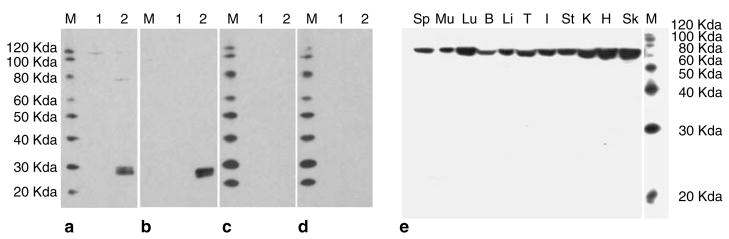
To detect the MPZL3 protein, we generated affinity-purified rabbit polyclonal antibodies against a peptide (DKLTIDWTYRPPSSSRT) in the predicted extracellular domain of the murine MPZL3 protein. To test the specificity of these antibodies, we transfected NIH/3T3 cells with an expression vector encoding a fusion protein between the murine MPZL3 and a Myc epitope tag. We analyzed total cell extracts by Western blot analysis using the anti-MPZL3 antibodies, detecting two bands at approximately 27 and 29 kDa (Figure 7a). These bands were also recognized by the anti-Myc antibody (Figure 7b). The size difference may be due to different post-translational modifications. When the anti-MPZL3 antibodies were preabsorbed with the DKLTIDWTYRPPSSSRT peptide at a 1:3, 1:10, or 1:32 molar ratio, binding to the fusion protein became much weaker at a 1:3 molar ratio (Figure 7c; very faint bands at 27 and 29 kDa were visible with a much longer exposure, data not shown), and could not be detected at a 1:10 (Figure 7d) or 1:32 molar ratio (data not shown). These results suggest that the anti-MPZL3 antibodies are specific for the antigen. The anti-MPZL3 antibodies also detected 27 and 29 kDa bands in NIH/3T3 cells transfected with a plasmid encoding the rc mutant MPZL3 protein fused to the Myc tag (data not shown), as expected. In addition, using the anti-MPZL3 antibodies, we detected bands that were of much higher molecular weight (~80 and ~110 kDa), which were not detected with preabsorbed anti-MPZL3 antibodies or the anti-Myc antibody (Figure 7a–d). These bands could be endogenous MPZL3 with different post-translational modifications or from different transcripts.
Figure 7. Characterization of the anti-MPZL3 antibodies and expression of MPZL3 in adult mouse organs.
(a–d) Western blot analysis of total cell extracts from control NIH/3T3 cells (lane 1) and NIH/3T3 cells transfected with pEF/myc/cyto containing wild-type murine Mpzl3 cDNA (lane 2). The primary antibodies used were: (a) rabbit anti-MPZL3 peptide DKLTIDWTYRPPSSSRT affinity-purified antibodies, (b) mouse anti-Myc tag antibody, rabbit anti-MPZL3 affinity-purified antibodies preabsorbed with (c) 1:3 or (d) 1:10 molar ratio of peptide DKLTIDWTYRPPSSSRT. Blot c showed very faint bands of 27 and 29 kDa after a much longer exposure (data not shown). (e) Western blot analysis of total protein extracts from adult mouse organs detected by rabbit anti-MPZL3 affinity-purified antibodies. Sp: spleen; Mu: muscle; Lu: lung; B: brain; Li: liver; T: testis; I: intestine; St: stomach; K: kidney; H: heart; Sk: skin. M: MagicMark XP.
In all the adult mouse organs analyzed by Western blot, we detected a single band at approximately 70 kDa (Figure 7e). When we used preabsorbed antibodies to analyze skin and kidney extracts, we detected no signal (data not shown), suggesting the antibody binding was specific. The band detected is much larger than the size predicted from the amino-acid sequence (237 amino acids before cleavage of predicted signal peptide), which may be a result of post-translational modifications, such as glycosylation or dimerization or from translation of different transcripts. Interestingly, the bands detected in NIH/3T3 cells (~80 and ~110 kDa) were not detected in any mouse organs analyzed, suggesting they may be unique for cultured NIH/3T3 cells.
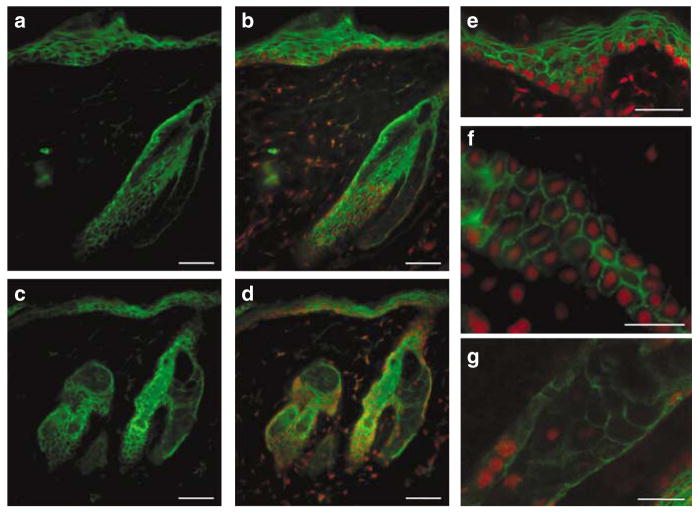
We then carried out indirect immunofluorescence of adult mouse skin sections to determine MPZL3 protein expression and localization. We detected MPZL3 expression in keratinocytes of the epidermis and hair follicles (Figure 8). By examining staining at high magnifications, it was clear that the staining was strong around the plasma membrane (Figure 8e and f), consistent with the prediction of a transmembrane protein involved in cell adhesion. We also detected staining in the cytoplasm, but not in the nuclei. We did not detect differences in MPZL3 protein distribution pattern between normal and rc/rc mouse skin. Using rc/rc skin sections with hypertrophic sebaceous glands, we also detected MPZL3 protein expression in the sebocytes (Figure 8c, d, and g). For negative control, indirect immunofluorescence using normal goat serum instead of anti-MPZL3 antibodies did not show any specific staining (data not shown).
Figure 8. Expression of MPZL3 in mouse skin sections detected by indirect immunofluorescence.
(a–f) MPZL3 (green) was detected in the keratinocytes of the epidermis and hair follicles as well as (a–d, g) the sebocytes. (b, d–g) Sections were counterstained with propidium iodide (red). (a, b): +/+ skin; (c–g): rc/rc skin. Bar = 50 μm in (a–d), 20 μm in (e–g).
Sequence analysis of the Mpzl3 gene in ruf/ruf mice
The ruf/ruf mice develop skin abnormalities similar to the rc/rc mice, such as “unkempt” and “wet” looking hair coats and sebaceous gland hypertrophy (Park et al., 2001), and the locus has been mapped to Chromosome 9 (Sweet et al., 1990). Although allelism between these two spontaneous mutations has not been tested directly, it is possible that they are caused by mutations in the same gene. However, our sequence analysis of the Mpzl3 gene coding sequences and splice sites did not detect any difference between the parental strain C3H/HeJ and C3H/HeJ-ruf genomic DNA purchased from the Jackson Laboratory (Bar Harbor, ME). Therefore, our results suggest that ruf and rc are likely not allelic.
DISCUSSION
In this study, we carried out positional cloning of the gene mutated in the rc mice. We mapped the rc locus to a 246-kb interval by high-resolution linkage analysis, and identified a missense mutation, which would result in an Arg100 → Gln substitution in a novel open reading frame within this interval. Based on the predicted domain structure of the encoded polypeptide, we named this novel gene Mpzl3, encoding a full-length MPZL3 polypeptide of 237 amino acids. Arg100 is a highly conserved residue within the conserved Ig-like V-type domain, and its substitution likely alters MPZL3 protein function in the rc/rc mice.
The MPZL3 protein
Through BLAST search, the highest levels of sequence homology with the MPZL3 protein were identified in the MPZ and MPZL2 (also called EVA1) proteins (Figure 5). Within the Ig V-type domain, the murine MPZL3 protein shares a 40.0% identity and 54.2% similarity at the amino-acid level with murine MPZ, and a 36.1% identity and 60.5% similarity at the amino-acid level with murine EVA1. All the consensus residues within the Ig V-type domain, such as the cysteines and the N-glycosylation site, as well as the Arg corresponding to Arg100 in the murine MPZL3 protein, are conserved between these three proteins. Based on the similarity of its domain structure to MPZ and EVA1 and its localization to the plasma membrane, the MPZL3 protein is likely involved in cell adhesion as reported for MPZ and EVA1 (Guttinger et al., 1998). Interestingly, the Mpzl3 gene is located approximately 1 kb 3′ to the Eva1 gene, and the sizes of exons 2 and 3 are identical between the two genes (167 and 211 bp, respectively). It is possible that one of these two genes arose through tandem duplication.
Whereas the Mpzl3 gene encodes a short peptide of 96 amino acids through an alternatively spliced transcript in both mice and rats, there is no EST evidence that such a transcript exists in humans. Both the mouse and rat 96-amino acid polypeptides possess a signal peptide but have only a portion of the Ig-like domain, and lack a transmembrane domain downstream. In addition, mouse EST evidence suggests the existence of other forms of Mpzl3 transcripts (Figure 4b). It is not known whether such transcripts are expressed at significant levels in mice or rats, and the subcellular location and possible functions of the encoded polypeptides remain to be determined.
The rc mutation
The rc mutation arose spontaneously without mutagens in 1966, and the allele has been maintained in cryopreserved embryos during most of the past four decades. Hence, we expected a “simple” mutation without gross chromosomal rearrangement (such as those induced by X-ray irradiation). It is therefore not surprising that we detected a single-nucleotide transition, which resulted in an amino-acid substitution. The mutated Mpzl3 transcripts and protein were detected at significant levels in the skin of rc/rc mice, suggesting that the effect of the mutation is likely post-translational. Substitution of the highly conserved Arg100 in the conserved Ig-like domain required for adhesion may result in decreased cell adhesion. Shedding of hair at the telogen phase in the rc/rc mice may be one of the effects of such reduced adhesion.
In this study, we did not detect any mutations/polymorphisms in the Mpzl3 gene in other mouse strains we analyzed. However, there are at least five documented variations in the mouse Mpzl3 gene between mouse strains: four of them being synonymous, and one being non-synonymous. This non-synonymous polymorphism results in an Ala27 → Val substitution within the signal peptide in the 129X1/SvJ strain. It likely has no functional consequence on the mature protein, as the mice appear normal.
The abnormalities in the rc/rc mice are not limited to the skin (Hayashi et al., 2004), and the mutated gene likely plays a role in the normal functioning of multiple organ systems. Not surprisingly, we detected the Mpzl3 transcripts and protein in a variety of organs. However, no other alleles of the Mpzl3 mutation have been described. It is possible that the mutation we identified in the rc/rc mice is a “hypomorph”. A more drastic change in the Mpzl3 gene, such as a large deletion or a frame-shift mutation resulting in the loss of functional domains, may lead to much more severe abnormalities in other organ systems and result in a lethal phenotype, and would therefore have never been documented as a natural mutation. Future functional analyses such as knockout or knockdown experiments will be able to address such a possibility.
Mutations in genes encoding adhesion molecules
Mutations in a number of genes involved in cell adhesion, particularly components of the desmosomes, both transmembrane and intracellular, have been associated with defects of the skin and heart where there are high levels of mechanical stress (McMillan and Shimizu, 2001). The Mpzl3 gene expression was detected at high levels in the skin and heart, where the rc/rc mice develop severe abnormalities (Hayashi et al., 2004), and the MPZL3 protein is localized to the plasma membrane, consistent with the assumption that it may be involved in cell adhesion. Whereas there have been no reports of mutations in the MPZL2 (EVA1) gene in human diseases, substitutions of conserved residues such as Thr124 → Met in the Ig domain in the MPZ protein have been identified in patients with Charcot-Marie-Tooth disease (Chapon et al., 1999; De Jonghe et al., 1999; Misu et al., 2000). Similarly, substitution of the highly conserved Arg100 in the Ig domain required for adhesion likely results in abnormal MPZL3 protein function in the rc/rc mice.
Interestingly, transgenic mice overexpressing c-myc in the keratinocyte stem cells also show sebaceous hypertrophy, hair loss, and spontaneous wounds (Arnold and Watt, 2001; Waikel et al., 2001; Frye et al., 2003), although the phenotype is dominant and much more severe than in the rc/rc mice. Overexpression of c-myc has been shown to drive the keratinocyte stem cells out of the stem cell compartment (Arnold and Watt, 2001; Waikel et al., 2001), and gene expression profiling has revealed that 40% of all down-regulated genes in the c-myc transgenic mice encoded cell adhesion molecules or cytoskeleton proteins, resulting in the reduced adhesive interactions of keratinocyte stem cells with the local microenvironment or niche (Frye et al., 2003). It is possible that the rc mutation in Mpzl3 leads to similarly compromised interactions of the keratinocytes with their microenvironment and manifestation of the rc phenotype.
MATERIALS AND METHODS
Mice
All animal procedures were approved by the University of Hawaii Institutional Animal Care and Use Committee (IACUC). Mice were maintained in a temperature-, humidity-, and light cycle (12:12)-controlled vivarium under specific pathogen-free conditions. One male and three female heterozygous rough coat (+/rc) mice in the C57BL/6J (B6/J) strain background were purchased from the Jackson Laboratory (Bar Harbor, ME) to establish our own rough coat mouse colony. Female BALB/cJ and CAST/Ei mice were also purchased from the Jackson Laboratory (Bar Harbor, ME) for backcross studies.
Histological analysis
The dorsal skin of euthanized B6/J-rc/rc and age- and sex-matched B6/J- +/rc mice was shaved, and skin biopsies were collected. They were embedded in OCT embedding medium or fixed in phosphate-buffered formalin and dehydrated and cleared in xylene before being embedded in paraffin. The cryosections were stained with oil red O for lipids, and the paraffin sections were stained with hematoxylin and eosin.
Backcross
B6/J-rc/rc mice were outcrossed with both female BALB/cJ and CAST/Ei mice to obtain F1 hybrids (+/rc). Because female rc/rc mice do not breed well (Hayashi et al., 2004), we used male B6/J-rc/rc mice and F1 females for our backcross experiment to obtain F2 hybrids for linkage analysis.
Genotyping
Genomic DNA of F2 hybrids was extracted from tail tip biopsy at the time of weaning using Proteinase K (Invitrogen Corporation, Carlsbad, CA) digestion and ethanol precipitation. PCR reactions were carried out to amplify microsatellites polymorphic between the parental strains, and the amplified DNA fragments (amplicons) were analyzed using 4% Metaphor agarose (Cambrex, Rockland, ME) gel electrophoresis. In cases when there were no more published microsatellite polymorphisms between the parental strains in the Mouse Genome Informatics “Strains and Polymorphisms” database (www.informatics.jax.org) or the Ensembl Mouse Genome Database (www.ensembl.org/Mus_musculus), we designed primers to detect novel polymorphisms between B6/J and BALB/cJ or between B6/J and CAST/Ei. Microsatellites with at least 15 CA or TG repeats based on sequence information in the Ensembl Mouse Genome Database (C57BL/6 strain) were amplified by PCR from these strains and the amplicons were analyzed by agarose gel electrophoresis. Those with detectable polymorphisms (at least 8 bp on a 4% Metaphor agarose gel) were used for linkage analysis.
Linkage analysis
Backcross offspring were scored for recombination events that segregate the microsatellite markers contributed by the two parental alleles and the rc locus (indicated by the phenotype). The distances between the loci and the LOD scores were calculated using the QTXb20 software (Manly et al., 2001).
Mutation detection
Primers for sequence analysis were designed based on the C57BL/6 genomic sequence and exon structure in the Ensembl Mouse Genome Database (www.ensembl.org/Mus_musculus). Primers were located in the introns, 5′ and 3′ UTRs or 5′ and 3′ flanking sequences, so that mutations in the coding sequences of exons as well as the splice donor, acceptor, and branch sites could be detected. Primers were synthesized at Integrated DNA Technologies (Coralville, IA), and PCR reactions were carried out using both wild-type B6/J and B6/J-rc/rc mouse genomic DNA as templates. The amplicons were analyzed by agarose gel electrophoresis, and the DNA was recovered from the gel using GeneClean Spin Kit (Q-Biogene, Carlsbad, CA). Sequences from both strands were obtained using the BigDye sequencing kit and an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA) at sequencing core facilities (MBSR, CGPBRI, and GMBF) at University of Hawaii at Manoa. Once the mutation was identified, it was confirmed in four more mice each of both +/+ and rc/rc genotypes from different parents, and in heterozygous (+/rc) mice. We also confirmed the mutation in B6/J-rc genomic DNA purchased from the Jackson Laboratory (Bar Harbor, ME) and in PCR-amplified cDNA generated from rc/rc mouse skin RNA. Sequence analysis of the mutated gene was also carried out on genomic DNA extracted from BALB/cJ, CAST/Ei, C3H/HeJ mice (The Jackson Laboratory, Bar Harbor, ME), B6C3F1, B6D2F1, CD1, and SwissGP mice (NCI-Frederick Animal Production Area, Frederick, MD), and C3H/HeJ-ruf genomic DNA purchased from the Jackson Laboratory (Bar Harbor, ME).
RNA extraction and RT-PCR analysis
The dorsal skin of euthanized B6/J-rc/rc and age- and sex-matched B6/J- +/+ mice was shaved, and skin biopsies as well as the internal organs were collected. Samples were frozen immediately in liquid nitrogen or immersed in RNAlater (Qiagen, Valencia, CA). Total RNA was extracted using TriReagent (Molecular Research Center Inc., Cincinnati, OH) following the manufacturer’s manual. Total RNA was reverse transcribed using the SuperScript III Reverse Transcriptase Kit (Invitrogen Corp., Carlsbad, CA). The first strand cDNA was used as templates for PCR amplification. The quality of the cDNA was confirmed by PCR amplification of glyceraldehyde-3-phosphate dehydrogenase cDNA analyzed by agarose gel electrophoresis.
Antibody generation
Two rabbits were immunized with synthesized peptide DKLTIDWTYRPPSSSRT (amino acids 63–79, in the predicted extracellular domain of the murine MPZL3 protein), and the serum was affinity purified for antibodies against the peptide (Bethyl Laboratories, Montgomery, TX).
Western blot analysis and indirect immunofluorescence
The full coding sequences of the 6-exon wild-type murine Mpzl3 cDNA, as well as Mpzl3 cDNA harboring the rc mutation, were subcloned into the PstI/NotI sites of pEF/myc/cyto (Invitrogen Corp., Carlsbad, CA), so that the Myc tag was at the carboxyl terminus of the fusion protein. The resulting plasmids were transfected into NIH/3T3 cells (ATCC, Manassas, VA) using LipofectAmine 2000 (Invitrogen Corp., Carlsbad, CA), and the total cellular proteins were extracted using modified radioimmunoprecipitation buffer (50 mM Tris-Cl, pH 7.4, 1 mM each of EDTA, phenylmethylsulfonylfluoride, Na3VO4, and NaF, 1% NP-40, 62.5 mM each of ALLN and N-ethylmaleimide, and 1:15 diluted protease inhibitor cocktail). Biopsies of adult mouse skin were homogenized and the proteins extracted as described (He et al., 2002). Protein extracts were analyzed by gradient (Invitrogen Corp., Carlsbad, CA) or non-gradient SDS-PAGE and blotted onto Immobilon transfer membrane (Millipore Corporation, Billerica, MA) for Western blot analysis following standard protocols. The molecular weight marker used was MagicMark XP (Invitrogen Corp., Carlsbad, CA). The rabbit anti-MPZL3 antibodies were diluted 1:400 and mouse anti-Myc tag antibody (Covance Research Products, Princeton, NJ) was diluted 1:200. For antibody preabsorption, the rabbit anti-MPZL3 antibodies were incubated with excess peptide DKLTIDWTYRPPSSSRT at 1:3, 1:10, or 1:32 molar ratio for 1 hour before being added to the blot. Antibody binding was detected with ECL Western Blot Detection Reagents (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) and BioMax film (Eastman Kodak Company, Rochester, NY).
Cryosections of back skin biopsies of normal and rc/rc mice were incubated with 1:200 dilutions of rabbit anti-MPZL3 antibodies or normal goat serum. Antibody binding was detected with goat anti-rabbit IgG conjugated with FITC, and the slides were mounted in VectorShield with propidium iodide (Vector Laboratories, Burlingame, CA). The sections were examined and photographed on a Zeiss AxioSkop 2 Plus fluorescent microscope or a Zeiss 5 PASCAL LSM confocal microscope (Carl Zeiss MicroImaging Inc., Thornwood, NY).
Acknowledgments
This work was supported by Grants AR047713 (KC) and AR050487 (TC) from NIH/NIAMS and G12RR003061 (KC, TC) from NIH/NCRR (RCMI program), and the Ingeborg v.F. McKee Fund from the Hawai’i Community Foundation (20040450 and 20050401 to TC). We thank Scott Lozanoff, Charles D. Boyd, Yusuke Marikawa, Olivier Le Saux, Athula Wikramanayake’s laboratory, Yvonne Tatsumura, Keith S Fong, Xiao-Jing Wang’s laboratory (Oregon Health and Science University), Dennis R Roop and Paul A Overbeek (Baylor College of Medicine) for helpful suggestions, Shannon Bennett and Durrell Kapan for help in linkage analysis, and Darlene Ramones and Marisa Tricas for technical assistance. Sequencing analyses were carried out at the CGPBRI, GMBF, and MBSR Sequencing Core Facilities at UHM, and microscopic images were captured and analyzed at the RCMI Imaging Core Facility. TC would like to dedicate this work to her PhD mentor, Dr Laurence D Etkin (University of Texas MD Anderson Cancer Center).
Abbreviations
- C57BL/6J
B6/J
- EST
expressed sequence tag
- EVA1
epithelial V-like antigen 1 (also called MPZL2)
- Mb
megabase
- MPZ
myelin protein zero
- MPZL2
myelin protein zero-like 2 (also called EVA1)
- MPZL3
myelin protein zero-like 3
- rc
rough coat
- RT
reverse transcriptase
- ruf
rough fur
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
References
- Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001;11:558–68. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- Chapon F, Latour P, Diraison P, Schaeffer S, Vandenberghe A. Axonal phenotype of Charcot-Marie-Tooth disease associated with a mutation in the myelin protein zero gene. J Neurol Neurosurg Psychiatry. 1999;66:779–82. doi: 10.1136/jnnp.66.6.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonghe P, Timmerman V, Ceuterick C, Nelis E, De Vriendt E, Lofgren A, et al. The Thr124Met mutation in the peripheral myelin protein zero (MPZ) gene is associated with a clinically distinct Charcot-Marie-Tooth phenotype. Brain. 1999;122(Part 2):281–90. doi: 10.1093/brain/122.2.281. [DOI] [PubMed] [Google Scholar]
- Dickie MM. Rough coat. Mouse News Lett. 1966;34:30. [Google Scholar]
- Eicher EM, Fox S, Reynolds S. Rough coat on Chromosome 9. Mouse News Lett. 1977;56:42. [Google Scholar]
- Fernandez-Gonzalez A, La Spada AR, Treadaway J, Higdon JC, Harris BS, Sidman RL, et al. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science. 2002;295:1904–6. doi: 10.1126/science.1068912. [DOI] [PubMed] [Google Scholar]
- Frye M, Gardner C, Li ER, Arnold I, Watt FM. Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development. 2003;130:2793–808. doi: 10.1242/dev.00462. [DOI] [PubMed] [Google Scholar]
- Guttinger M, Sutti F, Panigada M, Porcellini S, Merati B, Mariani M, et al. Epithelial V-like antigen (EVA), a novel member of the immunoglobulin superfamily, expressed in embryonic epithelia with a potential role as homotypic adhesion molecule in thymus histogenesis. J Cell Biol. 1998;141:1061–71. doi: 10.1083/jcb.141.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Cao T, Passmore H, Saux CJ, Fogelgren B, Khan S, et al. Progressive hair loss and myocardial degeneration in rough coat mice: reduced lysyl oxidase-like (LOXL) in the skin and heart. J Invest Dermatol. 2004;123:864–71. doi: 10.1111/j.0022-202X.2004.23436.x. [DOI] [PubMed] [Google Scholar]
- He W, Li AG, Wang D, Han S, Zheng B, Goumans MJ, et al. Overexpression of Smad7 results in severe pathological alterations in multiple epithelial tissues. EMBO J. 2002;21:2580–90. doi: 10.1093/emboj/21.11.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly KF, Cudmore RH, Jr, Meer JM. Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome. 2001;12:930–2. doi: 10.1007/s00335-001-1016-3. [DOI] [PubMed] [Google Scholar]
- McMillan JR, Shimizu H. Desmosomes: structure and function in normal and diseased epidermis. J Dermatol. 2001;28:291–8. doi: 10.1111/j.1346-8138.2001.tb00136.x. [DOI] [PubMed] [Google Scholar]
- Misu K, Yoshihara T, Shikama Y, Awaki E, Yamamoto M, Hattori N, et al. An axonal form of Charcot-Marie-Tooth disease showing distinctive features in association with mutations in the peripheral myelin protein zero gene (Thr124Met or Asp75Val) J Neurol Neurosurg Psychiatry. 2000;69:806–11. doi: 10.1136/jnnp.69.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YG, Hayasaka S, Takagishi Y, Inouye M, Okumoto M, Oda S. Histological characteristics of the pelage skin of rough fur mice (C3H/HeJ-ruf/ruf) Exp Anim. 2001;50:179–82. doi: 10.1538/expanim.50.179. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Chertkov O, Borue XV, Agulnik SI, Gibson-Brown JJ, Lyle SR, et al. Genetics analysis of mouse mutations abnormal feet and tail and rough coat, which cause developmental abnormalities and alopecia. Mamm Genome. 2002;13:675–9. doi: 10.1007/s00335-002-2191-6. [DOI] [PubMed] [Google Scholar]
- Searle AG, Spearman RI. “Matted”, a new hair mutant in the house mouse: genetics and morphology. J Embryol Exp Morphol. 1957;5:93. [Google Scholar]
- Sweet HO, Oda SI, Taylor BA, Rowe L, Davisson MT, Cook S, et al. Rough fur (ruf) Mouse Genome. 1990;86:236–7. [Google Scholar]
- Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet. 2001;28:165–168. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]