Abstract
Membrane vesicle (MV) release remains undefined, despite its conservation among replicating Gram-negative bacteria both in vitro and in vivo. Proteins identified in Salmonella MVs, derived from the envelope, control MV production via specific defined domains that promote outer membrane protein-peptidoglycan (OM-PG) and OM protein-inner membrane protein (OM-PG-IM) interactions within the envelope structure. Modulation of OM-PG and OM-PG-IM interactions along the cell body and at division septa, respectively, maintains membrane integrity while coordinating localized release of MVs with distinct size distribution and protein content. These data support a model of MV biogenesis, wherein bacterial growth and division invoke temporary, localized reductions in the density of OM-PG and OM-PG-IM associations within the envelope structure, thus releasing outer membrane as MVs.
Keywords: surface organelle, membrane vesicle, outer membrane
INTRODUCTION
The release of membrane vesicles (MVs) is conserved among Gram-negative bacteria (Kuehn and Kesty, 2005), including many pathogens (Stephens et al., 1982; Brandtzaeg et al., 1992; Garcia-del Portillo et al., 1997; Fiocca et al., 1999; Hellman et al., 2000; Keenan et al., 2000; Namork and Brandtzaeg, 2002; Rosenberger et al., 2004; Marsollier et al., 2007; Necchi et al., 2007). MVs originate from the bacterial surface by an undefined process, and are composed of outer membrane (OM) and periplasmic constituents, including proteins, phospholipids, and lipopolysaccharides (McBroom and Kuehn, 12 May 2005, posting date; Kuehn and Kesty, 2005; Mashburn-Warren and Whiteley, 2006). MVs from bacteria have been observed in many environments, including in vivo, where the function of MVs is likely multifaceted: MVs act as primary delivery vehicles for bacterial toxins lacking typical signal sequences (Horstman and Kuehn, 2000; Wai et al., 2003), promote cell-cell communication via transit of signaling molecules (Mashburn and Whiteley, 2005), inhibit phagosome-lysosome fusion during macrophage infection (Fernandez-Moreira et al., 2006), and are rich in antigens that serve as initial targets for innate and adaptive immune recognition (Bergman et al., 2005), generating protective immunity against bacterial challenge when used as an immunogen (Sexton et al., 2004; Alaniz et al., 2007).
The presence and importance of MVs released by bacteria growing on solid agar (Tetz et al., 1990), within biofilms (Beveridge et al., 1997; Schooling and Beveridge, 2006), in vitro (Garcia-del Portillo et al., 1997; Mashburn and Whiteley, 2005; Fernandez-Moreira et al., 2006), and in vivo (Stephens et al., 1982; Brandtzaeg et al., 1992; Fiocca et al., 1999; Marsollier et al., 2007; Necchi et al., 2007) have become increasingly evident. Although observed for decades (Knox et al., 1966; Work et al., 1966; Chatterjee and Das, 1967), the process by which Gram-negative organisms produce MVs is unknown. Several groups have investigated this question, proposing a range of models describing MV release. Early models suggested that MVs result from OM growth exceeding that of the peptidoglycan (Wensink and Witholt, 1981), or when fewer OM lipoprotein linkages to underlying layers are present (Hoekstra et al., 1976). However, direct supporting evidence of these proposed models, such as electron microscopic or quantitative methods to determine, for example, the rate of OM/peptidoglycan growth or the role or location of OM lipoprotein linkages, was not provided in these publications. Increased MV release has been observed in mutants lacking components of the tol-pal system, a group of envelope proteins exploited for entry of filamentous bacteriophages and group A colicins in E. coli (Webster, 1991; Bernadac et al., 1998). Drawing definitive conclusions about the role of these proteins in MV release, however, is confounded by comparing non-isogenic strains with multiple mutations, qualitative assessment of MV production, and electron microscopy of organisms during stationary phase (during which MV release is limited (Hoekstra et al., 1976; Bauman and Kuehn, 2006)). Furthermore, the suggestion that wild-type (WT) organisms do not produce MVs (Bernadac et al., 1998) is in direct conflict with observations of MV production by a variety of Gram-negative bacteria, including pathogens growing in vivo (Stephens et al., 1982; Brandtzaeg et al., 1992; Garcia-del Portillo et al., 1997; Fiocca et al., 1999; Hellman et al., 2000; Keenan et al., 2000; Namork and Brandtzaeg, 2002; Rosenberger et al., 2004; Marsollier et al., 2007; Necchi et al., 2007), and genetic evidence suggesting MV release cannot be abolished (McBroom and Kuehn, 12 May 2005, posting date; McBroom et al., 2006)). Subsequent investigations have proposed that MV release can act as an envelope stress response able to quickly rid the cell surface of misfolded proteins (McBroom and Kuehn, 2007), or that expression of modified forms of LPS (Kadurugamuwa and Beveridge, 1997; Nguyen et al., 2003) or interaction of hydrophobic molecules with the OM, such as the cell-cell communication molecule pqs (Mashburn and Whiteley, 2005; Mashburn-Warren et al., 2008), can stimulate MV production.
Multiple factors, some organism-specific, are apparently capable of inducing the release of MVs, yet the mechanism by which MVs are produced remains unknown (McBroom and Kuehn, 12 May 2005, posting date; Kuehn and Kesty, 2005; Mashburn-Warren and Whiteley, 2006), and quantitative approaches to this question have been underutilized to date. The conservation of this process among Gram-negative bacteria suggests the possibility of unifying factors or processes utilized by organisms demonstrating release of MVs. We used a multiphasic and quantitative approach to gain insight into the biogenesis of MVs, and demonstrate here that the release of MVs from actively dividing bacteria is specifically modulated by the density and distribution of highly conserved envelope protein interconnections.
RESULTS
Influence of protein constituents on MV release
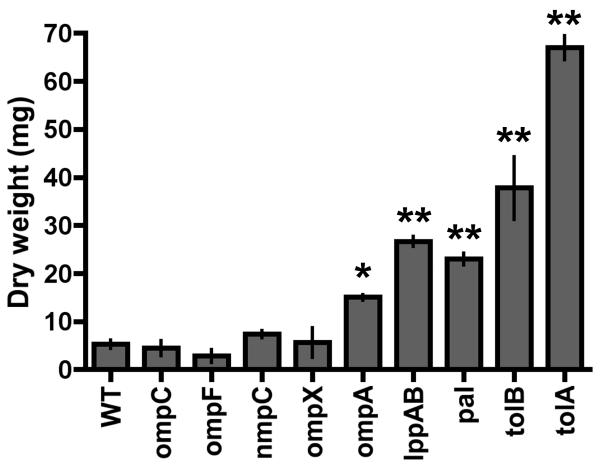
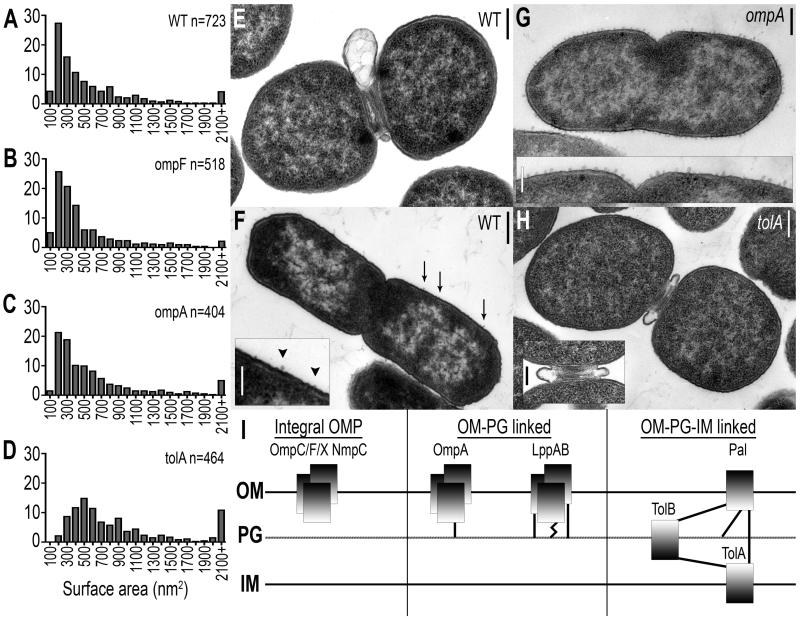
The release of MVs by Gram-negative bacteria has been investigated for at least 40 years (Knox et al., 1966; Work et al., 1966; Chatterjee and Das, 1967), but only recently have more global approaches been applied to understanding the key components of this process (McBroom et al., 2006). In general, it is known that MVs contain OM and periplasmic (PP) proteins, phospholipids, and lipopolysaccharide (Kuehn and Kesty, 2005). Therefore, we used proteomics as an unbiased approach to define the major protein constituents of MVs produced by WT bacteria, and then tested the hypothesis that these proteins were involved in MV release by developing quantitative methods to compare WT bacteria with isogenic mutants, each lacking one of the major MV proteins identified. Using 2D SDS-PAGE and MALDI-TOF mass spectrometry, we identified the major WT MV proteins as OmpC, OmpF, NmpC, OmpX, OmpA, LppAB, Pal, and TolB. We constructed targeted in-frame deletions of coding sequence of each corresponding gene and the mass of MVs produced by WT Salmonella and each mutant strain was quantified by a combination of tangential flow diafilatration and ultracentrifugation (see Experimental Procedures; Figure 1). MV release by mutants lacking OmpC, OmpF, NmpC, or OmpX was similar to WT (p>0.05), while mutants lacking OmpA, LppAB, Pal, TolB, or TolA (which closely associates with Pal and TolB) significantly increased MV production (p<0.01 for ompA, p<0.001 for lppAB, pal, tolB, and tolA). More in depth examination of MV size, reported to range from 10-200nm in diameter (Kuehn and Kesty, 2005), revealed that the size distribution of MVs released by WT S. typhimurium (Figure 2A), was unchanged in the absence of OmpC, OmpF, NmpC, or OmpX (Figure 2B, p>0.05). While the size distribution of MVs released by the ompA mutant was not significantly different than WT (Figure 2C, ompA, p=0.059), the population of MVs released by the lppAB mutant was shifted toward smaller MVs (Figure S1 lppAB, p<0.001). The pal, tolB, and tolA mutants, however, released MVs significantly larger than WT (Figure 2D, p<0.001).
Figure 1. Major MV proteins influence MV production.
MV production is quantitatively altered in bacteria lacking major MV proteins. Dry weight of MVs harvested from WT Salmonella and mutant strains was quantified (mean +/- standard error). *p<0.01 **p<0.001
Figure 2. WT MV release localized to division septa and cell body; MV proteins control size and localization of MV release.
(A) Size distribution of WT Salmonella MVs; x-axis represents MV size ranges (100 represents 1-100nm2). Compared to WT MVs, size distribution of (B) ompF MVs and (C) ompA MVs is similar (p>0.05; also ompC, nmpC, and ompX MVs, data not shown), while (D) tolA MV size is significantly increased (p<0.001; also pal and tolB MVs, data not shown). (E) WT MVs are released at constricted division septa and (F) along the cell body. (G) MVs are released along the cell body in the absence of OmpA (shown) and LppAB (Figure 3A), and (H) MV release occurs at division septa in tolA (shown), pal (Figure 3D), and tolB (data not shown) mutant strains. (I) Major MV proteins classified by envelope interconnections: Integral OM proteins OmpC, OmpF, OmpX, and NmpC lack extensive connectivity to envelope components, Lpp and OmpA bind PG (OM-PG linked), and Pal, TolB, and TolA form membrane-spanning protein complexes (OM-PG-IM linked). Dark shading represents N-termini, straight lines represent non-covalent interactions, and zig-zag line denotes covalent interaction. Bars = 200nm (except F inset, bar = 100nm), n = number of individual vesicles examined.
Although MV release has been described in general terms as occurring at bacterial surfaces (McBroom and Kuehn, 12 May 2005, posting date), MV release by Salmonella had not been previously visualized. We sought to capture nascent MV formation by extensively surveying electron micrographs of actively dividing cells. Strikingly, WT bacteria released MVs at constricted division septa and at locations distributed along the cell body (Figures 2E, F, S2); similar release was seen in cells lacking OmpC, OmpF, NmpC, or OmpX (data not shown). However, the absence of OmpA or LppAB induced localization of MV release along the cell body at regularly spaced intervals (Figures 2G, 3A, S3A). Conversely, increased MV release was localized to constricted division septa in the absence of Pal, TolB, or TolA (Figures 2H, 3D, S3B).
Figure 3. Envelope protein domains modulate MV production.
(A) MV release along cell body in the lppAB strain is (B) complemented with full length Lpp1-58, but (C) expression of abundant mutated Lpp1-57 unable to bind PG retains lppAB mutant MV production. (D) MV release at division septa in pal strain is (E) restored to WT MV release upon expression of full length Pal1-153, but (F) cannot be complemented by expression of truncated Pal1-123 unable to bind TolA. Bar = 200nm
Subsets of major MV proteins, therefore, similarly impact WT MV production. Integral outer membrane proteins (OMPs) OmpC, OmpF, NmpC, and OmpX did not alter WT MV release (Figures 1, 2B). Expression of LppAB and OmpA, OM proteins which bind PG via covalent (Braun and Sieglin, 1970) and non-covalent (De Mot and Vanderleyden, 1994; Koebnik, 1995) interactions, respectively (Figure 2I, middle panel, OM-PG linked), minimized cell body MV production (Figures 1, 2C,G). Pal, TolB, and TolA bridge the IM and OM via protein-protein and protein-PG interactions (Clavel et al., 1998; Walburger et al., 2002; Cascales and Lloubes, 2004; Parsons et al., 2006) (Figure 2I, right panel, OM-PG-IM linked), restricting MV release and MV size at division septa (Figures 1, 2D,H). Therefore, OM-PG linked proteins appear to control WT MV production at cell body locations (Figure 2G), supported by an even distribution of OM-PG linked proteins Lpp and OmpA along the cell body (Braun and Rehn, 1969; Lai et al., 2004), while OM-PG-IM complexes modulate MV production at division septa (Figure 2H), where the proteins Pal, TolA, and TolB, bridging the OM-PG-IM space, migrate during cell division (Gerding et al., 2007).
Interconnecting protein domains modulate MV release
One possible explanation for the impact of OM-PG and OM-PG-IM linked proteins on MV production is that their biophysical properties, resulting from their abundance in the envelope, contribute significantly to maintaining membrane stability. Their absence would result in envelope defects manifest as MV release. This seems unlikely, however, as MV release is unchanged in integral OMP mutant strains (Figures 1, 2B), which lack proteins of similar abundance to OM-PG and OM-PG-IM linked proteins in the envelope (Chai and Foulds, 1977). Alternatively, OM-PG and OM-PG-IM linked proteins may act specifically via protein-PG and/or protein-protein interactions to modulate MV production. We investigated whether changes in MV production resulted from the absence of an abundant envelope protein, or the specific envelope interconnection, using Lpp and Pal as representative OM-PG and OM-PG-IM linked proteins (Figure 2I).
Lpp, the most abundant protein in the cell envelope, forms trimers in vivo (Braun et al., 1970; Inouye et al., 1972). One-third of each trimer covalently binds PG via the C-terminal lysine residue (Braun and Rehn, 1969), and removal of this single amino acid (aa) renders Lpp incapable of covalent PG interaction. Full length Lpp, as well as mutated Lpp protein lacking only the C-terminal lysine residue, were expressed in the lppAB mutant strain (Figure S4A) in the OM (Figure S4B). As in the ompA mutant (Figure 2G), a strain which also lacks OM-PG connections (Figure 2I), MV release was similarly increased along the cell body in the lppAB mutant (Figure 3A). Expression of full length Lpp (Lpp1-58) restored covalent Lpp-PG interactions (Braun and Sieglin, 1970), WT MV production (Figures 3B, S4C, S5A), and WT MV size (Figure S5B; p=0.15 vs. WT (n.s.), p=2.8×10-5 vs. lppAB). While pLpp1-58 expression did not fully regain WT MV production levels in these conditions (Figure S5A; p=0.006 vs. WT), the effect on MV mass was nonetheless a significant alteration from that of the lppAB mutant (p=0.006 vs. lppAB). However, expression of Lpp lacking the single PG-interacting lysine residue (Lpp1-57) resulted in MV production that mimicked the lppAB strain (Figures 3C, S4C, S5A; p=0.0252 vs. WT, p=0.8012 vs. lppAB (n.s.)), and was unable to restore WT MV size distribution (Figure S5C; p=0.02 vs. WT, p=0.16 vs. lppAB (n.s.)), even when complemented to higher protein levels in the OM than Lpp1-58 (Figure S4B).
Interaction between Pal and TolA is mediated by the C-terminal 30 aa of Pal, removal of which abolishes Pal-TolA bridges (Cascales and Lloubes, 2004). Full length Pal and mutated Pal protein incapable of interacting with TolA were expressed in the pal mutant strain (Figure S4A) in the OM (Figure S4B). Investigation of the pal mutant, lacking membrane-spanning OM-PG-IM interactions, revealed increased septal MV production as in the tolA mutant (Figures 2H, 3D). Expression of the full length Pal protein (Pal1-153) restored OM-PG-IM complexes, returning septal MV release to WT levels (Figure 3E). Cells expressing the truncated form of Pal unable to interact with TolA (Pal1-123) released large MVs at division septa as in the pal mutant (Figure 3F), despite abundant expression of Pal1-123 in the OM (Figure S4B). Therefore, engagement of envelope tethers such as Lpp-PG and Pal-TolA (Figure 3B,E), and not simply the abundance of envelope proteins (Figure 3C,F), specifically impacts MV release along the cell body and at division septa.
Influence of envelope linkages on membrane integrity
Many pathogens release MVs in vivo (Stephens et al., 1982; Brandtzaeg et al., 1992; Garcia-del Portillo et al., 1997; Fiocca et al., 1999; Hellman et al., 2000; Keenan et al., 2000; Namork and Brandtzaeg, 2002; Rosenberger et al., 2004; Marsollier et al., 2007; Necchi et al., 2007), where growth and survival depend on surface modifications to maintain envelope integrity and enable them to resist a variety of host defense mechanisms (Gunn, 2000; Ernst et al., 2001). We therefore investigated whether major MV proteins influence envelope integrity by challenging WT and mutant strains to grow in the presence of the membrane chaotropic agent deoxycholate (DOC). DOC is a relevant environmental stimulus, as Salmonella must survive exposure to bile in vivo (Gunn, 2000). Both qualitative (Figure 4A,B) and quantitative (Figure 4C,D) methods of probing membrane integrity during growth in the presence of DOC demonstrated that WT Salmonella is resistant to DOC (Figure 4A,C, (Leifson, 1935)), which was not dependent on individual integral OMPs (Figure 4A,C, p>0.05 vs. WT, Figure S6A). pal, tolA, and tolB mutants displayed DOC-sensitivity (Figure 4A,C, p<0.0001 vs. WT, Figure S6B). Resistance was restored only by expression of full length, but not truncated, forms of OM-PG-IM linked proteins (Figure 4B,D, Figure S6B; pTolA complementation was partial but significant compared to the tolA mutant, p<0.0001), indicating that OM-PG-IM tethers were essential for DOC resistance.
Figure 4. Envelope interconnections necessary for membrane integrity.
Qualitative (A,B) and quantitative (C,D) analyses of membrane integrity. (A,C) WT Salmonella and mutants lacking integral OM proteins (OmpC, OmpF) are resistant to deoxycholate (DOC). OmpA-PG is dispensable for DOC-resistance, whereas loss of Lpp-PG and OM-PG-IM complexes (Pal, TolB, TolA) induces DOC-sensitivity. (B,D) DOC-sensitivity is dependent upon envelope linkages. Expression of full length, but not truncated, proteins complement deletions (complementation with pTolA is partial but significant compared to the tolA mutant; p<0.0001). For (A,B), three 10-fold dilutions shown from left to right. For (C,D), growth on DOC was quantified and adjusted to growth on LB alone; mean % growth +/- standard error is reported from at least three replicate experiments per strain. * p<0.001, ** p<0.0001
However, while cells lacking the OM-PG linked protein OmpA retained DOC-resistance (Figure 4A,C, p>0.05 vs. WT), the lppAB mutant strain exhibited a quantitatively intermediate level of DOC-resistance; lppAB mutant cells were less resistant to DOC than WT Salmonella, but more DOC-resistant than strains devoid of OM-PG-IM linked proteins (Figure 4A,C, lppAB vs. WT p=0 vs. WT p=0.0006, lppAB vs. pal, tolB, and tolA p<0.0001). This phenotype was dependent upon covalent Lpp-PG interactions, as expression of full length Lpp was required to completely restore resistance to DOC (Figure 4B,D; Lpp1-57 p<0.001, Lpp1-58 p>0.05 vs. WT). Covalent Lpp-PG connections (Braun and Sieglin, 1970), formed by 1/3 of Lpp molecules in the envelope (Inouye et al., 1972), are 2.5-times more abundant in the OM than non-covalent OmpA-PG linkages (Braun et al., 1970; Koebnik et al., 2000). Consequently, engagement of the more abundant, covalent Lpp-PG linkages (Braun and Rehn, 1969; Braun and Sieglin, 1970) is essential forcomplete DOC-resistance, while tethering of the less abundant, non-covalent OmpA-PG interactions (De Mot and Vanderleyden, 1994; Koebnik, 1995) is dispensable (Figure 4). Similarly, the contribution of covalent Lpp-PG linkages in minimizing MV release was greater than that of the non-covalent OmpA-PG linkages, as the lppAB mutant released more MVs than the ompA mutant (Figure 1, ompA vs. lppAB p=0.0121). Finally, expression of Lpp1-58 in the ompA mutant strain was unable to restore WT MVproduction (Figure S7), demonstrating that OmpA and LppAB uniquely contribute to MV release, and reaffirming that both abundance of protein linkages and localized tethering modulate MV release (Figures 1, 2, 3, S7) and membrane stability (Figure 4). These data demonstrate bacteria can balance membrane loss and maintenance of envelope integrity: cells devoid of non-covalent OmpA-PG envelope interactions maintain membrane stability upon challenge (Figure 4A,C) and MV size is unaltered (Figure 2A,C), yet MV production is increased over WT (Figure 1, p<0.01).
MVs released from division septa and cell body
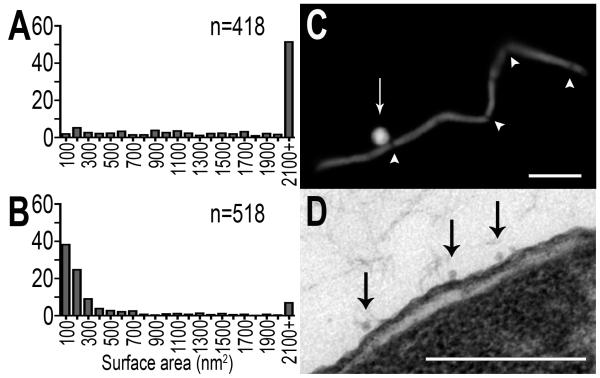
The ability of bacteria to replicate and for pathogens to cause disease is dependent upon growth and division, and constriction of division septa requires specifically coordinated protein redistribution and purposeful uncoupling of envelope linkages (Weiss, 2004; Gerding et al., 2007). As the Pal-TolB-TolA complex migrates to division septa during cell division (Gerding et al., 2007), and these OM-PG-IM linkages modulate MV production at septa (Figures 2H, 3D-F), we hypothesized that distribution of envelope connections, in addition to their abundance and nature of interaction, may influence MV release. Protein distribution was experimentally controlled by inducing filamentation to either promote (Figures S8A, S9A (Spratt, 1977; Botta and Park, 1981; Schmidt et al., 1981)) or prevent (Figure S8B (de Boer et al., 1989)) the generation of constricted division septa. MV populations were harvested from filamentous cells in each condition, which permits observation of MV formation in the absence of any envelope protein mutations or changes in total MV mass released (Figure S10). Sustained presence of division septa induced the release of an MV population enriched in large MVs (surface area ≥ 2100nm2) (Figure 5A, p=1.1 × 10-92 vs. WT; Figure S9B, p=3.0 × 10-64 vs. WT), and MVs released from non-septate filamentous Salmonella were enriched for small MVs (surface area ≤ 100nm2) (Figure 5B, p = 1.3 × 10-49 vs. WT). This biased size distribution of MVs released by septate and non-septate filamentous cells suggested that the size distribution of WT MVs (Figure 2A) may be the result of directed release of large and small MVs from specific locations. Visualization of MV release by filamentous Salmonella revealed that large MVs were released at constricted septa (Figures 5C, S9A), and small MVs originated from the cell body (Figure 5D). Therefore, the distribution of proteins containing OM-PG-IM and OM-PG linkages directs WT MV release at septum (Figure 2E) and cell body (Figure 2F): large MVs are derived primarily from constricted septa during cell division as modulated by localized OM-PG-IM connections (Figures 2H, 5C, S9A), and small MVs originate most frequently along the cell body controlled by OM-PG associations (Figures 2G, 5D).
Figure 5. Quantitatively distinct septal- and cell body-derived MV populations.
(A) Septal-derived MVs, harvested from septate filamentous cells, are significantly larger than WT MVs (p=1.1×10-92), and (B) non septal-derived MVs, harvested from non-septate filamentous cells, are significantly smaller than WT MVs (p=1.3×10-49). (C) Large MVs are released at constricted septa (Bar = 5um), and (D) small MVs are cell body derived (Bar=200nm). Arrowheads highlight constricted septa and arrows highlight MV release; n = number of individual vesicles measured.
Localization of MV formation, modulated by the differential spatial distribution of envelope protein linkages, dictates MV size, and the integral involvement of protein movement in this process suggests the site of nascent MV release may also manipulate MV protein constituents. Proteins in septal-and cell body-derived MVs (harvested from filamentous cells; Figures 5, S8) were identified by LC-MS/MS and quantified (Fu et al., 2008). Significant differences in the abundance of specific proteins in each MV population were identified at a 90% confidence level. Representative results are reported from two independent experiments (Table 1), demonstrating that specific proteins are differentially represented in MVs released at septa or from cell body locations. For example, TolB, known to migrate to division septa during constriction (Gerding et al., 2007), was highly enriched in septal MVs. Cell body-derived MVs, however, were enriched in the flagellar cap and hook/filament junction proteins FliD and FlgK, as peritrichous flagella are manifest at locations other than the division site (Aizawa and Kubori, 1998). We also identified enrichment of metabolic (AceE, SucA, RNaseE) and transcriptional/translational (RpoD, RplS, RpsO, RpsP) proteins in MV populations. Although typically annotated as cytoplasmic constituents, these proteins have been identified previously in MVs (Vaughan et al., 2006; Vipond et al., 2006; Lee et al., 2007), and ribosomal proteins are thought to be at the cell surface to permit translation of envelope proteins simultaneously with their incorporation into the membrane (Herskovits et al., 2002; Lee et al., 2007; Chevance and Hughes, 2008). We have demonstrated specificity in size and protein content of MVs released at distinct cellular sites, dependent upon purposeful, not random, localization of envelope linkages. Therefore, quantity (Figure 4), quality (Figures 1, 3, 4), and distribution (Figure 5, Table 1) of envelope linkages all contribute to directing MV biogenesis.
Table 1.
Quantitative comparison of protein composition of septal- and cell body derived MVs
| Average # of peptides | ||
|---|---|---|
| Septal MV proteins | Septal MVs | Body MVs |
|
| ||
| TolB | 25.33 | 2.67 |
| AceE | 158.67 | 10.33 |
| SucA | 16.67 | 0.00 |
| Rne | 8.67 | 0.67 |
| RpoD | 7.33 | 0.00 |
|
| ||
| Cell body MV proteins | ||
|
| ||
| FliD | 0.00 | 15.33 |
| FlgK | 4.33 | 47.67 |
| RplS | 3.00 | 28.33 |
| RpsO | 1.33 | 8.33 |
| RpsP | 0.00 | 12.33 |
Average number of peptides identified from each protein in MVs from septum and cell body.
DISCUSSION
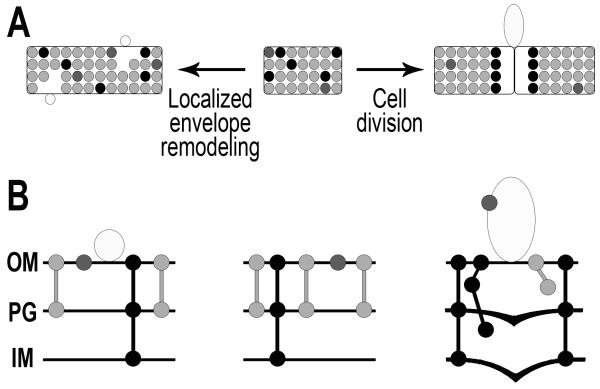
Our data support a model of MV biogenesis (Figure 6) in which MV release occurs at cell envelope regions where the density of specific conserved protein associations has temporarily decreased. In wild-type Salmonella (middle panel), envelope proteins connect the OM to the PG and IM. During growth (left panel), localized envelope remodeling briefly induces regions with fewer OM-PG connections along the cell body, resulting in the release of small MVs. During the regulated process of cell division (right panel), IM and PG layers actively grow into the division septum (Weiss, 2004), temporarily disrupting OM-PG-IM linkages circumferentially at the septum. Prior to reattachment of OM-PG-IM connections and completion of cell division, the unassociated OM is able to be released as an MV. MV production, therefore, is not due to random membrane instability, but rather is the result of the essential processes of cell growth and division, supporting the observation that this process is conserved among Gram-negative bacteria.
Figure 6. Model of MV biogenesis.
(A) Whole-cell view, (B) close-up membrane view. Middle panel: Major MV proteins in the WT envelope: Integral OM proteins (dark gray), OM-PG linked proteins (light gray) and OM-PG-IM complex proteins (black). Left panel: Localized envelope remodeling induces release of small MVs at regions of lower density OM-PG connections along cell body. Right panel: Active invagination of IM and PG during cell division causes temporary disruption of septal OM-PG-IM complexes. OM release (MVs) occurs circumferentially at the septum due to lower density OM-PG-IM protein interconnections.
The phenomenon of MV release has been recognized for decades (Knox et al., 1966; Work et al., 1966; Chatterjee and Das, 1967), prompting several groups of investigators to suggest models in which proteins and LPS were proposed to influence MV production. Many of these studies, however, qualitatively examined MV release in bacterial strains with multiply marked backgrounds and/or non-isogenic mutants, confounding interpretation of results. While our model shares elements of previously proposed mechanisms of MV production, our studies quantitatively and directly address the mechanism of MV release. We present here the first demonstration of quantitative analysis of MV mass released by a truly WT Gram-negative organism and a panel of isogenic mutants harboring targeted deletions of the coding sequence of important envelope proteins (Figure 1). Quantitative alterations in MV mass, as well as size frequency distribution (Figures 2A-D, S1) and localization of MV release determined through electron microscopic analysis (Figures 2E-H, 3, 5, S2, S3, S7), implicated interconnected envelope proteins (Figure 2I) in modulation of MV production. We determined that the involvement of envelope proteins was specific to protein-PG and/or protein-protein interacting domains, as complementation with the full length, but not truncated, proteins regained WT MV production (Figures 3, S5) and resistance to the membrane chaotropic agent DOC (Figures 4, S6). In addition, attempts to complement the MV over-producing mutant ompA with expression of another OM-PG linked protein (Lpp1-58) was unable to rescue the ompA mutant phenotypes (Figure S7), emphasizing the specificity of envelope interconnections modulating MV release. In bacteria without mutations in any envelope proteins, the redistribution of proteins during cell elongation and division resulted in localized MV release via two routes (Figures 5, 6, S8, S9), generating two MV populations with unique protein constituents (Table 1), determined by quantitative mass spectrometry. We have shown, therefore, that the influence of envelope proteins on MV release is specific and dependent upon not merely the presence or absence of an abundant protein in the membrane, but the quality, density, and spatial distribution of protein-PG and protein-protein interactions.
Analysis of the degree of envelope protein conservation in Gram-negative bacteria with a wide range of environmental niches, genera, and pathogenicity supports our conclusions that these proteins are likely a unifying factor in Gram-negative MV release. Thirty one Gram-negative genera encode at least one, if not all, of the interconnected proteins Lpp, OmpA, Pal, TolB, and TolA (Table S2, (BLAST; CMR)). While the overall identity of these protein sequences, as compared to that of Salmonella, varies from 10 to 100% (Tables S3-S7), there is extensive conservation of the domains known to be important for protein-PG and/or protein-protein interactions in the envelope. For example, Vibrio sp. express the OmpA protein, which has only 29% overall identity to Salmonella OmpA; however, the 4 amino acid residues known to interact non-covalently with PG are fully conserved (Table S4). Similarly, Legionella pneumophila expresses a Pal homologue with 100% conserved PG- and TolA-interacting domains, but only 33% overall identity to that of Salmonella Pal (Table S5). Thus, extensive conservation of protein-PG and protein-protein interacting domains exists among a wide range of Gram-negative organisms, including many well known to be prodigious MV-producers in vitro and in vivo (Kadurugamuwa and Beveridge, 1995, 1997; Fiocca et al., 1999; Hellman et al., 2000; Schooling and Beveridge, 2006)
Further examination of the well studied MV-producing Gram-negative organism P. aeruginosa supports the involvement of Lpp, OmpA, Pal, TolB, and TolA as potentially unifying factors in MV release. P. aeruginosa, which is known to produce high levels of MVs from the cell surface (McBroom and Kuehn, 12 May 2005, posting date; Schooling and Beveridge, 2006) expresses the interconnected envelope proteins examined in our studies (Table S2). However, the Lpp homologue of P. aeruginosa, OprI, does not covalently interact with the PG via its C-terminal lysine residue (Hancock et al., 1981). Therefore, the observation that MV production occurs at high levels and is distributed over the cell body of P. aeruginosa (similarly to the Salmonella lppAB mutant in Figure 3A) further supports the idea that these envelope proteins modulate MV production among Gram-negative microbes.
While our data support the likelihood of interconnected envelope proteins as a unifying factor in the modulation of MV release, additional organism-specific factors may also contribute to MV production, including bacterial cell size, shape, and LPS modifications. Cell size and shape have been integral in organism identification throughout the history of microbiology, yet the factors that determine and maintain these characteristics through many generations remain incompletely understood (Cabeen and Jacobs-Wagner, 2007; Osborn and Rothfield, 2007; Pichoff and Lutkenhaus, 2007). Although not yet explored, the biophysical properties of a curved (cocci) versus straight (bacilli) membranous surface should be considered as a potentially important organism-specific factor influencing nascent MV release from differently shaped Gram-negative organisms. This is especially intriguing given the knowledge that achieving the membrane curvature necessary to form an MV can be influenced by changes in envelope structures, such as LPS (Mashburn-Warren et al., 2008). For example, N. meningitidis is not only a Gram-negative diplococcus which produces abundant MVs in vitro and in vivo (Devoe and Gilchrist, 1973, 1974, 1975; Brandtzaeg et al., 1992; Nassif and So, 1995; Bjerre et al., 2000; Namork and Brandtzaeg, 2002), but does not encode homologues of Lpp, OmpA, Pal, TolA, or TolB (BLAST; CMR; Sturgis, 2001). We propose that the lack of OM-PG and OM-PG-IM interactions, combined with the yet to be defined effects of coccoid cell shape, may influence the abundance of MVs produced by N. meningitidis.
The influence of LPS-related factors on MV release has also been explored. The Pseudomonas aeruginosa quorum sensing molecule pqs stimulates MV release via alterations in LPS fluidity (Mashburn and Whiteley, 2005; Mashburn-Warren et al., 2008), and particular LPS species have been shown to be enriched in MVs (i.e. A band vs. B band LPS in P. aeruginosa MVs (Kadurugamuwa and Beveridge, 1995; Nguyen et al., 2003)). While modified LPS structures may influence MV release in organism-specific ways, the enrichment of particular LPS types in MVs does not exclude the possibility that particular LPS species are predominant in regions with fewer envelope interconnections and subsequently are released in MVs at these locations. In contrast to the conservation of interconnected envelope protein domains, analysis of LPS acyl chain number and length alone, in a wide range of Gram-negative bacteria, reveals little conservation (Table S8). Additional unexplored organism-specific factors likely to influence MV release are the extensive diversity of O-antigen structure and composition throughout Gram-negative bacteria (Smit et al., 1975; Lerouge and Vanderleyden, 2002), as well as other structural modifications, such as phosphorylation, to lipid A (Trent et al., 2006). Again, the widespread homology of envelope protein domains linking the OM-PG and OM-IM (Table S2-S7) among a wide variety of Gram-negative organisms supports the idea that interconnected envelope proteins play a conserved role in the release of MVs.
Many pathogenic bacteria handicap the immune response by actively inhibiting and/or killing host cells responsible for processing and presenting antigens (Brennan and Cookson, 2000; van der Velden et al., 2003; Alaniz et al., 2006; Tobar et al., 2006). However, MVs exist separately from live bacteria in vivo (Fiocca et al., 1999; Marsollier et al., 2007). Therefore, release and circulation of non-cytotoxic, antigen-rich MVs may represent important sources of bacterial antigens, betraying the presence of the bacterium to the host and making MVs a liability. Salmonella has evolved mechanisms to avoid immune recognition, which potentially includes restricting release of antigen-rich MVs (Figure 1). However, growth and division also rely on the directed redistribution of proteins (Weiss, 2004; Gerding et al., 2007), which not only results in MV release (Figures 5, 6), but leads to modulation of MV content (Figure 6, Table 1). The potential exists, therefore, for bacteria to hijack this inevitable physiologic process for its own means, such as toxin and/or cell signaling molecule secretion or inhibition of host cell processes (Horstman and Kuehn, 2000; Wai et al., 2003; Mashburn and Whiteley, 2005; Fernandez-Moreira et al., 2006).
Complete loss of OmpA-PG interactions increases MV release (Figures 1, 2G) without loss of membrane integrity (Figure 4A,C). Interestingly, MV release by an ompA lppAB mutant is significantly increased above either single mutant (data not shown; p=0.0002 vs. ompA, p=0.0011 vs. lppAB), and Salmonella down-regulates lpp and ompA transcripts during growth in macrophages (Eriksson et al., 2003). As we have demonstrated that Salmonella can balance MV release and maintenance of envelope stability (ompA mutant, Figures 1, 4), these data suggest that modulation of envelope proteins, through gene regulation, could result in different ratios of interconnected proteins such as OmpA and Lpp. Such coordination would provide Salmonella with a tunable system to facilitate MV production in response to changing environments (pH, ionic strength, temperature, etc.), contributing to other well defined mechanisms of surface modifications (Ernst et al., 2001).
EXPERIMENTAL PROCEDURES
Bacterial strains and media
Bacterial strains and plasmids are listed in Table S1. Strains were grown in LB (Teknova) or Tryptic Soy Broth (VWR), with carbenicillin (100ug/ml) or kanamycin (50ug/ml) when necessary to retain plasmids, and IPTG (100uM) to induce gene expression.
Construction of mutants
Targeted chromosomal deletions of coding sequence were constructed via the Lambda Red recombination system as described previously (Datsenko and Wanner, 2000) (See Supplemental Experimental Procedures).
Plasmid construction
Chromosomal genes, either full-length or specific truncations, were PCR amplified and cloned into IPTG-inducible pTrc99a vector (Amann et al., 1988). Plasmids pDB173 (expressing minCD) and pTB6 (expressing periplasmic GFP) have been previously described (de Boer et al., 1989; Bernhardt and de Boer, 2004).
MV harvest and quantification
Bacteria were grown to late log phase (OD600=0.6-0.8), cells were removed by centrifugation (45min @3800rpm), and culture supernatant filter-sterilized (0.22μm, Corning). Sterile supernatant was concentrated by molecular weight (100kDa MWCO, Pall), retentate ultracentrifuged (2hrs @35,000rpm), pellets resuspended in sterile water, and sterilized (0.22μm Spin-X column, Costar). Dry weight was measured by lyophilization and normalized to optical density of parent bacterial culture.
Mass spectrometry
WT MV proteins were separated by 2D SDS-PAGE (BioRad). Coomassie-stained protein spots were excised, trypsin digested, and identified by LC-MS/MS. Proteins in septal and cell-body MVs (50ug protein/sample) were separated by 15% Tris-glycine SDS-PAGE (BioRad) in triplicate, trypsin digested, and identified by LC-MS/MS (Proteomics Resource, Fred Hutchinson Cancer Research Center, Seattle, WA). Protein abundance was determined by a label-free method combining spectral index calculation with permutation analysis as previously described (Fu et al., 2008), with statistically significant cutoff values at the 90th percentile confidence interval.
Electron microscopy
MVs were resuspended in 10 mM MgCl2 and negatively stained with a 2% phosphotungstic acid solution (pH 7.3). Cells were fixed and embedded for TEM analysis of thin sections (See Supplemental Experimental Procedures). Samples were observed with a JEM-1200EXII transmission electron microscope (JEOL-TEM) operated at 80KV.
MV size measurement
MV size was measured from at least 3 separate electron micrographs of 2 independent MV preparations per strain in Adobe Photoshop using the ruler tool. Long and short axes were measured for 400-750 individual MVs per strain (n noted on figures represents number of individual MVs measured), and surface area of each MV was calculated (long axis/2 * short axis/2 * pi). Data is presented in surface area ranges of 100nm2, where 100 represents 1-100nm2, and all MVs larger than 2100nm2 group in the final category.
Sensitivity assays
Sensitivity to deoxycholate (DOC) was qualitatively measured as previously described (Gerding et al., 2007). For quantitative measurement, backdiluted cultures were grown to late-log phase (OD600=0.6-1.0), adjusted to equal OD600 values, and plated to LB +/- 0.1% DOC. CFU were quantified following overnight incubation at 37°C. Ability to grow in the presence of each agent was expressed as a percentage of each strain’s growth on LB alone.
Filamentation
Treatment with a sub-lethal concentration of azlocillin (10ug/ml in water, Sigma) specifically inhibits FtsI (PBP3, (Botta and Park, 1981; Schmidt et al., 1981)) and induces septated filamenting bacteria. ftsIts (Spratt, 1977) pTB6 were grown initially at 30°C and then at 42°C to induce filamentation. Filamentation without septa was induced by deletion of minCDE and inducible expression of minCD from pDB173 (See above, Table S1, and Supplemental Experimental Procedures (de Boer et al., 1989)).
Cell fractionation
Cell fractions were harvested from cultures in late exponential phase as previously described (Bergman et al., 2005). Briefly, cells were washed in Tris/sucrose and EDTA/lysozyme solutions to harvest periplasm, lysed in a French pressure cell to separate cytoplasm, and treated with Sarkosyl to separate IM and OM (See Supplemental Experimental Procedures).
Confocal microscopy
Filamentous cultures were spotted onto glass slides and allowed to air dry, then treated with ProLong Gold antifade reagent (Molecular Probes) and covered for incubation overnight in the dark. Cover slips were sealed prior to viewing. GFP fluorescence was visualized with a Leica SL confocal microscope in the W. M. Keck Center for Advanced Studies in Neural Signaling (University of Washington, Seattle, WA).
Western blotting
Expression of Lpp and Pal in sonicated bacteria and OM fractions were compared by Western blot using standard techniques.
Statistical analysis
MV production and quantification of growth on DOC were analyzed using the Student’s t test (unpaired samples, two-tailed), and the Chi-square test was used to analyze MV size distributions using GraphPad Prism version 4.0 for Macintosh (GraphPad Software, San Diego, California).
Supplementary Material
ACKNOWLEDGMENTS
We thank Ferric Fang, Sam Miller, and both past and current members of the Cookson laboratory for insightful discussions and manuscript review, and Lawrence Rothfield and Thomas Bernhardt for reagents. This work was supported by National Institutes of Health Grants AI47242, National Institutes of Health Interdisciplinary Training Grant in Bacterial Pathogenesis AI0055396 (to B.L.D.), and National Institute of General Medical Sciences Public Health Service National Research Service Award Grant T32 GM07270 (to T.B.).
REFERENCES
- Aizawa SI, Kubori T. Bacterial flagellation and cell division. Genes Cells. 1998;3:625–634. doi: 10.1046/j.1365-2443.1998.00219.x. [DOI] [PubMed] [Google Scholar]
- Alaniz RC, Cummings LA, Bergman MA, Rassoulian-Barrett SL, Cookson BT. Salmonella typhimurium coordinately regulates FliC location and reduces dendritic cell activation and antigen presentation to CD4+ T cells. J Immunol. 2006;177:3983–3993. doi: 10.4049/jimmunol.177.6.3983. [DOI] [PubMed] [Google Scholar]
- Alaniz RC, Deatherage BL, Lara JC, Cookson BT. Membrane vesicles are immunogenic facsimiles of Salmonella typhimurium that potently activate dendritic cells, prime B and T cell responses, and stimulate protective immunity in vivo. J Immunol. 2007;179:7692–7701. doi: 10.4049/jimmunol.179.11.7692. [DOI] [PubMed] [Google Scholar]
- Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- Bauman SJ, Kuehn MJ. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect. 2006;8:2400–2408. doi: 10.1016/j.micinf.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman MA, Cummings LA, Barrett SL, Smith KD, Lara JC, Aderem A, Cookson BT. CD4+ T cells and toll-like receptors recognize Salmonella antigens expressed in bacterial surface organelles. Infect Immun. 2005;73:1350–1356. doi: 10.1128/IAI.73.3.1350-1356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubes R. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol. 1998;180:4872–4878. doi: 10.1128/jb.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol. 2004;52:1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge TJ, Makin SA, Kadurugamuwa JL, Li Z. Interactions between biofilms and the environment. FEMS Microbiol Rev. 1997;20:291–303. doi: 10.1111/j.1574-6976.1997.tb00315.x. [DOI] [PubMed] [Google Scholar]
- Bjerre A, Brusletto B, Rosenqvist E, Namork E, Kierulf P, Ovstebo R, et al. Cellular activating properties and morphology of membrane-bound and purified meningococcal lipopolysaccharide. J Endotoxin Res. 2000;6:437–445. [PubMed] [Google Scholar]
- BLAST Basic Local Alignment Search Tool.
- Botta GA, Park JT. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J Bacteriol. 1981;145:333–340. doi: 10.1128/jb.145.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandtzaeg P, Bryn K, Kierulf P, Ovstebo R, Namork E, Aase B, Jantzen E. Meningococcal endotoxin in lethal septic shock plasma studied by gas chromatography, mass-spectrometry, ultracentrifugation, and electron microscopy. J Clin Invest. 1992;89:816–823. doi: 10.1172/JCI115660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Rehn K. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem. 1969;10:426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- Braun V, Rehn K, Wolff H. Supramolecular structure of the rigid layer of the cell wall of Salmonella, Serratia, Proteus, and Pseudomonas fluorescens. Number of lipoprotein molecules in a membrane layer. Biochemistry. 1970;9:5041–5049. doi: 10.1021/bi00828a001. [DOI] [PubMed] [Google Scholar]
- Braun V, Sieglin U. The covalent murein-lipoprotein structure of the Escherichia coli cell wall. The attachment site of the lipoprotein on the murein. Eur J Biochem. 1970;13:336–346. doi: 10.1111/j.1432-1033.1970.tb00936.x. [DOI] [PubMed] [Google Scholar]
- Brennan MA, Cookson BT. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol Microbiol. 2000;38:31–40. doi: 10.1046/j.1365-2958.2000.02103.x. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Skin and bones: the bacterial cytoskeleton, cell wall, and cell morphogenesis. J Cell Biol. 2007;179:381–387. doi: 10.1083/jcb.200708001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Lloubes R. Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol Microbiol. 2004;51:873–885. doi: 10.1046/j.1365-2958.2003.03881.x. [DOI] [PubMed] [Google Scholar]
- Chai TJ, Foulds J. Escherichia coli K-12 tolF mutants: alterations in protein composition of the outer membrane. J Bacteriol. 1977;130:781–786. doi: 10.1128/jb.130.2.781-786.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee SN, Das J. Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J Gen Microbiol. 1967;49:1–11. doi: 10.1099/00221287-49-1-1. [DOI] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel T, Germon P, Vianney A, Portalier R, Lazzaroni JC. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol Microbiol. 1998;29:359–367. doi: 10.1046/j.1365-2958.1998.00945.x. [DOI] [PubMed] [Google Scholar]
- CMR Comprehensive Microbial Resource.
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- Devoe IW, Gilchrist JE. Release of endotoxin in the form of cell wall blebs during in vitro growth of Neisseria meningitidis. J Exp Med. 1973;138:1156–1167. doi: 10.1084/jem.138.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoe IW, Gilchrist JE. Ultrastructure of pili and annular structures on the cell wall surface of Neisseria meningitidis. Infect Immun. 1974;10:872–876. doi: 10.1128/iai.10.4.872-876.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVoe IW, Gilchrist JE. Pili on meningococci from primary cultures of nasopharyngeal carriers and cerebrospinal fluid of patients with acute disease. J Exp Med. 1975;141:297–305. doi: 10.1084/jem.141.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- Ernst RK, Guina T, Miller SI. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 2001;3:1327–1334. doi: 10.1016/s1286-4579(01)01494-0. [DOI] [PubMed] [Google Scholar]
- Fernandez-Moreira E, Helbig JH, Swanson MS. Membrane vesicles shed by Legionella pneumophila inhibit fusion of phagosomes with lysosomes. Infect Immun. 2006;74:3285–3295. doi: 10.1128/IAI.01382-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiocca R, Necchi V, Sommi P, Ricci V, Telford J, Cover TL, Solcia E. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J Pathol. 1999;188:220–226. doi: 10.1002/(SICI)1096-9896(199906)188:2<220::AID-PATH307>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Fu X, Gharib SA, Green PS, Aitken ML, Frazer DA, Park DR, et al. Spectral index for assessment of differential protein expression in shotgun proteomics. J Proteome Res. 2008;7:845–854. doi: 10.1021/pr070271+. [DOI] [PubMed] [Google Scholar]
- Garcia-del Portillo F, Stein MA, Finlay BB. Release of lipopolysaccharide from intracellular compartments containing Salmonella typhimurium to vesicles of the host epithelial cell. Infect Immun. 1997;65:24–34. doi: 10.1128/iai.65.1.24-34.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn JS. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2000;2:907–913. doi: 10.1016/s1286-4579(00)00392-0. [DOI] [PubMed] [Google Scholar]
- Hancock RE, Irvin RT, Costerton JW, Carey AM. Pseudomonas aeruginosa outer membrane: peptidoglycan-associated proteins. J Bacteriol. 1981;145:628–631. doi: 10.1128/jb.145.1.628-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman J, Loiselle PM, Zanzot EM, Allaire JE, Tehan MM, Boyle LA, et al. Release of gram-negative outer-membrane proteins into human serum and septic rat blood and their interactions with immunoglobulin in antiserum to Escherichia coli J5. J Infect Dis. 2000;181:1034–1043. doi: 10.1086/315302. [DOI] [PubMed] [Google Scholar]
- Herskovits AA, Shimoni E, Minsky A, Bibi E. Accumulation of endoplasmic membranes and novel membrane-bound ribosome-signal recognition particle receptor complexes in Escherichia coli. J Cell Biol. 2002;159:403–410. doi: 10.1083/jcb.200204144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra D, van der Laan JW, de Leij L, Witholt B. Release of outer membrane fragments from normally growing Escherichia coli. Biochim Biophys Acta. 1976;455:889–899. doi: 10.1016/0005-2736(76)90058-4. [DOI] [PubMed] [Google Scholar]
- Horstman AL, Kuehn MJ. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J Biol Chem. 2000;275:12489–12496. doi: 10.1074/jbc.275.17.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M, Shaw J, Shen C. The assembly of a structural lipoprotein in the envelope of Escherichia coli. J Biol Chem. 1972;247:8154–8159. [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J Antimicrob Chemother. 1997;40:615–621. doi: 10.1093/jac/40.5.615. [DOI] [PubMed] [Google Scholar]
- Keenan J, Day T, Neal S, Cook B, Perez-Perez G, Allardyce R, Bagshaw P. A role for the bacterial outer membrane in the pathogenesis of Helicobacter pylori infection. FEMS Microbiol Lett. 2000;182:259–264. doi: 10.1111/j.1574-6968.2000.tb08905.x. [DOI] [PubMed] [Google Scholar]
- Knox KW, Vesk M, Work E. Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J Bacteriol. 1966;92:1206–1217. doi: 10.1128/jb.92.4.1206-1217.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik R. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol Microbiol. 1995;16:1269–1270. doi: 10.1111/j.1365-2958.1995.tb02348.x. [DOI] [PubMed] [Google Scholar]
- Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- Lai EM, Nair U, Phadke ND, Maddock JR. Proteomic screening and identification of differentially distributed membrane proteins in Escherichia coli. Mol Microbiol. 2004;52:1029–1044. doi: 10.1111/j.1365-2958.2004.04040.x. [DOI] [PubMed] [Google Scholar]
- Lee EY, Bang JY, Park GW, Choi DS, Kang JS, Kim HJ, et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7:3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- Leifson E. New culture media based on sodium desoxycholate for the isolation of intestinal pathogens and for the enumeration of colon bacilli in milk and water. Journal of Pathology and Bacteriology. 1935;40:581–599. [Google Scholar]
- Lerouge I, Vanderleyden J. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol Rev. 2002;26:17–47. doi: 10.1111/j.1574-6976.2002.tb00597.x. [DOI] [PubMed] [Google Scholar]
- Marsollier L, Brodin P, Jackson M, Kordulakova J, Tafelmeyer P, Carbonnelle E, et al. Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog. 2007;3:e62. doi: 10.1371/journal.ppat.0030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, et al. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol. 2008;69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol. 2006;61:839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- McBroom AJ, Kuehn MJ. 2.2.4, Outer Membrane Vesicles. In: A Böck, Kaper JB, Neidhardt FC, Nyström T, Rudd KE, Squires CL., editors. EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. [Online.] ASM Press; Washington, D.C.: [12 May 2005]. posting date. http://www.ecosal.org. [Google Scholar]
- McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol. 2006;188:5385–5392. doi: 10.1128/JB.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namork E, Brandtzaeg P. Fatal meningococcal septicaemia with “blebbing” meningococcus. Lancet. 2002;360:1741. doi: 10.1016/S0140-6736(02)11721-1. [DOI] [PubMed] [Google Scholar]
- Nassif X, So M. Interaction of pathogenic neisseriae with nonphagocytic cells. Clin Microbiol Rev. 1995;8:376–388. doi: 10.1128/cmr.8.3.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necchi V, Candusso ME, Tava F, Luinetti O, Ventura U, Fiocca R, et al. Intracellular, intercellular, and stromal invasion of gastric mucosa, preneoplastic lesions, and cancer by Helicobacter pylori. Gastroenterology. 2007;132:1009–1023. doi: 10.1053/j.gastro.2007.01.049. [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Saxena A, Beveridge TJ. Effect of surface lipopolysaccharide on the nature of membrane vesicles liberated from the Gram-negative bacterium Pseudomonas aeruginosa. J Electron Microsc (Tokyo) 2003;52:465–469. doi: 10.1093/jmicro/52.5.465. [DOI] [PubMed] [Google Scholar]
- Osborn MJ, Rothfield L. Cell shape determination in Escherichia coli. Curr Opin Microbiol. 2007;10:606–610. doi: 10.1016/j.mib.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Parsons LM, Lin F, Orban J. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry. 2006;45:2122–2128. doi: 10.1021/bi052227i. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Overview of cell shape: cytoskeletons shape bacterial cells. Curr Opin Microbiol. 2007;10:601–605. doi: 10.1016/j.mib.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger CM, Gallo RL, Finlay BB. Interplay between antibacterial effectors: a macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc Natl Acad Sci U S A. 2004;101:2422–2427. doi: 10.1073/pnas.0304455101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LS, Botta G, Park JT. Effects of furazlocillin, a beta-lactam antibiotic which binds selectively to penicillin-binding protein 3, on Escherichia coli mutants deficient in other penicillin-binding proteins. J Bacteriol. 1981;145:632–637. doi: 10.1128/jb.145.1.632-637.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooling SR, Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol. 2006;188:5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton K, Lennon D, Oster P, Crengle S, Martin D, Mulholland K, et al. The New Zealand Meningococcal Vaccine Strategy: a tailor-made vaccine to combat a devastating epidemic. N Z Med J. 2004;117:U1015. [PubMed] [Google Scholar]
- Smit J, Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studies with lipopolysaccharide mutants. J Bacteriol. 1975;124:942–958. doi: 10.1128/jb.124.2.942-958.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt BG. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J Bacteriol. 1977;131:293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DS, Edwards KM, Morris F, McGee ZA. Pili and outer membrane appendages on Neisseria meningitidis in the cerebrospinal fluid of an infant. J Infect Dis. 1982;146:568. doi: 10.1093/infdis/146.4.568. [DOI] [PubMed] [Google Scholar]
- Sturgis JN. Organisation and evolution of the tol-pal gene cluster. J Mol Microbiol Biotechnol. 2001;3:113–122. [PubMed] [Google Scholar]
- Tetz VV, Rybalchenko OV, Savkova GA. Ultrastructural features of microbial colony organization. J Basic Microbiol. 1990;30:597–607. doi: 10.1002/jobm.3620300819. [DOI] [PubMed] [Google Scholar]
- Tobar JA, Carreno LJ, Bueno SM, Gonzalez PA, Mora JE, Quezada SA, Kalergis AM. Virulent Salmonella enterica serovar typhimurium evades adaptive immunity by preventing dendritic cells from activating T cells. Infect Immun. 2006;74:6438–6448. doi: 10.1128/IAI.00063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent MS, Stead CM, Tran AX, Hankins JV. Diversity of endotoxin and its impact on pathogenesis. J Endotoxin Res. 2006;12:205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- van der Velden AW, Velasquez M, Starnbach MN. Salmonella rapidly kill dendritic cells via a caspase-1-dependent mechanism. J Immunol. 2003;171:6742–6749. doi: 10.4049/jimmunol.171.12.6742. [DOI] [PubMed] [Google Scholar]
- Vaughan TE, Skipp PJ, O’Connor CD, Hudson MJ, Vipond R, Elmore MJ, Gorringe AR. Proteomic analysis of Neisseria lactamica and N eisseria meningitidis outer membrane vesicle vaccine antigens. Vaccine. 2006;24:5277–5293. doi: 10.1016/j.vaccine.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Vipond C, Suker J, Jones C, Tang C, Feavers IM, Wheeler JX. Proteomic analysis of a meningococcal outer membrane vesicle vaccine prepared from the group B strain NZ98/254. Proteomics. 2006;6:3400–3413. doi: 10.1002/pmic.200500821. [DOI] [PubMed] [Google Scholar]
- Wai SN, Lindmark B, Soderblom T, Takade A, Westermark M, Oscarsson J, et al. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115:25–35. doi: 10.1016/s0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- Walburger A, Lazdunski C, Corda Y. The Tol/Pal system function requires an interaction between the C-terminal domain of TolA and the N-terminal domain of TolB. Mol Microbiol. 2002;44:695–708. doi: 10.1046/j.1365-2958.2002.02895.x. [DOI] [PubMed] [Google Scholar]
- Webster RE. The tol gene products and the import of macromolecules into Escherichia coli. Mol Microbiol. 1991;5:1005–1011. doi: 10.1111/j.1365-2958.1991.tb01873.x. [DOI] [PubMed] [Google Scholar]
- Weiss DS. Bacterial cell division and the septal ring. Mol Microbiol. 2004;54:588–597. doi: 10.1111/j.1365-2958.2004.04283.x. [DOI] [PubMed] [Google Scholar]
- Wensink J, Witholt B. Outer-membrane vesicles released by normally growing Escherichia coli contain very little lipoprotein. Eur J Biochem. 1981;116:331–335. doi: 10.1111/j.1432-1033.1981.tb05338.x. [DOI] [PubMed] [Google Scholar]
- Work E, Knox KW, Vesk M. The chemistry and electron microscopy of an extracellular lipopolysaccharide from Escherichia coli. Ann N Y Acad Sci. 1966;133:438–449. doi: 10.1111/j.1749-6632.1966.tb52382.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.