Summary
This work describes the identification and characterization of SurR, P. furiosus sulfur (S0) response regulator. SurR was captured from cell extract using promoter DNA of a hydrogenase operon that is down-regulated in the primary response of P. furiosus to S0, as revealed by DNA microarray experiments. SurR was validated as a sequence-specific DNA binding protein, and characterization of the SurR DNA binding motif GTTn3AAC led to the identification of several target genes that contain an extended motif in their promoters. A number of these were validated to contain upstream SurR binding sites. These SurR targets strongly correspond with ORFs and operons both up- and down-regulated in the primary response to S0. In vitro transcription revealed that SurR is an activator for its own gene as well as for two hydrogenase operons whose expression is down-regulated during the primary S0 response; it is also a repressor for two genes up-regulated during the primary S0 response, one of which encodes the primary S0-reducing enzyme NAD(P)H sulfur reductase. Herein we give evidence for the role of SurR in both mediating the primary response to S0 and controlling hydrogen production in P. furiosus.
Keywords: regulatory transcription factor, archaea, Pyrococcus furiosus, sulfur response, hydrogen production, SurR
Introduction
Archaeal organisms, while representing the third domain of life, have similarities to both bacteria and eukarya. Of particular interest is their transcriptional apparatus that shares both eukaryal and bacterial features. Transcription in archaea is carried out by a single type of RNA polymerase that is highly homologous to the eukaryal RNA polymerase II, and basal transcription in archaea can be achieved in vitro with a minimal complement of eukaryal-like transcription factors: TBP (TATA binding protein), TFB (transcription factor B), and occasionally TFE (transcription factor E) (Cramer, 2002, Hickey et al., 2002, Bartlett, 2005). Archaea also use eukaryal-like DNA promoter elements, TATA box and BRE (TFB recognition element), to direct the binding of basal transcription factors for transcription initiation (Bartlett, 2005). TBP and TFB bind cooperatively to the TATA box and BRE, respectively, and recruit RNA polymerase (Bartlett, 2005). TFB and RNA polymerase together define the transcription start site position, 25 to 30 bp downstream of the TATA box (Smale and Kadonaga, 2003, Kornberg, 2007). In contrast to the eukaryal-like basal transcription apparatus, regulation of transcription appears to occur predominantly along a bacterial theme, with bacterial-like transcriptional repressors dominating the group of archaeal regulatory transcription factors (rTFs) characterized so far (Bell, 2005, Geiduschek and Ouhammouch, 2005).
The hyperthermophile Pyrococcus furiosus is a model archaeal organism, and because of the substantial information already known about its biology, investigation of transcriptional regulators in this organism will shed light on mechanisms of archaeal transcriptional control as they relate to metabolic pathways. P. furiosus, originally isolated by Fiala and Stetter from a hydrothermal vent community off the coast of Italy (Fiala and Stetter, 1986), is a member of the genus Thermococcus which currently contains 24 species of obligately organotrophic fermentative anaerobes (Miroshnichenko and Bonch-Osmolovskaya, 2006). P. furiosus has an optimal growth temperature of 100°C and can utilize both carbohydrates and peptides as carbon sources, via fermentation to organic acids, CO2 and H2 (Fiala and Stetter, 1986). P. furiosus is somewhat unique among archaeal hyperthermophiles in that it can grow in the presence or absence of elemental sulfur (S0), depending on the available carbon source (Fiala and Stetter, 1986, Adams, 1994, Adams et al., 2001). This trait presents an opportunity to study the regulation of S0 metabolism by investigating the changes that occur in the absence or presence of S0 in a growing culture.
When maltose is used as the carbon source, P. furiosus growth is comparable both in the presence and absence of S0 (Adams et al., 2001); however, significant changes in gene expression are evident from DNA microarray analyses (Schut et al., 2001, Schut et al., 2007). A kinetic approach was used to identify the genes directly affected by the presence of S0 (Schut et al., 2007). Ten min after S0 addition, a defined ‘primary’ response is observed, followed by a ‘secondary’ response occurring after 30 min which entails up-regulation of many gene clusters involved in amino acid metabolism (Schut et al., 2007). A major part of the primary response involves down-regulation of the three major hydrogenase operons, and thus the response of P. furiosus to S0 is intimately connected with its ability to produce hydrogen. Concurrent with the down-regulation of genes involved in hydrogen production is the up-regulation of the recently characterized cytoplasmic NAD(P)H-dependent sulfur reductase (Schut et al., 2007). Identification of transcription factors involved in mediating the response to S0 will improve our understanding of this unique system and the associated transcriptional network.
The dramatic down-regulation of the operon that encodes the hydrogen-evolving membrane-bound hydrogenase (Schut et al., 2001, Schut et al., 2007) led us to choose this gene cluster as a target for transcription factor discovery utilizing a DNA-affinity protein capture approach. This approach brought about the discovery of a protein that regulates ORFs and operons involved in primary S0 response, and we have therefore named this protein SurR for sulfur response regulator. Herein we show the involvement of this regulatory transcription factor in controlling the metabolism of both S0 and hydrogen by P. furiosus.
Results
Discovery of SurR
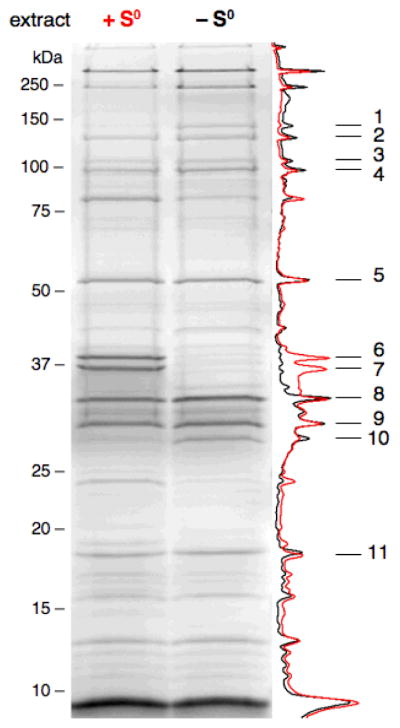
To identify transcription factors involved in the response of P. furiosus to S0, a promoter DNA affinity capture method was employed. The P. furiosus membrane-bound hydrogenase operon was selected as a target for transcription factor discovery because of its dramatic down-regulation during the primary response to S0 observed in DNA microarray expression profiles (Schut et al., 2007). The bait DNA was designed to contain ~200 bp of the membrane-bound hydrogenase operon promoter and ~100 bp of the first ORF in the operon, mbh1 (PF1423). Incidentally, a divergently transcribed ORF, PF1422, is located 150 bp upstream of the mbh1 ORF, and therefore the bait DNA most likely included the promoter region of this ORF as well. DNA affinity protein capture with the mbh1 promoter bait DNA was carried out using P. furiosus soluble cell extracts obtained from cultures grown in the presence and absence of S0. SDS PAGE analysis of the eluted DNA-binding proteins is presented in Figure 1. Eleven proteins were identified by mass spectrometry, and sequence analysis revealed that three of them (bands 8–10) were predicted transcriptional regulators: PF1827, PF0496 and PF0095. Of these, only PF0095 was represented differentially in the captured proteins from the two cell extracts (band 10). PF0095 contains an N-terminal helix-turn-helix domain related to the DNA-binding domain of the prokaryotic ArsR family of transcriptional regulators, and it is also a member of a small cluster of uncharacterized archaeal regulatory transcription factors (COG1777). The three other members of the Thermococcaceae family with sequenced genomes contain homologs of PF0095 with high sequence identity: PH0180 (72%), PAB0108 (72%), and TK1086 (65%). The PF0095 protein identified from the mbh1 bait DNA will hereafter be referred to as SurR.
Fig. 1.
Identification of SurR from cell extract with mbh1 bait DNA. Silver-stained denaturing gel of eluted proteins from DNA affinity capture with mbh1 bait DNA incubated in soluble cell extracts with (red) and without (black) S0 showing the corresponding densitometry scans of each lane. Arrows indicate identified proteins (NCBI annotations): 1, reverse gyrase, PF0495; 2, DNA-directed RNA polymerase subunit b, PF1564; 3, DNA-directed RNA polymerase subunit a′, PF1563; 4, Cell division control protein 48, aaa family, PF0963; 5, methylmalonyl-CoA decarboxylase, subunit alpha, PF0671; 6, methionine synthase vitamin B12-independent isozyme, PF1269; 7, conserved hypothetical protein, PF1268; 8, conserved hypothetical protein, PF1827; 9, conserved hypothetical protein, PF0496; 10, conserved hypothetical protein (SurR), PF0095; 11, conserved hypothetical protein, PF1572. Protein Bands 1, 6 and 7 can be challenged off with heparin, suggesting that they are likely non-specific DNA-binding proteins, and bands 4 and 5 are bead-binding proteins not removed by DNase digestion. A control protein capture experiment using a DNA probe from an ORF not regulated during the primary S0 response can be seen in Supplementary Fig. S1.
SurR binds mbh1 promoter DNA in a sequence-specific manner
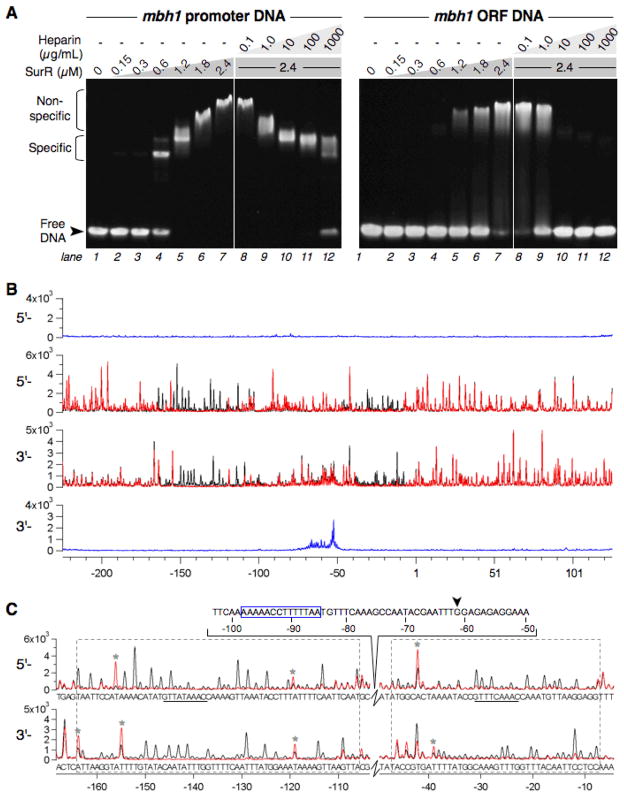
EMSA was used to determine the sequence specificity of the binding of recombinant SurR to an 81-bp region of the mbh1 promoter (+5 to −76 bp relative to the translation start) compared to an 80-bp region of the mbh1 ORF (Fig. 2A). The results indicate that SurR binds with sequence specificity to the mbh1 promoter DNA, completely shifting the DNA at 1.2 μM protein (left panel, lane 5), whereas for the mbh1 ORF DNA, the DNA is still not completely shifted at 2.4 μM (right panel, lane 7). Because a DNA-staining agent was used to visualize the DNA in the EMSA experiments, the anionic polysaccharide heparin was used as a competitor in place of DNA. Heparin has electrostatic properties similar to DNA so that it can be used as a non-specific competitor in EMSA (Moxley and Jarrett, 2005). Addition of heparin revealed that the association of SurR with promoter DNA was indeed sequence-specific while the binding to ORF DNA was of a nonspecific nature. At heparin concentrations higher than 10 μg/mL (approximately 3-fold mass excess of competitor over ORF DNA), the binding of SurR to the mbh1 ORF at 2.4 μM was completely blocked (right panel, lanes 10–12). Given this finding, it is not surprising that at high protein concentrations, SurR associates nonspecifically even with the mbh1 promoter DNA, as evidenced by the shift to very low mobility complexes at 1.8 to 2.4 μM protein (left panel, lanes 6–7). Heparin is able to remove these non-specific interactions of SurR with the promoter DNA at concentrations of 10 to 100 μg/mL (left panel, lanes 10–11), corresponding to approximately 3 to 30-fold mass excess of competitor over promoter DNA. It is also worth noting that heparin at sufficiently high concentrations can also challenge off sequence-specific DNA binding as evidenced by its effect on the binding of SurR to the mbh1 promoter at a concentration of 1 mg/mL, corresponding to a >270-fold higher mass concentration over the DNA (left panel, lane 12). These data conclusively show that SurR is a sequence-specific DNA-binding protein, and furthermore, that one of its targets appears to be in the promoter region upstream of mbh1.
Fig. 2.
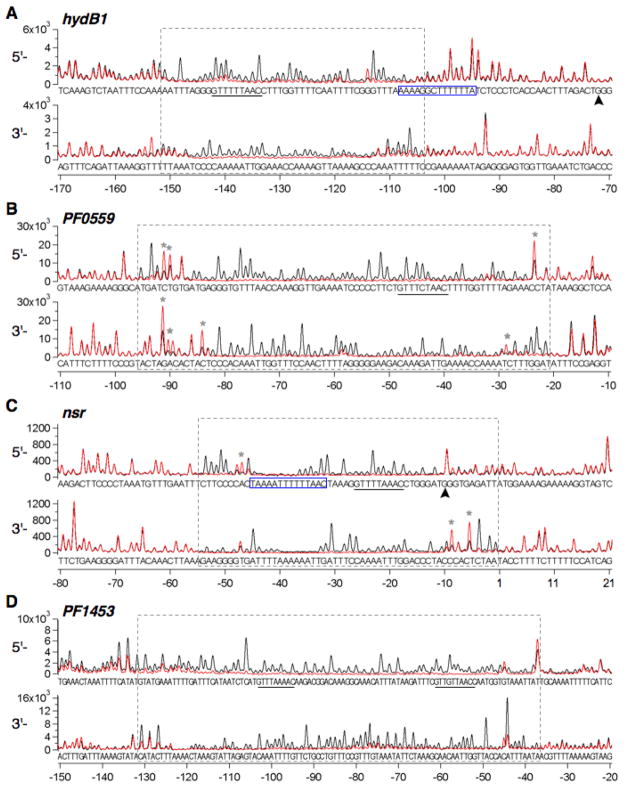
SurR binds to the membrane-bound hydrogenase promoter region. A, EMSA of SurR with mbh1 promoter (81 bp, 75 nM in each lane) and ORF (80 bp, 75 nM in each lane) DNA probes using heparin as a nonspecific DNA competitor (lanes 8–12 in each panel). Protein amounts are indicated at the top of the gel. All samples were run on one gel (stained with SYBR Green I nucleic acid gel stain). The horizontal dashed line suggests that nonspecifically bound SurR results in larger shifts. B, Fluorescence DNase I footprint of SurR (0.3 μM) on mbh1 probe DNA (10 nM). The y-axes are shown in fluorescence intensity units and the x-axis shows the base position relative to the mbh1 translation start site (+1). Traces for both strands of undigested probe are shown in blue, and traces for digested probe with (red) and without (black) protein are overlaid. The mbh1 coding strand is shown in the top two traces (5′-3′) and the noncoding strand in the bottom two traces (3′-5′). C, Expanded view of SurR footprint regions shown in B with corresponding nucleotide sequence. The footprint regions for both strands are boxed, GTTn3AAC motifs (see Fig. 4) are underlined, DNase I hypersensitive sites are marked by an asterisk (*), transcription start site is marked with an arrow, and BRE/TATA region is boxed in blue (located in the ‘inter-footprints sequence shown above the trace for the mbh1 coding strand).
To further define the region in the mbh1 promoter bound specifically by SurR, fluorescence-based DNase I footprinting was performed. The DNA probe was labeled at the 5′ ends with two distinct fluorophores so that both strands from one footprinting experiment could be analyzed simultaneously using a capillary electrophoreses automated DNA sequencer. The footprinting results of SurR (0.3 μM) with mbh1 probe DNA clearly show two regions of DNase protection extending from approximately −8 to −47 (length ~40 bp) and −106 to −164 (length ~59 bp) relative to the mbh1 translation start site (Fig. 2B, C). These footprints are most visible at protein/DNA molar ratios of 20 to 30; however, the mbh1 distal site is bound first by SurR as evidenced by footprinting experiments comparing SurR concentrations of 12 μM and 24 μM (see Supplementary Fig. S2A). At very high protein concentrations, all peaks disappear, indicating that the protein is binding non-specifically along the entire length of the DNA, outside its sequence-specific footprint site (data not shown). These results are consistent with the observation in EMSA that non-specific association with DNA occurs at high protein concentrations, resulting in very low-mobility protein-DNA complexes (see Fig. 2A). Furthermore, some peaks in the vicinity of the footprints have increased fluorescence intensity in the presence of SurR, signifying that nucleotides at these positions are hypersensitive to DNase when the protein is bound to the DNA (see asterisks, Fig. 2C).
SurR binds to its own promoter region shared with S0-regulated ORF PF0094
Interestingly, the surR gene (PF0095) is positioned 132 bp downstream and is divergently oriented from pdo (PF0094), a gene that is regulated in the primary response to S0. The pdo gene encodes a biochemically and structurally characterized enzyme, protein disulfide oxidoreductase (Ren et al., 1998, Pedone et al., 2004). In contrast to the down-regulation of the membrane-bound hydrogenase operon whose promoter was the target DNA from which SurR was discovered, pdo expression is up-regulated in the presence of S0 in microarray expression profiling experiments (Schut et al., 2007).
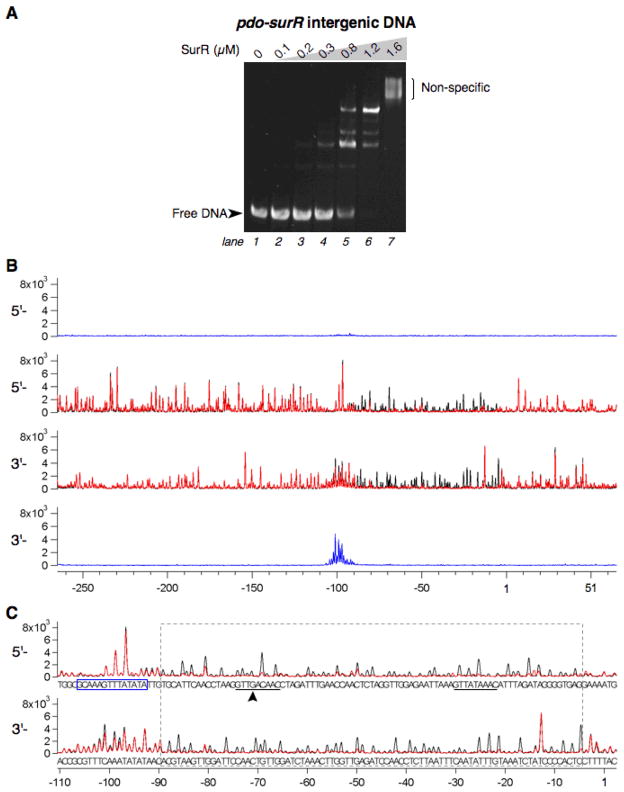
EMSA results with the pdo-surR intergenic DNA display a complete shift of the DNA probe at 1.2 μM protein (Fig. 3A, lane 6). As with the mbh1 promoter DNA, there are multiple bands of protein-DNA complexes; however, the number of resulting bands differs (compare Figs. 2Aleft panel, lanes 3–5 and 3A, lanes 4–6). DNase footprinting results of SurR (0.3 μM) on the pdo-surR promoter region reveal one large footprint (~85 bp) extending from approximately −5 to −89 relative to the pdo translation start site (Fig. 3B, C), or −44 to −128 relative to the surR translation start site. Footprinting experiments comparing different concentrations of SurR with the pdo-surR probe indicate that SurR does not bind preferentially to one part of the large footprint region, but instead binds across the entire footprint region, to a fraction of the probe DNA at 12 μM and to nearly all of the probe DNA at 24 μM (see Supplementary Fig. S2B). DNase I hypersensitive sites are not as evident for this DNA probe as compared to the mbh1 probe.
Fig. 3.
SurR binds with sequence-specificity to the pdo-surR promoter region. A, EMSA of SurR with pdo-surR promoter DNA (138 bp, 50 nM in each lane). Protein amounts are indicated at the top of the gel. The gel was stained with SYBR Green I nucleic acid gel stain. B, Fluorescence DNase I footprint of SurR (0.3 μM) on pdo-surR probe DNA (10 nM), with expanded view of footprint region shown in C. See caption of Fig. 1B,C for further explanations of footprint traces and annotations.
The SurR DNA recognition site contains the palindrome GTTn3AAC
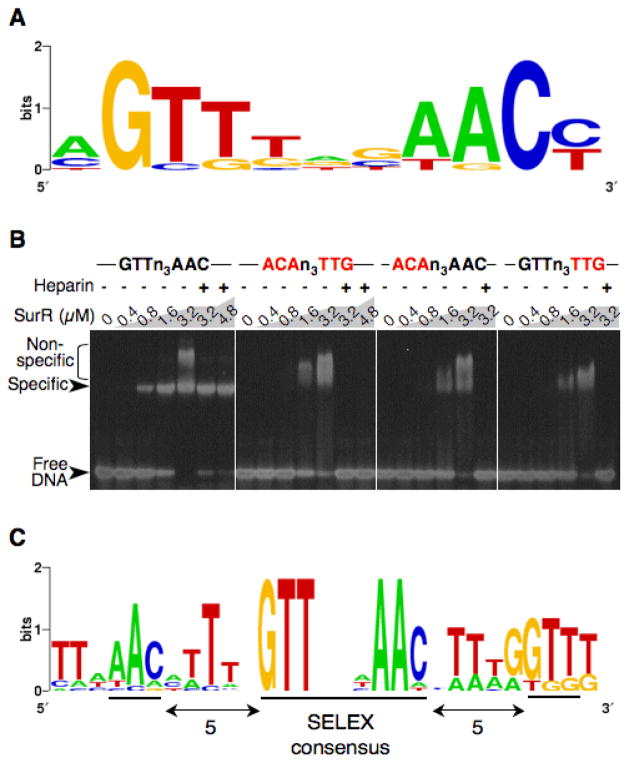
To determine the consensus DNA motif recognized by SurR, the artificial selection method SELEX was employed (Oliphant et al., 1989, Tuerk and Gold, 1990). Starting with a pool of synthetic DNA containing central 30-bp randomized sequences, EMSA was used to select sequences bound with higher affinity by SurR. Each pool of selected sequences was PCR-amplified and used in EMSA again for further selection. After six rounds of selection, the consensus palindromic DNA sequence GTTn3AAC was uncovered from 19 total sequenced SELEX DNAs (Fig. 4A).
Fig. 4.
SurR recognizes the palindrome GTTn3AAC. A, SurR DNA recognition motif determined by SELEX for 11 out of 19 selected sequences. B, EMSA of SurR with 40-bp DNA probes derived from one f the selected SELEX sequences, either containing the GTTn3AAC motif or mutated motifs, as indicated (mutated bases in red), at a concentration of 200 nM (~5 μg/mL). Heparin was used at a concentration of 20 μg/mL. Protein amounts are indicated at the top of the gel. Gel was stained with SYBR Green I nucleic acid gel stain. C, Consensus motif present in 200-bp ORF upstream regions of 13 GTTn3AAC-containing ORFs that are up- or down-regulated with S0. Motif diagrams were generated using WebLogo (Crooks et al., 2004).
To validate the GTTn3AAC motif as the necessary prerequisite for sequence-specific binding of DNA by SurR, a series of DNA fragments were tested with EMSA. A 40-bp DNA probe was designed from one of the selected SELEX DNA sequences containing the GTTn3AAC motif, along with three additional DNA probes identical to the first except having the motif mutated as follows: ACAn3TTG, ACAn3AAC, and GTTn3TTG (mutated bases in italics). As can be seen in Fig. 4B, SurR binds to the GTTn3AAC-containing probe DNA with higher affinity than the DNA probes with mutated motifs. Furthermore, adding heparin at a 4-fold mass excess over probe DNA does not remove the binding of SurR to the GTTn3AAC-containing probe but completely removes the non-specific binding observed for the probes with mutated sequences. The non-specific interactions of SurR with DNA can be generally distinguished in EMSA by a less well-defined shift of low mobility (see also Fig. 2A). These results indicate that the GTTn3AAC motif uncovered by SELEX is required for sequence-specific binding of DNA by SurR.
Each DNase footprint site in the mbh1 promoter was verified to contain one perfect GTTn3AAC motif, and the 85-bp footprint in the pdo-surR intergenic region contained two perfect consensus motifs (see underlined motifs in Figs. 2C and 3C). The presence of the GTTn3AAC motifs in the SurR footprint regions confirmed that the consensus sequence derived from the synthetic SELEX library was representative of the SurR recognition site in the P. furiosus genome.
The GTTn3AAC motif appears upstream of most ORFs involved in primary S0 response
All P. furiosus ORF upstream regions of 200 bp in length were searched for other occurrences of the SurR consensus motif GTTn3AAC, and a total of 123 unique motifs were found. Analysis of the sequence regions surrounding the GTTn3AAC consensus motifs within the known SurR DNA footprints (Figs. 2C and 3C) suggested the existence of an extended motif that included a second palindrome half-site with a 5-bp gap: GTTn3AACn5GTT. Searching all P. furiosus 200-bp ORF upstream regions with this string returned only 16 unique motifs (in a total of 23 ORF upstream regions, Table 1). Over half of the corresponding ORFs (14 out of 23) were part of divergently oriented gene pairs that presumably share the same promoter region (including the pdo-surR divergent gene pair). For the ORFs predicted to be part of operons (Tran et al., 2007), the ORF containing the upstream motif was first in its operon in all cases except for one (PF0913).
Table 1.
GTTn3AACn5GTT motifs found in 200-bp ORF upstream DNA regions
| ORFa | Strandb | Motif | Startc | Stopc | ORF Annotationd |
|---|---|---|---|---|---|
| PF0094 | − | GTTGTCAACCTTAGGTT | −82 | −66 | Protein disulfide oxidoreductase, pdo (Pedone et al., 2004) |
| PF0095 | + | GTTGTCAACCTTAGGTT | −67 | −51 | Sulfur response regulator, surR (this work) |
| PF0531 | − | GTTGGTAACAAAATGTT | −84 | −68 | cobalamin biosynthesis protein m |
| PF0532 | + | GTTACCAACGTAAGGTT | −100 | −84 | hypothetical protein |
| PF0547 | − | GTTGAAAACTTCAAGTT | −190 | −174 | hypothetical protein |
| PF0548 | + | GTTGAAAACTTCAAGTT | −67 | −51 | hydrogenase expression/formation protein |
| PF0559 | + | GTTTCTAACTTTTGGTT | −48 | −32 | hydrogenase expression/formation regulatory protein |
| PF0568 | − | GTTTATAACGCTATGTT | −24 | −8 | hypothetical protein |
| PF0569 | + | GTTTATAACGCTATGTT | −34 | −18 | hypothetical protein |
| PF0891 | + | GTTTTTAACCTTTGGTT | −142 | −126 | Hydrogenase I subunit beta, hydB1 (Ma et al., 1993) |
| PF0913 | − | GTTCAAAACCAAAGGTT | −197 | −181 | Mo formylmethanofuran dehydrogenase related |
| PF0914 | − | GTTCAAAACCAAAGGTT | −81 | −65 | Mo formylmethanofuran dehydrogenase related |
| PF0915 | − | GTTAGAAACCTTAGGTT | −58 | −42 | hypothetical protein |
| PF0926 | − | GTTTGGAACCTATTGTT | −81 | −65 | hypothetical protein |
| PF1100 | + | GTTTGCAACTCGTAGTT | −108 | −92 | hypothetical protein |
| PF1186 | − | GTTTAAAACCTTTAGTT | −34 | −18 | NAD(P)H sulfur reductase, nsr (Schut et al., 2007) |
| PF1328 | − | GTTCAAAACCTAAGGTT | −87 | −71 | hydrogenase gamma subunit |
| PF1329 | + | GTTCAAAACCTAAGGTT | −88 | −72 | Hydrogenase II subunit beta (Ma et al., 2000) |
| PF1422 | − | GTTATAAACCAAAAGTT | −19 | −3 | thioredoxin reductase |
| PF1423 | + | GTTATAAACCAAAAGTT | −146 | −130 | hypothetical protein |
| PF1423 | + | GTTTCAAACCAAATGTT | −31 | −15 | Membrane-bound hydrogenase subunit A, mbh1 (Sapra et al., 2000) |
| PF1516 | − | GTTTACAACTTTATGTT | −58 | −42 | GMP synthase subunit B |
| PF1621 | − | GTTAGTAACTAAAAGTT | −78 | −62 | hypothetical protein |
| PF1622 | + | GTTAGTAACTAAAAGTT | −196 | −180 | n-type ATP pyrophosphatase superfamily |
The ORF from which the 200-bp upstream sequence was searched.
The orientation of the ORF in the genome is indicated, either on the forward (+) or the reverse (−) strand.
Start and stop positions are relative to the sequence of the ORF upstream region where −1 corresponds to the first nucleotide upstream from the ORF start.
Annotations are from REFSEQ and JCVI-CMR. Genes that have been experimentally studied are listed with the corresponding reference.
Furthermore, the ORFs found to contain this motif in their upstream regions overlapped significantly with ORFs involved in the primary S0 response. As can be seen in Table 2, nine out of twelve of the ORFs/operons down-regulated 10 min after S0 addition have the extended motif within 200 bp upstream of the ORF, and four out of five of the up-regulated ORFs/operons contain the GTTn3AAC consensus motif in their upstream DNA regions (two of these also contain the extended motif). It is worth noting that none of the ORFs which are up- or down-regulated after 30 min of S0 addition, those that are part of the secondary response to S0 (Schut et al., 2007), have the extended motif in their upstream DNA. These results clearly point to a relationship between SurR target genes and the primary response of P. furiosus to the presence of S0.
Table 2.
Motifs found upstream of ORFs involved in the primary response to S0
| ORF/operona | ORF description/operon functionb | GTTn3AACn5GTTc | GTTn3AACc |
|---|---|---|---|
| Up-regulated >5-fold 10 min after S0 addition (Schut et al., 2007) | |||
| PF0094 | Protein disulfide oxidoreductase (pdo) (Pedone et al., 2004) | 1 | 2 |
| PF0261–0262 | unknown transporter | 0 | 0 |
| PF1186 | NAD(P)H sulfur reductase (nsr) (Schut et al., 2007) | 1 | 1 |
| PF1453–1441 | Membrane-bound oxidoreductase (mbx) (Silva et al., 2000) | 0 | 2 |
| PF2051–2052 | putative transcriptional regulators | 0 | 1 |
| Down-regulated >5-fold 10 min after S0 addition (Schut et al., 2007) | |||
| PF0450 | Glutamine synthetase, catalytic region | 0 | 0 |
| PF0531–0529 | Cobalt transport | 1 | 1 |
| PF0559 | Hydrogenase maturation protein HypF | 1 | 1 |
| PF0736 | Conserved hypothetical protein | 0 | 0 |
| PF0736.1 | Conserved hypothetical protein | 0 | 0 |
| PF0891–0894 | Hydrogenase I (PF0891: hydIB) (Ma et al., 1993) | 1 | 1 |
| PF0913 | Formylmethanofuran dehydrogenase, subunit E | 1 | 2 |
| PF0915 | Cytochrome c biogenesis protein | 1 | 1 |
| PF0926–0925 | Unknown | 1 | 1 |
| PF1329–1332 | Hydrogenase II (Ma et al., 2000) | 1 | 1 |
| PF1423–1436 | Membrane-bound hydrogenase (PF1423: mbh1) (Sapra et al., 2000) | 2 | 2 |
| PF1621 | Fibronectin, type III-like fold | 1 | 1 |
Predicted operons are represented by hyphenated loci with the first ORF of the operon (from which the motif was found upstream) listed first, followed by the last ORF of the operon.
ORF descriptions and operon functions are essentially derived from (Schut et al., 2007). ORFs and operons that have been previously described or characterized are listed with the corresponding reference.
Motifs were identified from a search of 200-bp ORF upstream DNA of all P. furiosus ORFs. The number of motifs present is indicated.
To better understand the in vivo SurR binding site, MEME motif-finding software (Bailey and Elkan, 1994) was used to search for an extended motif in the forward and reverse strands of all the motif-containing 200-bp upstream sequences of ORFs involved in the primary S0 response (listed in Table 2). The resulting consensus motif found in all 13 input sequences is shown in Figure 4C and contains the GTTn3AAC SELEX motif with adjacent palindrome half-sites spaced 5 bases away on either side. This identified ‘genomic’ motif suggests that the GTTn3AAC motifs must occur in pairs or multiples at defined intervals in order to direct specific high-affinity binding of SurR. Furthermore, the genomic motif indicates the relevance of A/T tracts in the 5-bp gap regions. It is possible that the SELEX experiment may have uncovered this extended motif if more selection rounds had been completed or if the artificial selection DNA length had been greater than 30 bp.
SurR binds to promoter regions of other ORFs involved in primary S0 response
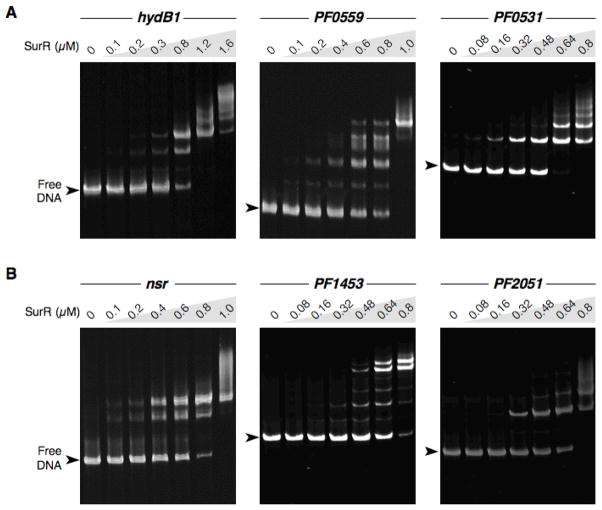
To verify that the identified motifs upstream of S0-regulated ORFs represent bona fide SurR binding sites, several of the putative promoter regions were selected as targets to test in EMSAs with SurR. Besides mbh1 (PF1423), the upstream DNA of three additional S0 down-regulated ORFs were chosen: PF0891 (hydB1), PF0559, and PF0531 (see Table 2). PF0891 is the first ORF in a four-ORF operon encoding soluble hydrogenase I of P. furiosus (Ma et al., 1993, Ma and Adams, 2001) and was selected because the primary response to S0 predominantly involves down-regulation of hydrogenase transcripts. The PF0559 ORF encodes a potential hydrogenase maturation protein and was also the most highly down-regulated ORF in the batch microarray experiment (Schut et al., 2001). PF0531 encodes a ‘cobalt transporter’ according to annotation, and it is likely the start of a four-ORF operon; however, this putative operon currently has no obvious connection with S0 metabolism.
With the exception of pdo (PF0094), which has already been shown to have a SurR binding site in its promoter region, all of the remaining up-regulated ORFs that contain the upstream GTTn3AAC motif were selected to test in EMSAs with SurR: PF1186 (nsr), PF1453, and PF2051 (see Table 2). The protein product of PF1186 has recently been identified as the sulfur reductase of P. furiosus, termed NSR for NAD(P)H sulfur reductase (Schut et al., 2007). PF1453 is the start of a 14-ORF operon (PF1453-PF1441) encoding a putative membrane-bound oxidoreductase that likely serves as a replacement for the membrane-bound hydrogenase when S0 is present (Silva et al., 2000, Schut, 2007 #144). PF2051 encodes a probable transcriptional regulator that may function in the secondary response to S0 (Schut et al., 2007).
EMSA experiments confirmed that SurR binds to all of these promoter-ORF DNA probes, exhibiting the characteristic multiple protein-DNA complexes for the DNA probes of both down- and up-regulated primary S0 response ORFs (Fig. 5). The number of shifted bands representing protein-DNA complexes varied for each probe, with approximately 3 bands observed for hydB1 (A, left panel), 4–5 for PF0559 (A, middle panel), 2–3 for PF0531 (A, right panel), 2 for nsr (B, left panel), 5–6 for PF1453 (B, middle panel), and 2 for PF2051 (B, right panel). The presence of multiple bands of protein-DNA complexes reinforces the notion that multiple SurR copies may bind to adjacent recognition sites and that the various promoter regions may have different numbers of these sites resulting in variability in the number of protein-DNA complexes observed in EMSA.
Fig. 5.
Validation of GTTn3AAC-containing upstream DNA regions of primary S0 response ORFs as SurR targets. EMSAs of SurR with probe DNA containing ORFs down-regulated (A) and up-regulated (B) during primary S0 response. Probe DNA concentrations (in each lane) are as follows: hydB1, 50 nM (326 bp); PF0559, 25 nM (282 bp); PF0531, 20 nM (488 bp); nsr, 25 nM (380 bp); PF1453, 20 nM (515 bp); PF2051, 20 nM (424 bp). Protein amounts are indicated at the top of the gel. Gel sizes were adjusted according to DNA standards such that free DNA positions are comparable among gels. Gels were stained with SYBR Green I nucleic acid gel stain.
The location of the SurR binding sites on hydB1 (PF0891), PF0559, nsr (PF1186) and PF1453 DNA probes were determined by fluorescence DNase I footprinting using the protein to DNA ratio that resulted in a nearly complete shift of the DNA probe in the corresponding EMSAs (see Fig. 5). A SurR footprint is visible on each probe (Fig. 6); however, the sizes of each and their positions relative to ORF starts vary greatly (for full-length footprints of these probes, see Supplementary Figs. S3 and S4). The SurR footprint on the hydB1 probe covers ~48 bp and extends from −104 to −151 from the hydB1 translation start site (Fig. 6A). The PF0559 footprint is ~75 bp in length and is located from −21 to −95 relative to the translation start site, as defined by NCBI (the JCVI-CMR annotated start site lies 18 bp downstream) (Fig. 6B). The footprints on the nsr and PF1453 probes extend from −1 to −54 (~54 bp) (Fig. 6C) and −37 to −131 (~95 bp) (Fig. 6D) from the translation start sites, respectively. The lengths of the SurR footprint on each probe correspond to some extent with the number of shifted EMSA bands for the same probe: essentially, the larger the footprint, the greater number of protein-DNA complexes observed in EMSA. The conserved GTTn3AAC motif is present in each footprint (see underlined sequences in Fig. 6A–D); however, its location within each footprint varies.
Fig. 6.
SurR footprints in promoters of primary S0 response ORFs. Expanded views of fluorescence DNase I footprints of SurR on probe DNA of primary S0 response ORFs: A, His6-SurR (0.32 μM) on hydB1 probe DNA (20 nM), B, His6-SurR (0.64 μM) on PF0559 probe DNA (20 nM), C, SurR (0.24 μM) on nsr probe DNA (10 nM), D, SurR (0.48 μM) on PF1453 probe DNA (10 nM). See fluorescence footprints of full-length undigested and digested probes in Supplementary Figs. S3 and S4. See caption of Fig. 1B,C for further explanations of footprint traces and annotations.
SurR is a transcriptional activator and repressor
To validate SurR as a regulatory transcription factor, its direct effect on basal transcription was tested using a well-established in vitro transcription system for P. furiosus (Hethke et al., 1996, Hethke et al., 1999). The in vitro transcriptional regulation profile of SurR was determined for two down-regulated ORFs in the primary S0 response, mbh1 (PF1423) and hydB1 (PF0891), and two up-regulated ORFs in the primary S0 response, pdo (PF0094) and nsr (PF1186). Additionally, the in vitro transcription start site for each promoter was examined by primer extension (sequencing gels are provided in Supplementary Fig. S5), and the regulatory effect of SurR on transcription of its own gene was also investigated.
The transcription start site of mbh1 (PF1423) was found to be positioned −61 bp from the translation start site, and therefore the SurR footprints on the mbh1 promoter do not overlap with either the transcription start site or the TATA box (see promoter elements relative to footprint in Fig. 2C). The BRE/TATA region matches the consensus sequence suggested by van de Werken (VRAAAnnTTWWWAW (van de Werken et al., 2006)). The transcription start site for hydB1 (PF0891) is also located some distance upstream from the translation start, at −73 bp. The SurR footprint is therefore located upstream from the BRE/TATA region but slightly overlaps the BRE (see promoter elements relative to footprint in Fig. 6A). The BRE and TATA sequences for this ORF also closely match the consensus.
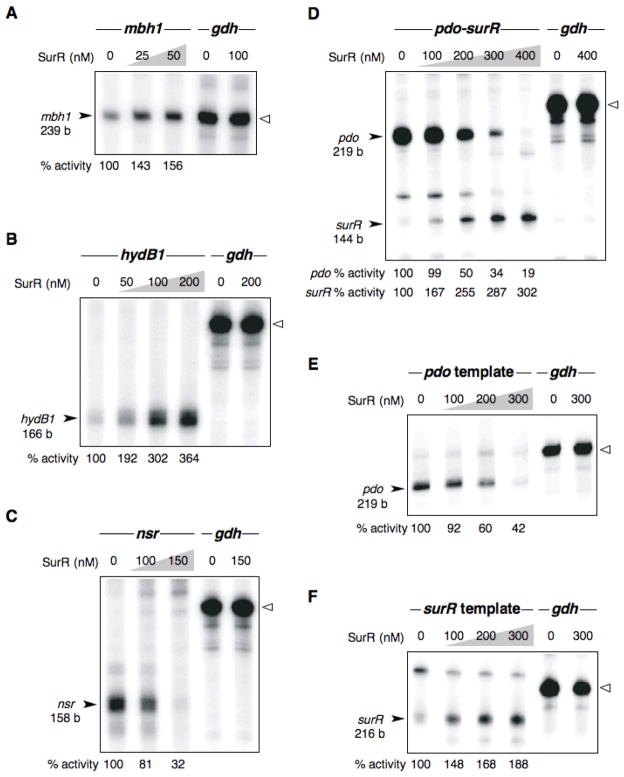
Basal transcription from the mbh1 probe produced a transcript only from mbh1 (PF1423) and not from the divergent ORF, PF1422. Addition of SurR resulted in a 56% increase in transcript level, demonstrating that SurR is a transcriptional activator for mbh1 (Fig. 7A). Transcription of hydB1 in the presence of SurR resulted in a dramatic increase in hydB1 transcript, up to ~260% (Fig. 7B). In both cases, SurR did not affect the basal transcription level from the gdh control template, indicating that SurR is a transcriptional activator for both mbh1 and hydB1, and presumably both of the corresponding operons, membrane-bound hydrogenase (14 ORFs) and soluble hydrogenase I (4 ORFs), respectively.
Fig. 7.
SurR regulates transcription of primary S0 response ORFs and its own ORF. In vitro transcription results for SurR showing activation of down-regulated ORFs involved in the primary S0 response, mbh1 (A) and hydB1 (B), and repression of up-regulated ORFs involved in the primary S0 response, nsr (C) and pdo in pdo-surR template (D) and a template containing only pdo and associated promoter elements (E). Activation of surR, divergently transcribed from and sharing a promoter region with pdo, is shown in D with the pdo-surR template and in F with a template containing only surR with associated promoter elements and part of the SurR binding site. The control gdh transcript (248 b) in each gel image is marked by an open triangle. Template amounts used are as follows: mbh1, 10 nM; hydB1, 25 nM; nsr, 25 nM; pdo-surR, 25 nM; pdo, 25 nM; surR, 25 nM; gdh, 10 nM. The % activity shown at the bottom of each gel was obtained by averaging the quantified transcripts (quantified with a Phosphor Imager) from three transcription experiments. The % activity for the control gdh transcript varied by less than 10% in each experiment.
The transcription start site for nsr (PF1186) is located at −10 bp from the translation start, placing both the transcription start and the BRE/TATA region within the SurR footprint for this promoter region (see promoter elements within footprint in Fig. 6C). As expected, SurR repressed transcription from nsr, resulting in a decrease in transcript level by nearly 70% (Fig. 7C).
The transcription start site for pdo (PF0094) is located at −71 bp from the translation start site and is within the footprint region occupied by SurR; however, the BRE sequence and TATA box are located just upstream from the footprint region (see promoter elements relative to footprint in Fig. 3C). Interestingly, pdo and surR appear to share a TATA box region since the surR transcription start site is located only −16 bp from the translation start, placing the surR TATA box in the same region as the pdo TATA box. The BRE and TATA box sequences for surR, however, appear to deviate substantially from the consensus and are probably less than optimal for initiation of basal transcription.
The pdo-surR transcription template contained portions of divergent ORFs pdo (PF0094) and surR (PF0095), and basal transcription produced transcripts from both ORFs, although the pdo transcript was much more abundant in comparison with the surR transcript (Fig. 7D). Addition of SurR resulted in an ~80% reduction of the pdo transcript with concomitant increase in its own transcript by ~200%. To determine if the increase in the surR transcript was the result of transcriptional activation or was simply due to increased availability of basal transcription components because of the repression of pdo, the transcriptional regulation of each ORF was examined independently using two templates. The first contained the BRE/TATA region and SurR footprint for pdo (pdo template) and the second contained the BRE/TATA region and part of the SurR footprint for surR (surR template). With the pdo template, SurR repressed transcription by almost 60% (Fig. 7E), and for the surR template, activation was confirmed, showing an increase in transcript level by approximately 90%, even though this template contained only one-third of the SurR footprint site and only one of the two GTTn3AAC motifs (Fig. 7F). Transcription of the gdh control template remained unaffected by SurR in each case.
Taken together, the in vitro transcription results show that SurR can both activate and repress transcription, depending on the gene and promoter context. These results are consistent with the positions of the core promoter elements relative to the SurR footprints in the promoter regions as summarized in Figure 8.
Fig. 8.
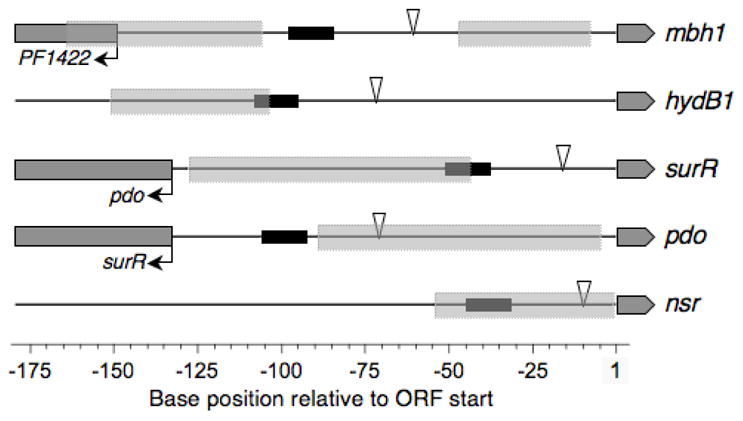
SurR footprint positions with respect to basal promoter elements indicate transcriptional regulatory mechanisms. Summary of SurR footprint positions (light grey boxes with broken lines), BRE/TATA regions (black boxes) and transcription initiation sites (open triangles) relative to target ORF start sites for genes regulated by SurR in in vitro transcription experiments (see Fig. 7). Promoter regions are aligned at the target ORF translation start position (dark grey box-arrows) and bent arrows indicate divergently transcribed ORFs (dark grey boxes). Note that the footprints defined by DNase I cleavage are likely larger than the actual protection region of SurR on the DNA due to the inability of the large DNase I enzyme to access the DNA just outside of the SurR binding region.
Discussion
SurR is a sequence-specific DNA binding protein and most likely interacts with DNA via its conserved helix-turn-helix motif located in the N-terminal ~60 amino acids of the protein. The purpose of the C-terminal region (~170 residues) is not evident from sequence analysis; however, it may serve as a dimerization domain, given that the protein recognizes a palindromic motif. EMSA and footprinting results for different target ORFs show different numbers of protein-DNA complexes (2 to 6) and various footprint lengths (40 to 95 bp), implying that SurR can bind to multiple adjacent sites that vary from the GTTn3AAC consensus sequence determined by SELEX. The presence of at least one perfect GTTn3AAC motif in each footprint suggests that this is the primary motif responsible for initial sequence recognition and binding by SurR. Perhaps SurR first recognizes the consensus motif GTTn3AAC and then facilitates binding of additional SurR proteins to adjacent, equally spaced degenerate motifs. Indeed, there is an extended motif present in the GTTn3AAC-containing promoters of primary S0 response ORFs (Fig. 4C) where motif elements repeat at 14-bp intervals, with A/T-rich tracts in the 5-bp gaps between the GTTn3AAC motif and adjacent palindrome half-sites. Although some degenerate motifs occurring at defined spacing (n3 and n5) near the conserved GTTn3AAC motifs can be found in several of the SurR footprint regions, cooperativity is not evident from analysis of footprint occupation with different protein concentrations on the mbh1 and pdo-surR promoter regions (Fig. S2)
SurR most likely serves as a transcriptional regulator of nearly all of the genes involved in the primary response of P. furiosus to S0. Our data show that SurR binds with sequence-specificity to promoters of ORFs for which the presence of S0 elicits either negative or positive transcriptional responses in vivo, according to DNA microarray expression profiles (Schut et al., 2007). A total of 53 ORFs are significantly regulated within 10 min of S0 addition, and when operon predictions are taken into account, the number of regulated gene clusters is reduced to 17 (Schut et al., 2007). Of these 17 ORFs/operons, only 4 of these do not contain the upstream GTTn3AAC motif within 200 bp of the translation start, and most contain the extended motif GTTn3AACn5GTT (Table 2).
Furthermore, of the 16 GTTn3AACn5GTT motifs found from a search of 200-bp upstream DNA sequences of all P. furiosus ORFs (Table 1), 12 occur in the upstream regions of ORFs and operons involved in the primary S0 response. The four additional motifs occur in the upstream regions of PF1100 and PF1516 and in the intergenic regions of the divergently transcribed gene pairs PF0547-PF0548 and PF0568-PF0569. Of these, all are annotated as ‘hypothetical proteins’ except for PF1516, which is annotated as GMP synthase, and PF0548, a putative hydrogenase expression/formation protein homologous to HypC. It is reasonable that the latter is a putative SurR target since PF0548 is likely involved in regulation of hydrogenase formation in P. furiosus as its homolog HypC is responsible for Fe insertion during hydrogenase maturation in E. coli and other organisms (Blokesch et al., 2004). A relationship is not obvious for the other apparent regulatory targets of SurR that are not among those genes part of the primary S0 response. It is possible that these genes have subtle transcriptional regulation that was not detectable in the microarray experiments.
SurR appears to play an activating role for at least two of the S0 down-regulated ORFs, both of which are part of hydrogenase operons. There are two SurR footprints in the mbh1 promoter region (of the membrane-bound hydrogenase operon), an mbh1-proximal site extending ~40 bp and an mbh1-distal site covering ~59 bp with a 59-bp ‘inter-footprint’ space in which the BRE/TATA region and the transcription initiation site are located. The larger mbh1-distal site is located just upstream of the BRE/TATA region, and presumably SurR activates transcription by recruiting TBP and/or TFB from this upstream site. The small mbh1-proximal site does not interfere with the transcription start site of mbh1; we hypothesize that it may serve to promote bending of the DNA, as suggested by the presence of DNase I hypersensitive sites (see peaks marked by asterisks in Fig. 2C), to form a structure more favorable to binding by the basal transcription apparatus or perhaps even aid in RNA polymerase recruitment. It is interesting that Ptr2 of M. jannaschii activates transcription in vitro from the rb2 promoter by binding to sites both upstream and downstream of the TATA box, even though the downstream site overlaps the transcription start site (Ouhammouch et al., 2005).
In contrast to mbh1, the hydB1 promoter contains only one SurR footprint directly upstream from the BRE/TATA region, and activation of this gene may be similar to the mechanism for mbh1 activation from the mbh1-distal site. Regulation of hydrogenase gene expression appears to be a key role for SurR as the recognition motif occurs in the promoter regions of all three P. furiosus hydrogenase operons, as well as at least two genes involved in hydrogenase maturation (PF0559, hypF, and PF0548, hypC). It is also likely involved in regulation of the handful of other genes that are down-regulated during primary S0 response, as most of these also contain SurR recognition motifs in their promoter regions (Table 2). The expression of these genes as detected by DNA microarray becomes highly down-regulated as part of the primary S0 response (Schut et al., 2007); therefore, the activation of these genes by SurR must be quickly counteracted in some way as part of the response to S0.
SurR has also been shown to be a transcriptional repressor for at least two genes involved in the primary S0 response of P. furiosus, nsr and pdo (Fig. 7C, D). We propose that SurR also functions as a transcriptional repressor for the two of three remaining gene clusters involved in primary S0 response. These are the 14-ORF membrane-bound oxidoreductase operon (mbx, PF1453-PF1441), thought to functionally replace the membrane-bound hydrogenase, and the two-ORF operon of putative regulators PF2051–PF2052, which may be involved in mediating the secondary response to S0. The consensus motif GTTn3AAC occurs in the upstream DNA regions of all four of these ORFs/operons (Table 2), SurR has been shown to bind to DNA probes of all of these in EMSAs (Figs. 3A and 5B), and a SurR footprint location has been verified for three of these (Figs. 3B–C and 6C, D). Furthermore, the locations of the SurR footprints upstream of nsr and pdo with respect to the identified transcription start sites indicate that the mechanism by which SurR represses transcription of these ORFs is by blocking access to basal promoter elements. In the case of nsr, the SurR footprint appears to overlap both the transcription start site and the BRE/TATA region, and SurR would therefore prevent the binding of TBP and TFB as well as the recruitment of RNA polymerase. The SurR binding site upstream of pdo overlaps just the transcription start site, and SurR would therefore prevent RNA polymerase recruitment but might allow simultaneous binding of TBP and TFB at the unobstructed BRE/TATA region. SurR regulation of pdo is particularly interesting since the divergent surR gene shares with it a short intergenic space (132 bp), and the two genes appear to share a TATA box region. We have demonstrated that SurR activates transcription of its own gene while repressing that of pdo, and the position of the SurR binding site in this promoter region sheds some light on this dual regulatory mechanism. SurR likely blocks RNA polymerase from being recruited to the pdo transcription initiation site while simultaneously recruiting TBP to the shared TATA region to allow the initiation of transcription from the surR transcription start site 16 bases upstream of the gene. Although SurR activates its own gene in vitro, surR is not among the genes regulated significantly during the primary or the secondary response to S0, as determined by microarray expression profiles; therefore, the in vivo effects of SurR on transcription of its own gene may presumably be more subtle.
The SurR footprint locations in relation to sequence elements give a good indication of the general mechanism of regulation imposed by SurR on transcription of various ORFs (Fig. 8). In all three cases of activation, the SurR footprint occurs just upstream from the BRE/TATA or slightly overlapping the distal side of the BRE, suggestive of a role for SurR in recruitment of basal transcription factors to the BRE/TATA elements to accelerate transcription. SurR may activate transcription by facilitating recruitment of TBP specifically, perhaps similar to the activation mechanism determined for Ptr2 (Ouhammouch et al., 2003). Repression by SurR appears to occur through known mechanisms, by blocking the binding of TBP and TFB as is the case for Lrs14 of S. solfataricus (Bell and Jackson, 2000) and/or the recruitment of RNA polymerase as is the mechanism for both LrpA (Brinkman et al., 2000, Dahlke and Thomm, 2002) and Phr (Vierke et al., 2003) of P. furiosus. Further experiments need to be performed to fully understand the activation and repression mechanisms employed by SurR.
Considering the in vitro transcription results, SurR should exert its transcriptional effect in the absence of S0, activating genes that are down-regulated during the primary S0 response and repressing genes that are up-regulated during the primary S0 response. This is also consistent with results from the DNA-affinity capture experiment in that the SurR protein band was present at a higher intensity in the –S0 extract lane (Fig. 1, band 10). A question that remains is how the transcriptional activity of SurR is itself mediated during the response to S0 since it appears to exert regulation when S0 is absent. Other factors or proteins may be involved in exerting an effect on SurR that would result in both the deactivation and derepression of target ORFs necessary for the response of P. furiosus to the presence of S0. This is currently under investigation.
Experimental Procedures
Cell growth and processing
P. furiosus (DSM 3638) was grown on maltose with and without S0, and the soluble cell extract was obtained as in (Schut et al., 2001), except that no DNase I was added. Aliquots of cell extract were stored in anaerobic vials at −80°C for use in DNA affinity protein capture experiments.
DNA affinity protein capture
The 314-bp mbh1 DNA probe (genome coordinates 1337201–1337513) used for protein capture was PCR-amplified from P. furiosus genomic DNA using the biotinylated primer 5′-ccctaacttggtggtgccta and the unlabeled primer 5′-ccagctatgagttcctgggta. To immobilize the bait DNA, excess mbh1 probe was incubated with ~200 μg magnetic DynaBeads M-280 Streptavidin (Invitrogen) per the manufacturer’s protocol (1 mg beads binds ~55 pmol DNA of length ~300 bp). The bead-bound DNA was then incubated at 55°C for 30 min with 2.5 mg/mL P. furiosus soluble cell extract obtained from cells grown either in the presence or absence of S0. Unbound proteins were removed by three successive washes with buffer B (50 mM EPPs, 100 mM KCl, 1 mM EDTA, 5% glycerol, 0.1% triton-X, 1 mM DTT, pH 7.5). DNA-bound proteins were eluted at 95°C for 5 min with 1× Laemmli buffer containing no β-mercaptoethanol, as this tended to strip streptavidin from the bead surface. Eluted proteins were analyzed by SDS-PAGE with silver staining.
In-gel digestion and peptide mass mapping
Bands of interest in SDS-PAGE lanes were excised and subjected to in-gel tryptic digestion (Shevchenko et al., 1996, Gharahdaghi et al., 1999). Peptide mass mapping was performed by the Chemical and Biological Sciences Mass Spectrometry Facility (University of Georgia, Athens, GA) on a Bruker Autoflex (TOF) mass spectrometer (Bruker Daltonics Inc., Billerica, MA). Proteins were identified by peptide mass fingerprinting using the MASCOT online search engine (www.matrixscience.com (Perkins et al., 1999)) to search the NCBI database of archaeal genomes and a local server hosting ProteinProspector (Clauser et al., 1999) to search a P. furiosus genome database.
Preparation of SurR clones
The PF0095 sequence encoding SurR (genome coordinates 103332 – 104030) was cloned into a pET24d vector modified to incorporate an N-terminal hexahistidine tag on the expressed protein (pET24dBAM, e.g., (18)). To generate a His-tag cleavable SurR protein construct, the PF0095 sequence was subcloned from pET24dBAM-PF0095 into a pET24dBAM-TEV vector modified to encode a TEV protease site (5′-gaaaacctgtacttccaagga) between the N-terminal His-tag and the protein sequence (His6-TEV-SurR). The pET24dBAM-PF0095 plasmid and the pET24dBAM-TEV vector were kind gifts from Francis Jenney.
Expression and purification of SurR
BL21-CodonPlus(DE3)-RIPL cells (Stratagene) were used for recombinant protein expression. His6-SurR was expressed in a 1-L culture induced with 0.4 mM IPTG at OD600 ~0.9–1 according to standard protocols. His-tag cleavable SurR was expressed using the auto-induction protein expression protocol established by Studier (Studier, 2005). Briefly, one 50-mL culture grown for 8–10 h at 37°C in P-0.5G media was used to inoculate 1 L of ZYP-5052 media which was then divided equally into two 2-L flasks. The large-scale cultures were grown for 18–20 h at 37ÜC. Cells were harvested at 6,000g at 4°C for 15 min. and resuspended in Binding Buffer (20 mM sodium phosphate, 0.5 M NaCl, pH 7.4) containing a protease inhibitor cocktail (Sigma). Soluble cell extract was prepared by sonication and centrifugation according to standard protocols. His6-SurR and His6-TEV-SurR were purified over 1-mL HisTrap affinity columns (GE Healthcare) and eluted with a gradient of Eluting Buffer (20 mM sodium phosphate, 0.5 M NaCl, 0.5 M imidazole, pH 7.4) using an AKTA system (GE Healthcare). Protein-containing fractions were concentrated and buffer exchanged into 20 mM HEPES, 200 mM NaCl, pH 7.6, using an Amicon Ultra-15 centrifugal filter device with a 10 kDa molecular weight cut-off (Millipore). The His6-TEV-SurR protein was subjected to His-tag cleavage using AcTEV protease (Invitrogen) according to the manufacturer’s instructions, except that less protease was used in conjunction with a longer incubation time. Following digestion, the protein sample was applied directly to a fresh HisTrap column (GE Healthcare) equilibrated with 20 mM HEPES, 200 mM NaCl, pH 7.6, using a 0.2 mL/min flow rate. The column flow-through containing the tagless protein was collected. Protein concentrations were determined using a Bio-Rad DC Protein Assay kit, and aliquots of protein were stored at −80°C.
Electromobility shift assay
DNA probes for EMSA were PCR-amplified from P. furiosus genomic DNA or from pUC18-cloned promoter-ORF DNA (see below) using slightly extended M13 forward and reverse sequencing primers (5′-ttgtaaaacgacggccagt and 5′-caggaaacagctatgaccatg, extended sequence in italics), followed by either PCR purification using a Qiagen PCR Purification Kit, gel purification using a Qiagen Qiaquick Gel Purification Kit, or ethanol precipitation. The 81-bp mbh1 promoter probe (genome coordinates 1337338–1337418) was PCR-amplified from P. furiosus genomic DNA using primers 5′-gccaatacgaatttggagagagg and 5′-gacattcgccaaacctcctt, the 80-bp mbh1 ORF probe (1337435–1337514) was amplified using primers 5′-atatgggcttacttcctctgg and 5′-ccagctatgagttcctgggta, and the 138-bp pdo-surR intergenic DNA probe (103197–103334) was amplified using primers 5′-cattttcctcacccctatc and 5′-catattcatcacctacccatatc. The probe for testing the GTTn3AAC motif uncovered by SELEX was derived from one of the selected SELEX DNAs and was prepared by annealing the following oligonucleotides: 5′-gcttcatccgtgacGTTcacAACcaaatagattgaggatc and 5′-gatcctcaatctatttgGTTgtgAACgtcacggatgaagc (selected 30-bp randomized region in italics with motif capitalized). Three additional probes were designed with the GTTcacAAC motif mutated to ACAcacTTG, ACAcacAAC, and GTTcacTTG (mutated bases in italics). EMSA reactions of probe DNA with varying amounts of protein were set up in 10-μL volumes in EMSA buffer (20 mM HEPES, 200 mM KCl, 5% glycerol, 1 mM EDTA, pH 7.5). Heparin was used as a non-specific competitor in some binding reactions. Reactions were incubated at 55 or 70°C for 20 min and immediately loaded onto a non-denaturing 5% polyacrylamide gel (BioRad). The resulting gel was stained with SYBR Green nucleic acid gel stain (Invitrogen) to stain the DNA followed by SYPRO Ruby protein gel stain (Bio-Rad) to verify the presence of protein in the shifted bands.
Cloning of P. furiosus promoter-ORF DNA
Promoter-ORF DNA fragments were cloned from P. furiosus genomic DNA into pUC18 (GenBank/EMBL accession number, L09136) for versatile PCR-amplification of probes for EMSA, footprinting and in vitro transcription experiments. The design of the DNA fragment to be cloned was optimized primarily for use as a transcription template, and secondarily as a footprinting probe. For a transcription template, at least ~100 bp of the ORF was included to allow for an adequate transcript in an in vitro transcription assay; for footprinting, the DNA length chosen was ~300 to 500 bp with the putative promoter region centered in the probe to allow for satisfactory visualization of the footprint. DNA was PCR-amplified from P. furiosus genomic DNA using a high-fidelity Pfu DNA polymerase (Stratagene) and cloned into pUC18 using standard procedures (see Supplementary Table S1 for primer and sequence details). Positive clones were selected by blue/white colony screening and verified by colony PCR and sequencing (Sequetech, Inc, Mountain View, CA).
Fluorescence DNase I footprinting
Fluorescence footprinting was performed as a modification from (Wilson et al., 2001) based on the DNase I footprinting method (Galas and Schmitz, 1978). Footprinting probes were PCR-amplified from pUC18-cloned promoter-ORF DNA (Table S1) using 5′ 6FAM and HEX labeled modified M13 primers for analysis on a 3730×1 automated DNA sequencer (Applied Biosystems). The standard M13 forward primer was extended at the 5′ end to ensure that the base adjacent to the fluorophore was not a guanine, as guanine can quench fluorescence at close proximity. Consequently, the slightly extended M13 forward and reverse primers used were 5′-ttgtaaaacgacggccagt (6FAM-labeled) and 5′-caggaaacagctatgaccatg (HEX-labeled), respectively. PCR-amplified probes were purified on non-denaturing 5% polyacrylamide gels (BioRad) using the crush-and-soak method (Sambrook and Russel, 2001). Resulting probe DNA was concentrated by ethanol precipitation and quantified using the Hoechst 33258 dye binding assay prior to use in footprinting reactions.
Footprinting reactions were composed of two parts, the protein-DNA binding reaction and the cleavage reaction. The protein-DNA incubation solutions were similar to the EMSA reactions except the reactions were set up in 50-μL volumes with ~150–200 ng DNA probe, incubations of individual reactions were separated by 2 or 3-min intervals, and the final footprinting buffer composition (from a 5×stock solution) was as follows: 20 mM HEPES, 100 mM KCl, 15 mM MgCl2, 5 mM CaCl2, 1 mM EDTA, 1 mM DTT, 5% glycerol, pH 8. Protein-DNA solutions were incubated for 20 min at 55°C or 70°C. For the DNase I cleavage reaction, 0.03–0.05 U of DNase I (from a 0.01 U/μL dilution in 10 mM Tris, pH 8.0) was added to each 50-μL protein-DNA mixture, and the solution was incubated for 1 min at room temperature. The reaction was stopped by adding 145 μL of Stop Solution (130 mM NaCl, 20 mM EDTA, 0.6% SDS) followed immediately by 200 μL of buffered phenol:chloroform:isoamyl alcohol (25:24:1) with vigorous vortexing. After centrifugation, 180 μL of the aqueous phase was removed and prepared for ethanol precipitation with 18 μL 3 M sodium acetate (pH 5.2), 1 μL glycogen (20 mg/mL, Roche), and 500 μL 100% ethanol. Samples were stored at −20°C in precipitation solution until they were prepared for analysis. DNA samples were precipitated, resuspended in 10 μL of HiDi deionized formamide (Applied Biosystems) premixed with GS-500 internal size standard (Applied Biosystems) (0.2 μL per sample), and assembled into a 96-well reaction plate (Applied Biosystems). Fragment analysis was performed at the Laboratory for Genomics and Bioinformatics (University of Georgia) using a 3730×1 automated DNA sequencer (Applied Biosystems) calibrated with the DS-30 dye set. Raw peak data were extracted from the result files using the BatchExtract program available for download from NCBI(ftp://ftp.ncbi.nih.gov/pub/forensics/BATCHEXTRACT). Raw peak data were analyzed, calibrated, and visualized using the graphing and analysis software IGOR Pro (Wavemetrics, Inc.).
SELEX
The SELEX probe was designed with a 30-nt randomized region flanked by priming regions each containing three restriction sites (XbaI, EcoRI, HindIII on the 5′ side and BamHI, EcoRI, SalI on the 3′ side of the ssDNA probe), ggtctagagaattcaagcttc-(n)30-ggatccgaattcgtcgac. The double-stranded SELEX probe was PCR-amplified from the synthetic single-stranded oligonucleotide with primers that slightly extended the original SELEX probe length (thereby eliminating the 3′ self-complementary region at the SalI site and improving the PCR specificity), 5′-actactgtcgacgaattcgga and 5′-gctcaggtctagagaattcaa. To synthesize the dsDNA probe, 100 pmol of the single-stranded SELEX oligonucleotide was amplified with 2 nmol of each primer for a total of 5 PCR cycles. The PCR-amplified double-stranded SELEX probe was polyacrylamide gel-purified using the crush-and-soak method (Sambrook and Russel, 2001). Selection rounds were set up essentially as for the EMSA reactions, except for the amount of SELEX probe used. For the first round of selection, 0.6 μM SELEX probe was used, and for all succeeding selection rounds, 0.1 μM was used; protein concentrations ranged from 0.6–1.2 μM. After each selection round, DNA was purified from shifted protein-DNA complexes, amplified with the SELEX primers using 15 cycles of PCR, and polyacrylamide gel-purified before proceeding to the next selection round. A total of 6 selection rounds were performed in this manner. The selected DNA was digested with EcoRI, concatemerized, and cloned into pUC18. Blue/white color screening and colony PCR were used to identify colonies that contained plasmids with the largest concatemers, and plasmid DNA was then amplified and sequenced. A total of 14 sequences were obtained from the 6th selection round, and a total of 5 sequences were obtained from the 5th selection round. These sequences were searched using MEME online motif searching software (Bailey and Elkan, 1994) to elucidate a common motif among the selected DNA.
Construction of a P. furiosus ORF upstream DNA database to be searched for the SELEX motif
A database of all ORF upstream regions was prepared by extracting sequences upstream of each ORF in the P. furiosus genome. First, all ORF definitions for P. furiosus from the REFSEQ, GENBANK, and JCVI-CMR databases were amassed. ORF definitions from the three databases were correlated using the stop position for each ORF definition as a unique identifier. An ORF corresponding to a given stop position was redefined according to the ORF definition that yielded the shortest ORF. This was done to ensure that the maximum length upstream sequence was used for searching. Next, 200-bp sequences just upstream of each ORF were extracted and recorded in the database. Using a Python script (http://www.python.org/) and regular expression functions, the resulting database was searched for candidate binding sites using both forward and reverse strands of the discovered SELEX motif (GTTn3AAC and extended motif GTTn3AACn5GTT, see Results).
In vitro transcription
The in vitro transcription was performed as described previously (Hethke et al., 1996, Hethke et al., 1999). Templates were PCR-amplified from cloned probe DNA (Table S1) with M13 forward and reverse primers except for the templates used to show regulation of pdo and surR. The pdo-surR template (408 bp) for in vitro transcription was amplified from the cloned pdo probe (Table S1) with the internal primer 5′-gccacagcagttgatgac and the M13 forward primer to generate a template which allowed for transcripts of different sizes. The two templates containing either only pdo and only surR were PCR-amplified from the cloned pdo probe (Table S1) using the internal primer 5′-ctgtggcgcaaagtttatatattg and the M13 forward primer for pdo (261 bp) and the internal primer 5′-ctaggttgtcaaccttaggttg and the M13 reverse primer for surR (271 bp). Each transcription reaction was assembled in a final volume of 50 μL. Transcription mixtures contained 40 mM HEPES buffer, pH 7.3; 2.5 mM MgCl2; 0.1 mM EDTA; 280 mM KCl; 0.44 mM each of ATP, GTP, CTP, 0.002 mM UTP and 2 μCi [α-32P]-UTP; 23 nM TFB (recombinant); 104 nM TBP (recombinant); 19 nM RNA polymerase; and different amounts (0–400 nM) of recombinant SurR. The reaction was incubated for 30 min at 70°C. Runoff transcripts were analysed on an 8% polyacrylamide urea gel in 1×TBE buffer. The transcription products were quantified and visualized by phosphorimaging (FLA-5000, Fuji, Japan).
Primer extension analyses of transcription start sites
For analyses of the in vitro transcriptional start sites, transcription reactions with unlabeled precursors (0.44mM each) were performed. The end-labelled primers used for the reactions were as follows: 5′-ggtaacttaacatacctcacac, complementary to nucleotides +72 to +93 of hydB1 (PF0891); 5′-ctctagaagcggcactcattcc, complementary to nucleotides +53 to +74 of PF1186; 5′-gctatgagttcctgggtactc, complementary to nucleotides +139 to +159 of mbh1 (PF1423); 5′-ccttaattaccttcttgtcagc complementary to nucleotides +90 to +111 of pdo (PF0094); and 5′-gcattccatacaagttagg, complementary to nucleotides +76 to + 94 of surR (PF0095). The primers were annealed with RNA products from a transcription reaction and extended with reverse transcriptase. The cDNA was analysed by electrophoresis on an 8% DNA sequencing gel. Primer extension products were quantified and visualized using a PhosphorImager (FLA-500, Fuji, Japan).
Supplementary Material
Acknowledgments
We thank Dr. Francis E. Jenney for providing the recombinant PF0095 clone as well as the pET24dBAM-TEV cloning vector. We also thank Dr. Mohamed Ouhammouch for his detailed protocols and useful suggestions for the footprinting and SELEX work. This work was supported by the National Science Foundation (MCB-9631093 to R. A. S.), the National Institutes of Health (GM042025 to R. A. S.), the Department of Energy (FG05-95ER20175 to M. W. W. A.), and by the priority program of the Deutsche Forschungsgemeinschaft for “Regulation of genome function and gene regulation in Archaea” (to M. T.).
References
- Adams MW. Biochemical diversity among sulfur-dependent, hyperthermophilic microorganisms. FEMS Microbiol Rev. 1994;15:261–277. doi: 10.1111/j.1574-6976.1994.tb00139.x. [DOI] [PubMed] [Google Scholar]
- Adams MW, Holden JF, Menon AL, Schut GJ, Grunden AM, Hou C, Hutchins AM, Jenney FE, Jr, Kim C, Ma K, Pan G, Roy R, Sapra R, Story SV, Verhagen MF. Key role for sulfur in peptide metabolism and in regulation of three hydrogenases in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 2001;183:716–724. doi: 10.1128/JB.183.2.716-724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bartlett MS. Determinants of transcription initiation by archaeal RNA polymerase. Curr Opin Microbiol. 2005;8:677–684. doi: 10.1016/j.mib.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Bell SD. Archaeal transcriptional regulation--variation on a bacterial theme? Trends Microbiol. 2005;13:262–265. doi: 10.1016/j.tim.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Bell SD, Jackson SP. Mechanism of autoregulation by an archaeal transcriptional repressor. J Biol Chem. 2000;275:31624–31629. doi: 10.1074/jbc.M005422200. [DOI] [PubMed] [Google Scholar]
- Blokesch M, Albracht SPJ, Matzanke BF, Drapal NM, Jacobi A, Bock A. The complex between hydrogenase-maturation proteins HypC and HypD is an intermediate in the supply of cyanide to the active site iron of [NiFe]-hydrogenases. J Mol Biol. 2004;344:155–167. doi: 10.1016/j.jmb.2004.09.040. [DOI] [PubMed] [Google Scholar]
- Brinkman AB, Dahlke I, Tuininga JE, Lammers T, Dumay V, de Heus E, Lebbink JH, Thomm M, de Vos WM, van Der Oost J. An Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus is negatively autoregulated. J Biol Chem. 2000;275:38160–38169. doi: 10.1074/jbc.M005916200. [DOI] [PubMed] [Google Scholar]
- Clauser KR, Baker P, Burlingame AL. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- Cramer P. Multisubunit RNA polymerases. Curr Opin Struct Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlke I, Thomm M. A Pyrococcus homolog of the leucine-responsive regulatory protein, LrpA, inhibits transcription by abrogating RNA polymerase recruitment. Nucleic Acids Res. 2002;30:701–710. doi: 10.1093/nar/30.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala G, Stetter KO. Pyrococcus-Furiosus Sp-Nov Represents a Novel Genus of Marine Heterotrophic Archaebacteria Growing Optimally at 100-Degrees C. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek EP, Ouhammouch M. Archaeal transcription and its regulators. Mol Microbiol. 2005;56:1397–1407. doi: 10.1111/j.1365-2958.2005.04627.x. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hethke C, Bergerat A, Hausner W, Forterre P, Thomm M. Cell-free transcription at 95 degrees: thermostability of transcriptional components and DNA topology requirements of Pyrococcus transcription. Genetics. 1999;152:1325–1333. doi: 10.1093/genetics/152.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hethke C, Geerling AC, Hausner W, de Vos WM, Thomm M. A cell-free transcription system for the hyperthermophilic archaeon Pyrococcus furiosus. Nucleic Acids Res. 1996;24:2369–2376. doi: 10.1093/nar/24.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey AJ, Conway de Macario E, Macario AJ. Transcription in the archaea: basal factors, regulation, and stress-gene expression. Crit Rev Biochem Mol Biol. 2002;37:537–599. doi: 10.1080/10409230290771555. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci U S A. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Adams MW. Hydrogenases I and II from Pyrococcus furiosus. Methods Enzymol. 2001;331:208–216. doi: 10.1016/s0076-6879(01)31059-5. [DOI] [PubMed] [Google Scholar]
- Ma K, Schicho RN, Kelly RM, Adams MW. Hydrogenase of the hyperthermophile Pyrococcus furiosus is an elemental sulfur reductase or sulfhydrogenase: evidence for a sulfur-reducing hydrogenase ancestor. Proc Natl Acad Sci USA. 1993;90:5341–5344. doi: 10.1073/pnas.90.11.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Weiss R, Adams MW. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J Bacteriol. 2000;182:1864–1871. doi: 10.1128/jb.182.7.1864-1871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnichenko ML, Bonch-Osmolovskaya EA. Recent developments in the thermophilic microbiology of deep-sea hydrothermal vents. Extremophiles. 2006;10:85–96. doi: 10.1007/s00792-005-0489-5. [DOI] [PubMed] [Google Scholar]
- Moxley RA, Jarrett HW. Oligonucleotide trapping method for transcription factor purification systematic optimization using electrophoretic mobility shift assay. J Chromatogr. 2005;1070:23–34. doi: 10.1016/j.chroma.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Oliphant AR, Brandl CJ, Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol Cell Biol. 1989;9:2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouhammouch M, Dewhurst RE, Hausner W, Thomm M, Geiduschek EP. Activation of archaeal transcription by recruitment of the TATA-binding protein. Proc Natl Acad Sci USA. 2003;100:5097–5102. doi: 10.1073/pnas.0837150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouhammouch M, Langham GE, Hausner W, Simpson AJ, El-Sayed NM, Geiduschek EP. Promoter architecture and response to a positive regulator of archaeal transcription. Mol Microbiol. 2005;56:625–637. doi: 10.1111/j.1365-2958.2005.04563.x. [DOI] [PubMed] [Google Scholar]
- Pedone E, Ren B, Ladenstein R, Rossi M, Bartolucci S. Functional properties of the protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus: a member of a novel protein family related to protein disulfide-isomerase. Eur J Biochem. 2004;271:3437–3448. doi: 10.1111/j.0014-2956.2004.04282.x. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Ren B, Tibbelin G, de Pascale D, Rossi M, Bartolucci S, Ladenstein R. A protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus contains two thioredoxin fold units. Nat Struct Biol. 1998;5:602–611. doi: 10.1038/862. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; NY: 2001. [Google Scholar]
- Sapra R, Verhagen MF, Adams MW. Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 2000;182:3423–3428. doi: 10.1128/jb.182.12.3423-3428.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schut GJ, Bridger SL, Adams MW. Insights into the Metabolism of Elemental Sulfur by the Hyperthermophilic Archaeon Pyrococcus furiosus: Characterization of a Coenzyme A-Dependent NAD(P)H Sulfur Oxidoreductase. J Bacteriol. 2007;189:4431–4441. doi: 10.1128/JB.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schut GJ, Zhou J, Adams MW. DNA microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus: evidence for anNew type of sulfur-reducing enzyme complex. J Bacteriol. 2001;183:7027–7036. doi: 10.1128/JB.183.24.7027-7036.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Silva PJ, van den Ban ECD, Wassink H, Haaker H, de Castro B, Robb FT, Hagen WR. Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur J Biochem. 2000;267:6541–6551. doi: 10.1046/j.1432-1327.2000.01745.x. [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Tran TT, Dam P, Su Z, Poole FL, 2nd, Adams MW, Zhou GT, Xu Y. Operon prediction in Pyrococcus furiosus. Nucleic Acids Res. 2007;35:11–20. doi: 10.1093/nar/gkl974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- van de Werken HJ, Verhees CH, Akerboom J, de Vos WM, van der Oost J. Identification of a glycolytic regulon in the archaea Pyrococcus and Thermococcus. FEMS Microbiol Lett. 2006;260:69–76. doi: 10.1111/j.1574-6968.2006.00292.x. [DOI] [PubMed] [Google Scholar]
- Vierke G, Engelmann A, Hebbeln C, Thomm M. A novel archaeal transcriptional regulator of heat shock response. J Biol Chem. 2003;278:18–26. doi: 10.1074/jbc.M209250200. [DOI] [PubMed] [Google Scholar]
- Wilson DO, Johnson P, McCord BR. Nonradiochemical DNase I footprinting by capillary electrophoresis. Electrophoresis. 2001;22:1979–1986. doi: 10.1002/1522-2683(200106)22:10<1979::AID-ELPS1979>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.