Abstract
Dengue viruses (DENV) cause significant morbidity and mortality worldwide and are transmitted by the mosquito Aedes aegypti. Mosquitoes become infected after ingesting a viremic bloodmeal, and molecular mechanisms involved in bloodmeal digestion may affect the ability of DENV to infect the midgut. We used RNA interference (RNAi) to silence expression of four midgut serine proteases and assessed the effect of each RNAi phenotype on DENV-2 infectivity of Aedes aegypti. Silencing resulted in significant reductions in protease mRNA levels and correlated with a reduction in activity except in the case of late trypsin. RNA silencing of chymotrypsin, early and late trypsin had no effect on DENV-2 infectivity. However, silencing of 5G1 or the addition of soybean trypsin inhibitor to the infectious bloodmeals significantly increased midgut infection rates. These results suggest that some midgut serine proteases may actually limit DENV-2 infectivity of Ae. aegypti.
INTRODUCTION
Dengue viruses (DENV) serotypes 1−4 have re-emerged during the last 30 years as a serious public health threat in many tropical and sub-tropical regions of the world. Currently, 50−100 million cases of dengue fever occur worldwide with an estimated three billion people at risk.1,2 The primary vector for DENV is Aedes aegypti, which is highly anthropophilic and endophagic.3,4 DENVs infect Ae. aegypti when the mosquito imbibes a bloodmeal from a viremic host.
Bloodmeal digestion in Ae. aegypti occurs in the midgut, which regulates the release of numerous endo- and exoproteolytic enzymes. Trypsins account for the majority of bloodmeal digestion in mosquitoes and are expressed in a biphasic manner.5 Early trypsin (ET, GenBank accession no. X64362), the principal enzyme during the early phase of digestion, is immediately translated from a pool of mRNAs, reaches maximal concentrations and activity by 3 hours post bloodmeal (hpbm), and then precipitously declines.6,7 There are two trypsins, late trypsin (LT, GenBank accession no. M77814) and 5G1 (GenBank accession no. X64363), associated with late phase digestion. LT and 5G1 are initially detected at 6−8 hpbm and reach peak concentrations by 24 hpbm; these two trypsins are thought to account for the majority of endoproteolytic activity during digestion.8–11 Furthermore, a chymotrypsin (CHYMO, GenBank accession no. U56423) enzyme has been characterized from Ae. aegypti. CHYMO is transcribed prior to feeding and immediately translated following bloodmeal uptake. The CHYMO mRNA, protein, and protease activity levels remain high through the duration of digestion.12 The significance of these enzymes in Ae. aegypti digestion is well documented, but little is known about their effect on vector competence for DENV.13–15
During the initial stages of infection of an arthropod, arboviruses infect and replicate in midgut epithelial cells. Others have shown that enteric viruses of vertebrates, such as rotaviruses, use the proteolytic enzymes of the gut for viral enhancement of infection.16,17 Similarly, previous studies have found evidence suggesting that the arboviruses La Crosse virus (LACV; Bunyaviridae) and blue tongue virus (BTV; Reoviridae) use vector midgut proteases for proteolytic processing to increase viral infectivity.18,19 Studies in Ae. aegypti suggest that midgut serine proteases condition vector competence for DENV-2. Genetic mapping experiments have identified multiple quantitative trait loci (QTL) that control Ae. aegypti vector competence at the stage of midgut infections for DENV-2. Three of these QTL are defined by the linkage map marker genes early trypsin and late (abundant) trypsin positioned at 22 and 32 cM on chromosome II, respectively, and 5G1 (late trypsin) at position 58 cM on chromosome III.20,21 Furthermore, in vivo studies, in which soybean trypsin inhibitor (STI) was fed in the presence of an infectious DENV-2 bloodmeal, demonstrated that this treatment resulted in suppression of late trypsin protein accumulation, reduced DENV-2 midgut titers, and delayed viral dissemination from the midgut to secondary tissues.22
In light of these findings, we hypothesized that midgut serine proteases individually and collectively enhance DENV-2 infectivity of Ae. aegypti and that by specific silencing of these proteases through RNA interference (RNAi), we should be able to reduce or block DENV-2 infection of the mosquito. Initially, we characterized the involvement of ET and LT during the course of a DENV-2 infection. Subsequently, this analysis was expanded to include 5G1 and CHYMO. We evaluated the effect of RNAi silencing of each of the gene products on the midgut infection (MIR) and dissemination rates (DR) of DENV-2 in Ae. aegypti. These data were then corroborated with trypsin inhibition experiments using exogenous trypsin inhibitors.
MATERIALS AND METHODS
Aedes aegypti rearing
The mosquitoes used for these experiments were obtained from our colonized Rexville D strain (RexD). They were maintained at a constant temperature of 27°C with a relative humidity of 80% and a 14:10 light: dark photoperiod.
Dengue virus Type 2 production
C6/36 cells were infected at a multiplicity of infection (MOI) of 0.01 with DENV-2 Jamaica 1409 high passage (JAM1409hp). Subsequently, the medium was replaced 7 days post infection (dpi), cells and supernatant were harvested 4 days later and used for infectious bloodmeals.23
RNA extraction and cDNA synthesis
Whole body RNA extractions were performed on pools of five unfed and five 24 hpbm fed female mosquitoes using TRIzol according to manufacturer's protocol (Invitrogen, Carlsbad, CA). The RNA was subsequently used for production of a cDNA pool with an oligo-d(T) primer and SuperScript II reverse transcriptase (Invitrogen).
Double stranded RNA (dsRNA) synthesis
The genes of interest (GOI) were PCR amplified using gene–specific primers designed to amplify ∼500 bp fragments from the 3′ region. Each primer set included a T7 promoter sequence in both the forward and reverse primers (Table 1). Products were amplified using the following settings: 95°C for 5 minutes, 30 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 45 seconds followed by a final 10 minute extension step at 72°C. PCR products were amplified with a 2 X Master Mix containing Taq polymerase (Promega, Madison, WI). The PCR fragments were purified and subsequently used for dsRNA production.
Table 1.
Primers used for dsRNA synthesis and Q-RT-PCR*
| Primer Name | 5′-Sequence-3′ | Position |
|---|---|---|
| T7-Bgal F | TAA TAC GAC TCA CTA TAG GGG TCG CCA GCG GCA CCG CGC GCC TTT C | |
| T7-Bgal R | TAA TAC GAC TCA CTA TAG GGC CGG TAG CCA GCG CGG ATC ATC GG | |
| T7-ET F | TAA TAC GAC TCA CTA TAG GGG CAT CAT AGT GAA AGT CAA ATC GG | Nucleotides 271−295 |
| T7-ET R | TAA TAC GAC TCA CTA TAG GGC TCG GAA ACC TCT CGG ATC CAT TG | Nucleotides 736−759 |
| T7-LT F | TAA TAC GAC TCA CTA TAG GGC ACC GCC ACG AGA AGT ACA ACC CAC | Nucleotides 307−331 |
| T7-LT R | TAA TAC GAC TCA CTA TAG GGC CCT GCT CAC AGT CCA GTC TTC TGC | Nucleotides 756−774 |
| T7−5G1 F | TAA TAC GAC TCA CTA TAG GGC GAA ATA CGA TGA AGT TAC CAC CGA AC | Nucleotides 345−369 |
| T7−5G1 R | TAA TAC GAC TCA CTA TAG GGA ATC ATT TCA TTT AAG ACA TGC AGT TC | Nucleotides 817−843 |
| T7-CHYMO F | TAA TAC GAC TCA CTA TAG GGA CAT CGT CCA CGA GGA CTA TCA AGG AGG | Nucleotides 324−350 |
| T7-CHYMO R | TAA TAC GAC TCA CTA TAG GGA CGA CGT AAT GGG AAA CTC CGG CAA ACA C | Nucleotides 751−778 |
| Actin QPCR F | CGC TCG TTG TCG ACA ATG G | Nucleotides 249−261 |
| Actin QPCR R | CAT ACC GAC CAT CAC ACC C | Nucleotides 352−370 |
| ET QPCR F | CCC AAA GCC AAC CAA CCT | Nucleotides −61 5′ NCR |
| ET QPCR R | CGA CCC TCC GCA GAA ATG | Nucleotides 184−201 |
| LT QPCR F | GTT CAC TTC AAC GGT GGT TTT C | Nucleotides 3−24 |
| LT QPCR R | AGA ACT TGG AAT GGG AAC TGA C | Nucleotides 104−125 |
| 5G1 QPCR F | CTT CCC ACT TCT GTG GAG GAT C | Nucleotides 185−206 |
| 5G1 QPCR R | GAT GGC GGT TGA CCT TCT TAA C | Nucleotides 322−343 |
| CHYMO QPCR F | GGT AGC TTT CCT GCT CGT TG | Nucleotides 18−37 |
| CHYMO QPCR R | GTA TTG GAG TAT GCG GTC TTG | Nucleotides 222−242 |
Underlined sequences correspond with the T7 promoter sequence. T7 primer sets were used for PCR amplification of genes used for dsRNA synthesis. The Q-RT-PCR primer sets were designed to amplify ∼150 bp fragment from each of the genes so as to not overlap with the regions used for dsRNA silencing of the genes.
dsRNA = double stranded RNA; Q-RT-PCR = quantitative reverse transcriptase polymerse chain reaction.
The dsRNA molecules were synthesized using the T7 Megascript kit according to the manufacturer's protocol (Ambion, Austin, TX). Samples were resuspended in 50 μL of phosphate-buffered saline (PBS), quantified and brought to a final concentration of 1 μg/μL in PBS. A fragment of the Escherichia coli β-galatosidase (B-gal) gene was used as the template to produce the control dsRNA.24
dsRNA injection and artificial bloodfeeds
Adult female mosquitoes were cold-anesthetized 4−5 days post emergence and intrathoracically injected with ∼500 ng of dsRNA. Following a three day recovery period, mosquitoes were given an artificial bloodmeal containing a 1:1 ratio of JAM1409hp cell culture medium and defibrinated sheep blood. The infectious bloodmeal titers ranged from 8 × 104 to 5.33 × 106 pfu/mL for the dsRNA injection experiments and 5.33 × 104 to 1.4 × 106 pfu/mL for trypsin inhibitor experiments. After feeding, mosquitoes were cold-anesthetized and five unfed mosquitoes were collected and placed at −80°C for analysis of ET and CHYMO transcript levels. At 24 hpbm five mosquitoes were collected and placed at −80°C for analysis of LT, 5G1, and CHYMO transcript levels. Seven dpi midguts from each dsRNA treatment group were collected and triturated. The midgut homogenates were filtered through a 0.2 μm pore size membrane syringe filter. Samples were subsequently titrated on LLC-MK2 cells by plaque assay. Finally, at 14 dpi paired midguts and heads were collected from each group for analysis by indirect immunofluorescence assay (IFA) to determine dissemination rates (DR). These experiments were completed in triplicate.
Quantitative reverse transcriptase polymerase chain reactions (Q-RT-PCR)
Total RNA was extracted from the five unfed and five 24 hpbm mosquitoes from each experimental group/replicate. RNA was resuspended in 50 μL of nuclease free H2O and quantified. Individual RNA concentrations were normalized within experimental replicates and stored at −80°C until Q-RT-PCR analysis was performed.
The Q-RT-PCR reactions were completed using an Opticon 2 Real-Time Thermocycler (Bio-Rad, Hercules, CA). Primers were designed to amplify ∼150 bp fragments from the 5′ of the cDNA (Table 1). Amplification products were chosen so as to not overlap with the dsRNA region used for silencing. Ae. aegypti actin (GenBank accession no. XM001655126) was chosen as the normalization control because it exhibited similar amplification efficiency to the target mRNAs, and because it did not fluctuate in the mosquito between experimental and control dsRNA treatments. The Qiagen SYBR Green Quantitect Q-RT-PCR kit was used for analysis (Qiagen, Valencia, CA). Each sample was analyzed in duplicate with control and GOI primer sets. Reactions were run according to manufacturer's protocols, except reaction volumes were reduced to 20 μL. The reaction parameters were 30 minutes 50°C RT reaction followed by 15 minutes at 95°C, and a 40 cycle PCR reaction with the following settings: 95°C for 15 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, and the plate was read at 76°C for 1 second. A final 10 minute extension period at 72°C was applied followed by melting curve analysis being read every 0.2°C/seconds between 70−95°C.
Data from the Q-RT-PCR reaction were analyzed using Opticon 3 software and the Comparative Ct analysis method (Bio-Rad). Melting curves were assessed to confirm sole amplification of target mRNAs and threshold values were set at 0.02. The ΔC(t) values were determined for each sample by subtracting the average actin control value from the average experimental value. These values were used to determine the statistical significance between the B-gal control group and experimental groups. The ΔΔC(t) values were then determined by subtracting the average ΔC(t) value of the B-gal group from the average ΔC(t) value of the experimental group. From this a normalized target value was determined using the equation 2−ΔΔCt and converted to percent decrease.
Enzymatic activity assays
Trypsin and chymotrypsin activity assays were performed using the colorimetric substrates Nα-benzoyl-D,L-arginine 4-nitroanilide hydrochloride (DL-BAPNA) and N-succinyl-Ala-Ala-Pro-Phe-P-nitroanilide (Suc-AAPF-pNA), respectively (Sigma-Aldrich, St. Louis, MO). These reagents were resuspended at a concentration of 200 mM in dimethyl sulfoxide (DMSO).
Three days after injection, mosquitoes were offered an artificial bloodmeal containing either 50% FBS, 40% PBS, 1 mM ATP, and 2 mg of phenol red or defibrinated sheep blood. The FBS meals were provided to experimental groups to be analyzed for early phase activity because hemoglobin in the blood routinely interfered with the absorbance readings. This did not seem to be a problem at the 24-hour time point, so defibrinated blood was provided to those groups to be analyzed for late phase protease activity.
For analysis of both early and late phase protease activity, 10 intact engorged midguts were collected either 2 hours post feeding (hpf) or 24 hpbm. One midgut equivalent was mixed with activity buffer (50 mM Tris-HCl and 10 mM CaCl2, pH 7.0) and the respective colorimetric substrate and incubated at 37°C for 30 minutes. Analysis of late phase proteolytic activity was performed on 1/10th of a midgut and reactions were incubated for 5 min at 37°C. Absorbance values were read at 405 nm. Each sample was tested in triplicate and each experiment was performed three times. Activity is reported as the percentage of activity relative to the B-gal dsRNA injected control groups.
Trypsin inhibitors
The trypsin inhibitors used for these experiments were soybean trypsin inhibitor (STI) (Fluka Biochemika) and a synthetic serine protease inhibitor 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) (Sigma-Aldrich). Each reagent was brought to a concentration of 1 mM in PBS and diluted to 100 μM in the bloodmeals. The control group contained an equal volume of PBS alone. Trypsin activity, MIR, and DR were evaluated in triplicate.
Indirect immunofluorescence assays (IFA)
The IFAs were performed according to the protocol outlined in Bennett and others.25 Briefly, dissected midguts and heads were fixed in either 4% paraformaldehyde or acetone, respectively. Midguts were washed and stained in PBS + 0.1% Triton X100, and PBS was used for the heads. Initially, samples were incubated with the flavivirus E-glycoprotein specific murine monoclonal antibody, 3H5. The samples were washed and the secondary stain applied, which included biotinylated sheep anti-mouse antibody (Amersham, Arlington Heights, IL) and 0.005% Evans blue counterstain. Finally, samples were developed with streptavidin-fluorescein (Amersham) and visualized for fluorescence.
Statistical analysis
Statistical analyses were carried out using SAS 9.1 (SAS Institute Inc., Cary, NC). Transcript and activity levels were analyzed using analysis of variance (ANOVA) with a randomized block design and a Fisher's exact test was used to analyze infection rates.
RESULTS
Suppression of mRNAs and enzymatic activity
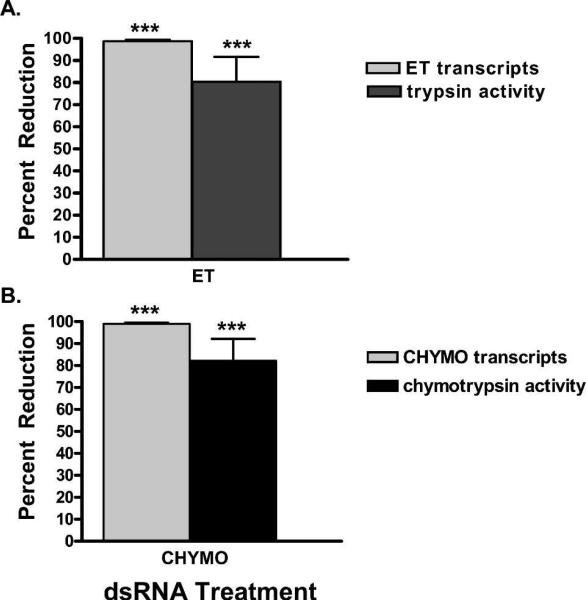
dsRNA silencing of the four target midgut serine proteases was assayed through reduction in mRNA transcript levels as determined by Q-RT-PCR and protease–specific activity assays. ET transcript levels were decreased by 98.7% (P < 0.0001), generating an 82% reduction in early phase tryptic activity compared with the dsRNA B-gal injected control (P < 0.0021) (Figure 1A). Furthermore, reduction of ET transcript levels had no effect on LT transcript levels, consistent with results previously reported (Figure 2A).26 Suppression of ET had no effect on late phase tryptic activity (data not shown).
Figure 1.
The effects of double stranded RNA (dsRNA) silencing on early trypsin (ET) and chymotrypsin (CHYMO) transcript levels and enzymatic activity. Percent reduction in respective transcript levels as determined by quantitative reverse transcriptase polymerse chain reaction (Q-RT-PCR) on unfed mosquitoes and percent enzymatic activity in mosquitoes 2 hpf. A, ET transcript levels and early phase tryptic activity after injection with ET dsRNA. B, CHYMO transcript levels and early phase chymotryptic activity after injection with CHYMO dsRNA. The quantitative levels of transcripts or enzymatic activity reduction are plotted relative to the B-gal control group. (*) indicate groups significantly different from the B-gal injected controls.
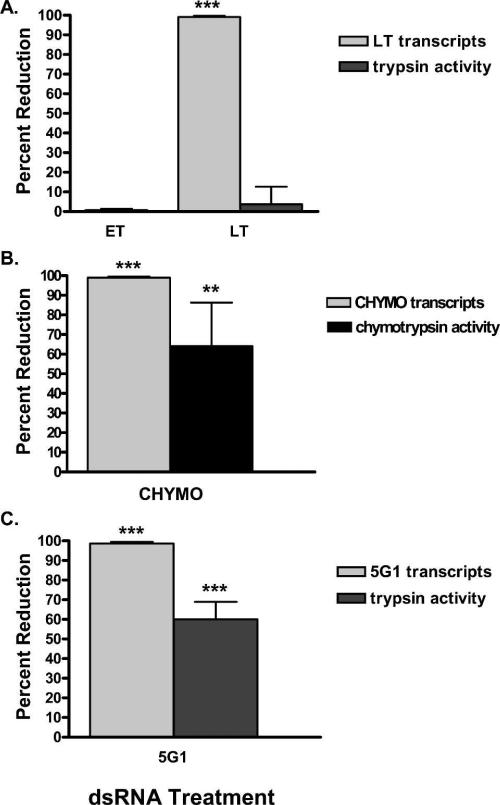
Figure 2.
The effects of double stranded RNA (dsRNA) silencing on late trypsin (LT), 5G1, and chymotrypsin (CHYMO) transcript levels and enzymatic activity 24 hpbm. Percent reduction in respective transcript levels as determined by quantitative reverse transcriptase polymerse chain reaction (Q-RT-PCR) and percent enzymatic activity in mosquitoes 24 hpbm. A, LT transcript levels and late phase tryptic activity after injection with either early trypsin (ET) or LT dsRNA. B, 5G1 transcript levels and late phase tryptic activity after injection with 5G1. C, CHYMO transcript levels and late phase chymotryptic activity after injection with CHYMO dsRNA. The quantitative levels of transcripts or enzymatic activity reduction are plotted relative to the B-gal control group. (*) indicate statistically significant differences compared with the B-gal control group.
A significant reduction in LT transcript levels was recorded in the LT (99.4%, P < 0.001) treatment group (Figure 2A). Despite near complete ablation of LT transcript levels in mosquitoes 24 hpbm we observed no effect on late phase tryptic activity (Figure 2A).
The effect of RNAi silencing of CHYMO transcript levels was measured in unfed and 24 hpbm mosquitoes. A reduction in CHYMO transcript levels of 99.1% (P < 0.0007) in unfed mosquitoes corresponded to an 83% decrease in chymotrypsin activity during early phase digestion (P < 0.0013) (Figure 1B). At 24 hpbm, during late phase digestion, CHYMO transcripts were depleted by 99.2% (P < 0.0015), which resulted in a 69% reduction in late phase chymotryptic activity compared with controls (P < 0.0117) (Figures 2B).
Finally, dsRNA targeting of 5G1 mRNA correlated with a 98.9% reduction (P < 0.0038) in transcript levels in mosquitoes 24 hpbm (Figure 2C). In contrast to LT, silencing of 5G1 mRNA correlated with > 50% (P < 0.0001) reduction in late phase tryptic activity (Figure 2C).
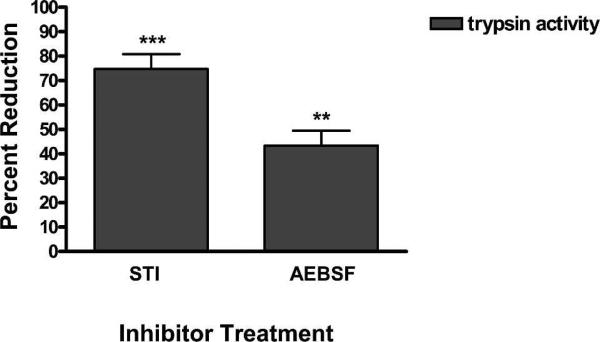
To supplement the above RNAi data, early and late phase tryptic activity was evaluated following the spiking of artificial bloodmeals with two different exogenous trypsin inhibitors. Trypsin activity was significantly reduced by > 70% in the presence of STI (P < 0.0004) and by > 40% with AEBSF (P < 0.0064) during the first two hours of digestion (Figure 3), but by 24 hpbm the inhibitory effects had become negligible (data not shown).
Figure 3.
The effects of trypsin inhibitors on early phase tryptic digestion in Ae. aegypti. Mosquitoes were fed either soybean trysin inhibitor (STI), 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), or an equal volume of phosphate-buffered saline (PBS). Mosquitoes were evaluated for early phase tryptic activity 2 hpf. Values are reported as percent reduction in enzymatic activity compared with the PBS fed control group. (*) indicate a significant reduction in tryptic activity.
Midgut infection and dissemination rates
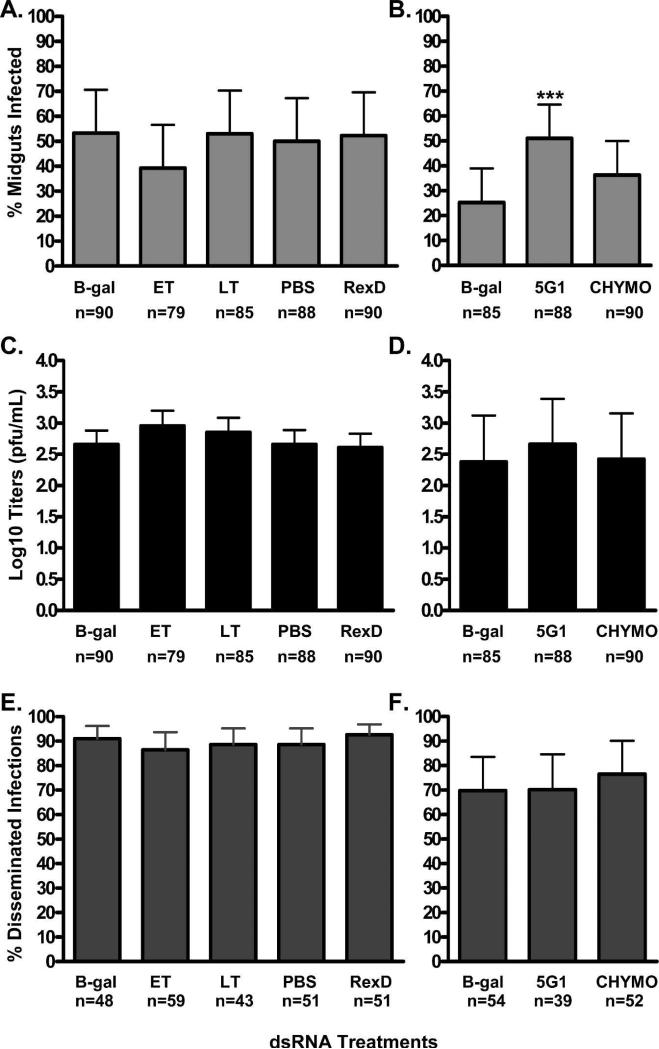
After confirmation of transcript suppression, MIR and DR were assessed for each of the experimental groups. Suppression of ET or LT had no effect on MIR or the intensity of infection as determined by plaque titration (Figure 4A and 4C, respectively). These experiments were then expanded to include 5G1 and CHYMO. We observed that suppression of 5G1 resulted in significantly higher MIR (P < 0.0005), although this did not correlate to an increase in infection intensities. Suppression of CYHMO had no significant effect on DENV-2 MIR and titers (Figure 4B and 4D, respectively). Dissemination rates were unaffected by suppression of any of the targets (Figure 4E and 4F).
Figure 4.
The effects of double stranded RNA (dsRNA) suppression of midgut serine proteases on DENV-2 infectivity. A and B, Midgut infection rates 7 dpi as determined by plaque titrations, C and D, titers of DENV-2 in midguts 7 dpi and E and F, DENV-2 dissemination rates 14 dpi. B-gal values can be found in duplicate because experiments on the right were performed at a later time point compared with the corresponding graph to the left. Sample size values under (E and F), correspond to the number of indirect immunofluorescence assays (IFA) positive midguts per each group analyzed. (*) indicate a significant difference compared with the B-gal control as determined by Fisher's exact test. The phosphate-buffered saline (PBS) control groups were injected with an equal volume of PBS alone and the RexD control groups were un-injected.
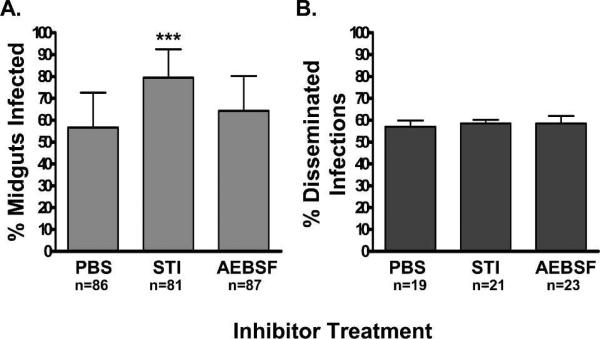
The effects of the exogenously-supplied trypsin inhibitors on DENV-2 infection rates were studied to complement the RNAi silencing results. Furthermore, it allowed us to repeat previous experiments in which STI was used and to expand upon the repertoire of inhibitors tested. Co-feeding DENV-2 with STI (P < 0.0002) significantly increased MIR, while AEBSF (P < 0.3525) had no effect, as determined by IFA (Figure 5A). There was no effect on DR for either group compared with the PBS fed control group (Figure 5B).
Figure 5.
The effects of trypsin inhibitors on DENV-2 infectivity. A, Midgut infection rates of DENV-2 7 dpi and B, dissemination rates 14 dpi as determined by indirect immunofluorescence assays (IFA). Sample size values under (B) correspond to the number of IFA positive midguts per each group analyzed. (*) indicate that the experimental groups were significantly different than the PBS control group by Fisher's exact test. The PBS control groups were injected with an equal volume of phosphate-buffered saline (PBS) alone and the RexD control groups were un-injected.
DISCUSSION
Mosquitoes acquire an arbovirus infection through acquisition of a bloodmeal from an infected vertebrate host. Virus in the bloodmeal encounters the highly proteolytic environment of the midgut lumen, which could lead to virus degradation. Although evidence exists that arboviral infections of the vector occur within the first hour post bloodfeeding before peritrophic matrix formation and accumulation of digestive enzymes, others have found that viable virions can remain in the midgut for hours.27,28 For example, St. Louis encephalitis virus (SLEV), another flavivirus, can persist in the midgut of Culex pipiens for at least 8 hpbm before infecting the midgut epithelium.28 In light of these data, midgut infection with certain arboviruses may be a continual process that occurs over a period of hours post-ingestion. To this end, arboviruses may have had to adapt to or potentially take advantage of this proteolytic environment. The involvement of midgut serine proteases in arbovirus infectivity of vectors has been previously implicated.18,19,22 The objective of this study was to determine if specific midgut serine proteases are required for DENV-2 to efficiently infect Aedes aegypti. Our results suggest that late phase tryptic activity in the midgut associated with 5G1 activity actually reduces midgut infection by DENV-2. The other serine proteases tested had no apparent affect on DENV-2 midgut infection or dissemination. In addition, the previously implicated trypsin-like enzyme (LT) may lack trypsin activity based on the dsRNA experiments and may not be a trypsin serine protease (Figure 2A), however, this needs to be tested directly using recombinant LT protein in a trypsin activity assay.
The use of RNAi allowed for highly specific analysis of each of the four identified midgut serine proteases. Suppression levels for each of the genes were determined at the time points corresponding to their peak levels of expression as previously reported.6,7,10–12 The potent and specific suppression of ET had no effect on LT transcript levels or late phase tryptic activity, validating a study by Lu and others that challenged previous data suggesting a link between ET activity and LT transcription (Figure 2A).26 Although early phase tryptic activity was only reduced by 82% in these experiments (Figure 1A), it could be argued that the remaining 18% of ET activity was enough to activate LT transcription. However, addition of di-isopropyl-fluorophosphate (a potent serine protease inhibitor) during trypsin activity assays of dsRNA ET treated mosquitoes did not further suppress tryptic activity.26 These results suggested that dsRNA silencing of ET was sufficient to reduce almost all early phase serine protease hydrolysis of DL-BAPNA and the residual trypsin activity may be accounted for by non-serine proteases capable of hydrolyzing the substrate.
The levels of LT suppression and late phase tryptic activity were analyzed by Q-RT-PCR and DL-BAPNA assays, respectively. Surprisingly, despite the significant reduction in LT transcript levels as recorded by Q-RT-PCR there was no reduction in late phase tryptic activity (Figure 2A). These results imply that either redundancy exists within late phase tryptic enzymes thereby masking the effects of specific LT suppression or LT is not a trypsin. Our data support the latter explanation. The first explanation is problematic because suppression of 5G1, another late phase tryptic enzyme, resulted in a 60% reduction in late phase tryptic activity (Figure 2C). Furthermore, alignment data showed that LT lacks the critical Asp 189 residue commonly associated with trypsins.11 The original authors concluded that LT may be a unique variant of the classic trypsin archetype, but our data suggest otherwise.10 Further studies are being conducted to identify the function of this protein.
As previously stated, suppression of 5G1 coincided with a significant decrease in transcript levels and late phase tryptic activity. However, there was still 40% activity compared with controls, suggesting that there may be other active trypsins during late phase digestion, which remain to be identified (Figure 2C). Likewise, suppression of CHYMO resulted in a significant reduction of early and late phase chymotryptic activity (Figure 1B and 2B), although because activity was not ablated, we cannot rule out the possibility that other chymotryptic enzymes may be present in the midgut.
The presence of 100 μM STI or AEBSF in the bloodmeal significantly reduced early phase tryptic activity (Figure 3). Others have demonstrated that each of these compounds could maintain a level of 50% suppression for up to 18 hpbm.26 However, our data showed that by 24 hpbm the inhibitory effects of STI and AEBSF had become negligible.
QTL mapping data suggests that several regions on chromosome II and III that include the linkage map marker genes, early trypsin, late (abundant) trypsin and 5G1 (late trypsin), are associated with midgut infection rates with DENV-2.20,21 It is important to note that while these QTLs are identified by these linkage map genes, there is low resolution of base pairs/centimorgan (cM) in the Ae. aegypti genome (1.0−3.4 Mbp/ cM), and the QTLs likely encode hundreds of genes that could also influence midgut infection rates.29 Similarly, these QTLs are only associated with midgut infection by DENV-2, but they do not predict whether the associations are negative or positive. We found that suppression of ET and LT resulted in no discernible effects on the ability of DENV-2 to infect the midgut or disseminate to secondary tissues (Figure 4A). However, these results may be a reflection of the low levels of ET accumulation (maximal concentration of 300 ng/midgut at 3 hpbm compared with LT and presumably 5G1, which achieve sub-maximal levels ten-times that of ET by 8 hpbm) and the lack of tryptic activity associated with LT.14 Comparatively, when we suppressed the third linkage map marker gene, 5G1 (late trypsin), we found a significant increase in midgut infection rates (Figure 4B). Similarly, general suppression of trypsin activity via STI resulted in enhanced midgut infections by DENV-2 (Figure 5A). Although the same was not true for AEBSF, we found that this chemical exhibited a weaker effect on early phase tryptic activity (42% reduction versus 78% reduction by STI, Figure 3) and thereby a weaker effect on early 5G1 activity 6−8 hpbm. This difference in trypsin inhibition may account for the differences observed in MIR between the inhibitors.
As a result of the inherent variability associated with having to grow DENV-2 fresh to infect Ae. Aegypti, there was considerable differences in bloodmeal titers and consequently MIRs between experimental replicates. Yet this has no effect within replicates because experimental and control groups within each replicate are offered the same bloodmeal. This is evident in the B-gal control groups MIR between two experiments (represented in Figures 4A and 4B). In experiment 1 (Figure 4A) bloodmeal titers were consistently high (∼1 × 106) resulting in B-gal MIR of 53%, whereas experiment 2 (Figure 4B) had highly variable bloodmeal titers across replicates resulting in B-gal MIR of 26%. The effects of the variability in bloodmeal titers of the three replicates in experiment 2 (5.33 × 106, 8.67 × 105 and 8.67 × 104) are evident by the correspondingly lower MIR for each experimental group; however, in each replicate 5G1 silenced mosquitoes consistently had higher MIR than the control B-gal group. These results suggest that silencing of 5G1 increases MIR DENV-2 in Ae. aegypti irrespective of bloodmeal titers.
Our results for the effects of STI on DENV-2 infectivity differ markedly from a previous report by Molina-Cruz and others, who observed a significant decrease in the viral RNA and E-glycoprotein accumulation in midguts 7 days after feeding in the presence of STI. Furthermore, they noticed delayed dissemination kinetics compared with controls.22 The discrepancy observed between these and the current experiments might be explained by the source of the STI used in the experiments. A recent report by Lu and others demonstrated that the STI purchased from Sigma and used in previous experiments designed to investigate the signal transduction cascade of LT may have contained a contaminate that altered Ae. aegypti physiology, resulting in reduced LT transcription, peritrophic matrix 1 (PM1) formation and oviposition rates, and expulsion of the bloodmeal.26 These same physiologic permutations were used as a determinant of trypsin inhibition by Molina-Cruz and others.22 Conversely, we used STI purchased from Fluka Biochemika and demonstrated trypsin inhibition using the DL-BAPNA assay. Furthermore, we observed that inhibitor-fed mosquitoes did not expel their bloodmeal or have a reduction in oviposition rates (data not shown).
Overall, these results suggest that serine proteases do not proteolytically activate the virus, but rather that proteolytic activity, specifically tryptic activity, within the midgut actually limits virus infectivity. Support for this conclusion can be found in a study using LACV, which found that pre-treatment of LACV with pronase prior to infection of cell culture or mosquitoes significantly reduced viral infectivity.30 We speculate that during the early periods of 5G1 expression, 6−8 hpbm, virus particles remaining in the bloodmeal are vulnerable to degradation by 5G1. This degradation would lead to a reduction in the number of viable virions able to eventually infect the midgut. These studies highlight the fact that each virus/mosquito system is unique, and the rate at which viruses infect the midgut is dependent upon the virus and mosquito species. In an effort to better understand the underlying physical barriers associated with Ae. aegypti vector competence, future studies should investigate the kinetics of DENV-2 infection and the role of 5G1 heterogeneity and expression levels in different Ae. aegypti populations.
The results presented in this paper more completely describe the specific contributions of serine proteases, specifically LT and 5G1, in Ae. aegypti bloodmeal digestion. Perhaps the more important outcome of this study is that it confirmed that proteins within the midgut directly interact with the virus and that novel control strategies (ie., transmission blocking vaccines or transgenic mosquitoes) targeting these proteins are a realistic control option once a suitable target is identified. The results also emphasize our poor understanding of the basic physiology of mosquito digestion and Ae. aegypti vector competence and signify the importance of future research in these areas.
Acknowledgments
We thank Erik Powers for his assistance with mosquito rearing, dissections, and plaque titrations, Cynthia Meredith for maintaining the insectaries, and Dr. James Zumbrunnen for his help with the statistical analysis.
Financial support: These studies were funded by the National Institute of Health (AI-25489) and the Fellowship Training Program (T01/CCT822307) provided by the Centers for Disease Control and Prevention.
REFERENCES
- 1.Gubler DJ. The global resurgence of arboviral diseases. Trans R Soc Trop Med Hyg. 1996;90:449–451. doi: 10.1016/s0035-9203(96)90286-2. [DOI] [PubMed] [Google Scholar]
- 2.Monath T. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson M. Aedes aegypti: Biology and Ecology. Pan American Health Organization; Washington DC: 1986. pp. 1–59. [Google Scholar]
- 4.Tinker M. Larval habitat of Aedes aegypti (L.) in the United States. Mosq News. 1964;24:426–432. [Google Scholar]
- 5.Felix CR, Betschart B, Billingsley PF, Freyvogel TA. Post-feeding induction of trypsin in the midgut of Aedes aegypti L (Diptera, Culicidae) is separable into 2 cellular phases. Insect Biochem. 1991;21:197–203. [Google Scholar]
- 6.Noriega FG, Wang XY, Pennington JE, Barillas-Mury CV, Wells MA. Early trypsin, a female-specific midgut protease in Aedes aegypti: isolation, amino-terminal sequence determination, and cloning and sequencing of the gene. Insect Biochem Mol Biol. 1996;26:119–126. doi: 10.1016/0965-1748(95)00068-2. [DOI] [PubMed] [Google Scholar]
- 7.Pennington JE, Noriega FG, Wells MA. The expression of early trypsin in Aedes aegypti. J Cell Biochem. 1995;21A(Suppl):211. [Google Scholar]
- 8.Graf R, Binz H, Briegel H. Monoclonal antibodies as probes for Aedes aegypti trypsin. Insect Biochem. 1988;18:463–470. [Google Scholar]
- 9.Graf R, Raikhel AS, Brown MR, Lea AO, Briegel H. Mosquito trypsin immunocytochemical localization in the midgut of bloodfed Aedes aegypti (L). Cell Tissue Res. 1986;245:19–27. doi: 10.1007/BF00218082. [DOI] [PubMed] [Google Scholar]
- 10.Barillas-Mury C, Graf R, Hagedorn HH, Wells MA. cDNA and deduced amino acid sequence of a blood meal-induced trypsin from the mosquito, Aedes aegypti. Insect Biochem. 1991;21:825–831. [Google Scholar]
- 11.Kalhok SE, Tabak LM, Prosser DE, Brook W, Downe AE, White BN. Isolation, sequencing and characterization of two cDNA clones coding for trypsin-like enzymes from the midgut of Aedes aegypti. Insect Mol Biol. 1993;2:71–79. doi: 10.1111/j.1365-2583.1993.tb00127.x. [DOI] [PubMed] [Google Scholar]
- 12.Jiang QJ, Hall M, Noriega FG, Wells M. cDNA cloning and pattern of expression of an adult, female specific chymotrypsin from Aedes aegypti midgut. Insect Biochem Mol Biol. 1997;27:283–289. doi: 10.1016/s0965-1748(97)00001-5. [DOI] [PubMed] [Google Scholar]
- 13.Black WC, Bennett KE, Gorrochotegui-Escalante N, Barillas-Mury CV, Fernandez-Salas I, Munoz MD, Farfan-Ale JA, Olson KE, Beaty BJ. Flavivirus susceptibility in Aedes aegypti. Arch Med Res. 2002;33:379–388. doi: 10.1016/s0188-4409(02)00373-9. [DOI] [PubMed] [Google Scholar]
- 14.Noriega FG, Wells MA. A molecular view of trypsin synthesis in the midgut of Aedes aegypti. J Insect Physiol. 1999;45:613–620. doi: 10.1016/s0022-1910(99)00052-9. [DOI] [PubMed] [Google Scholar]
- 15.Pennington J, Wells M. The adult midgut: structure and function. In: Marquardt W, Black WC IV, Freier J, Hagerdorn H, Hemingway J, Higgs S, James A, Kondratieff B, Moore C, editors. Biology of Disease Vectors. Elsevier Academic Press; Burlington, MA: 2005. pp. 289–295. [Google Scholar]
- 16.Espejo RT, Lopez S, Arias C. Structural polypeptides of simian rotavirus Sa11 and the effect of trypsin. J Virol. 1981;37:156–160. doi: 10.1128/jvi.37.1.156-160.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estes MK, Graham DY, Mason BB. Proteolytic enhancement of rotavirus infectivity—molecular mechanisms. J Virol. 1981;39:879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig GV, Christensen BM, Yuill TM, Schultz KT. Enzyme processing of La Crosse virus glycoprotein G1: a bunyavirus-vector infection model. Virology. 1989;171:108–113. doi: 10.1016/0042-6822(89)90516-3. [DOI] [PubMed] [Google Scholar]
- 19.Mertens PP, Burroughs JN, Walton A, Wellby MP, Fu H, O'Hara RS, Brookes SM, Mellor PS. Enhanced infectivity of modified bluetongue virus particles for two insect cell lines and for two Culicoides vector species. Virology. 1996;217:582–593. doi: 10.1006/viro.1996.0153. [DOI] [PubMed] [Google Scholar]
- 20.Bennett KE, Flick D, Fleming KH, Jochim R, Beaty BJ, Black WC., IV Quantitative trait loci that control dengue-2 virus dissemination in the mosquito Aedes aegypti. Genetics. 2005;170:185–194. doi: 10.1534/genetics.104.035634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosio CF, Fulton RE, Salasek ML, Beaty BJ, Black WC., IV Quantitative trait loci that control vector competence for dengue-2 virus in the mosquito Aedes aegypti. Genetics. 2000;156:687–698. doi: 10.1093/genetics/156.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molina-Cruz A, Gupta L, Richardson J, Bennett K, Black WC, IV, Barillas-Mury C. Effect of mosquito midgut trypsin activity on dengue-2 virus and dissemination in Aedes aegypti. Am J Trop Med Hyg. 2005;72:631–637. [PubMed] [Google Scholar]
- 23.Pierro DJ, Salazar MI, Beaty BJ, Olson KE. Infectious clone construction of dengue virus type 2, strain Jamaican 1409, and characterization of a conditional E6 mutation. J Gen Virol. 2006;87:2263–2268. doi: 10.1099/vir.0.81958-0. [DOI] [PubMed] [Google Scholar]
- 24.Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, Olson KE. RNA interference acts as a natural antiviral response to O'nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc Natl Acad Sci USA. 2004;101:17240–17245. doi: 10.1073/pnas.0406983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett K, Olson K, Munoz Mde L, Fernandez-Salas I, Farfan-Ale J, Higgs S, Black WC, IV, Beaty B. Variation in vector competence for dengue 2 virus among 24 collections of Aedes aegypti from Mexico and the United States. Am J Trop Med Hyg. 2002;67:85–92. doi: 10.4269/ajtmh.2002.67.85. [DOI] [PubMed] [Google Scholar]
- 26.Lu SJ, Pennington JE, Stonehouse AR, Mobula MM, Wells MA. Reevaluation of the role of early trypsin activity in the transcriptional activation of the late trypsin gene in the mosquito Aedes aegypti. Insect Biochem Mol Biol. 2006;36:336–343. doi: 10.1016/j.ibmb.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Houk EJ, Kramer LD, Hardy JL, Chiles RE. Western equine encephalomyelitis virus - In vivo infection and morphogenesis in mosquito mesenteronal epithelial cells. Virus Res. 1985;2:123–138. doi: 10.1016/0168-1702(85)90243-6. [DOI] [PubMed] [Google Scholar]
- 28.Whitfield SG, Murphy FA, Sudia WD. St. Louis encephalitis virus: An ultrastructural study of infection in a mosquito vector. Virology. 1973;56:70–87. doi: 10.1016/0042-6822(73)90288-2. [DOI] [PubMed] [Google Scholar]
- 29.Brown SE, Severson DW, Smith LA, Knudson DL. Integration of the Aedes aegypti mosquito genetic linkage and physical maps. Genetics. 2001;157:1299–1305. doi: 10.1093/genetics/157.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hacker JK, Volkman LE, Hardy JL. Requirement for the G1-protein of California encephalitis virus in infection in vitro and in vivo. Virology. 1995;206:945–953. doi: 10.1006/viro.1995.1017. [DOI] [PubMed] [Google Scholar]