Abstract
hCLCA2 is frequently downregulated in breast cancer and is a candidate tumor suppressor gene. We show here that the hCLCA2 gene is strongly induced by p53 in response to DNA damage. Adenoviral expression of p53 induces hCLCA2 in a variety of breast cell lines. Further, we find that p53 binds to consensus elements in the hCLCA2 promoter and mutation of these sites abolishes p53-responsiveness and induction by DNA damage. Adenoviral transduction of hCLCA2 into immortalized cells induces p53, CDK inhibitors p21 and p27, and cell cycle arrest by 24 hours, and caspase induction and apoptosis by 40 hours post-infection. Transduction of the malignant tumor cell line BT549 on the other hand does not induce p53, p21, or p27 but instead induces apoptosis directly and more rapidly. Knockout and knockdown studies indicate that growth inhibition and apoptosis are signaled via multiple pathways. Conversely, suppression of hCLCA2 by RNA interference enhances proliferation of MCF10A and reduces sensitivity to doxorubicin. Gene expression profiles indicate that hCLCA2 levels are strongly predictive of tumor cell sensitivity to doxorubicin and other chemotherapeutics. Because certain Cl- channels are proposed to promote apoptosis by reducing intracellular pH, we tested whether, and established that, hCLCA2 enhances Cl- current in breast cancer cells and reduces pH to ∼6.7. These results reveal hCLCA2 as a novel p53-inducible growth inhibitor, explain how its downregulation confers a survival advantage to tumor cells, and suggest both prognostic and therapeutic applications.
Keywords: DNA damage, p53, chloride channel regulator, breast cancer, tumor suppressor, intracellular acidification
Introduction
The evolution of tumor cell from primary cell requires the disabling of a succession of safeguards that enforce the differentiated state (1). One of the first obstacles to be overcome by a hyperplastic cell is the activation of the DNA damage checkpoint in response to telomere erosion (2). Activating this checkpoint in mammary epithelial cells culminates in permanent cell cycle arrest or death (3). DNA damage signaling also figures in arrest at later stages of tumor progression, for example in response to chromosome breaks and chemotherapy with agents such as doxorubicin (4).
DNA damage signals converge on p53, which then activates a battery of genes involved in DNA repair, cell cycle arrest, apoptosis, and other anti-neoplastic functions (5, 6). Loss of p53 is one of the most common mutations in cancer, allowing the cell to escape this safeguard mechanism (7). Consequently, p53 mutation is one of the most powerful prognostic markers in breast cancer (8). The restoration of p53 itself being technically formidable, there is great interest in identifying p53 target genes for therapeutic potential (9, 10).
The Chloride Channel Accessory (CLCA) protein family has four members in humans and at least seven in mouse, with various members expressed throughout epithelia, endothelia, and other cell types (11). The CLCA archetype is an ∼900 amino acid, Type I transmembrane protein that does not closely resemble any other protein type (12). Ectopic expression of CLCA proteins has consistently been shown to produce a novel Cl− current that can be activated by calcium and blocked by typical Cl− channel inhibitors; this led to their designation as chloride channels (13). However, we recently demonstrated that hCLCA2 has only a single, C-terminal transmembrane segment followed by a 16 amino acid cytoplasmic tail, a structure incompatible with function as a channel (14). Similar conclusions have been reached for other CLCA family members, suggesting that they regulate Cl− current rather than forming an ion-conductive pore (15-17).
Ion channels influence cell fate decisions by a number of avenues, e.g., by regulating cell volume, intracellular pH (pHi), concentration of signaling molecules such as calcium, and activity of protein kinases and phosphatases (18). Certain ion channels have been proposed to be either oncogenic or tumor suppressive (18-21). Recent observations suggest that the CLCA family may harbor tumor-suppressor genes. We reported previously that several mouse CLCA genes are transcriptionally downregulated in mammary tumor cell lines and that ectopic re-expression of these genes inhibits tumor cell growth and survival in vitro (22, 23). Downregulation of hCLCA2 itself was observed by in situ hybridization and RT-PCR of both human breast cancer specimens and breast cancer cell lines (24, 25). Methylation of the hCLCA2 promoter region appears at least partly responsible for downregulation; in addition, missense mutations were detected in some tumor specimens (25). A breast cancer cell line stably transfected with hCLCA2 cDNA produced smaller tumors in nude mice than vector-transfected controls, however no effect on in vitro proliferation was observed (24).
Certain members of the CLCA family are stress-inducible genes. For example, we recently determined that two mouse paralogs of hCLCA2 are upregulated during mammary involution and can be induced in vitro by stresses such as cell detachment (22, 23). To gain insight into human breast cancer, we asked whether human CLCA2 was also stress-inducible. We show here that hCLCA2 is potently induced by several DNA damaging agents and that the primary arbiter of stress-response, p53, binds to the hCLCA2 promoter and strongly activates its expression. We find that ectopic expression of hCLCA2 promotes arrest and apoptosis while knockdown has opposite effects, indicating that hCLCA2 is a novel effector of the p53 growth-inhibitory stress response.
Experimental Procedures
Cell Lines and Cell Culture
The human immortalized mammary epithelial cell line MCF-10A and transformed derivative MCF10CA1d (referred to herein as CA1d) were obtained from the Barbara Ann Karmanos Cancer Institute (Detroit, Mich.) and were grown in CM (1:1 DME/F-12), 5% FBS plus hEGF (Invitrogen, 1 ng/ml), insulin (Sigma, 10 μg/ml) and hydrocortisone (Sigma, 0.5 μg/ml) for all experiments. The human cancer cell lines MCF-7, BT-549, HCT116 and low passage HEK293 were grown in NM (DME/10% FBS). The HCT116 cell lines bearing homozygous deletions of p53, p21, or Bax were a generous gift from B. Vogelstein.
Quantification of mRNA in mammary cell lines
RNA was extracted and reverse-transcribed as described (23). Quantitative PCR was performed on an ABI 7500 instrument using the SybrgreenER pre-mix (Invitrogen). Primers for hCLCA2 were 5′-cagctagtctttggattccaggaa-3′ and 5′-tcagggctttgcagagaatgat-3′. Reactions were performed in triplicate and expression was normalized to beta-actin. Bax primers were 5′-tccggggagcagcccagaggc-3′ and 5′-agacacgtaaggaaaacgcatta-3′ and p21 primers were 5′-ggagactctcagggtcgaaa-3′ and 5′-ggattagggcttcctcttgg-3′.
Chromatin immunoprecipitation
For ectopically expressed p53, two 10 cm dishes of confluent CA1D cells were infected with Ad-p53 at an moi of 200 for 24 h. For ChIP of endogenous p53, MCF7 cells were treated with 1uM doxorubicin or DMSO for 24h. Chromatin cross-linking and immunoprecipitation (ChIP) were performed as described (26). Details are available upon request. The hCLCA2 promoter interval -359 to -157 containing the p53 binding consensus was amplified with the primers 5′- cagtccagatatactgatttc-3′ (forward) and 5′- ctgggacctgcctctacaag-3′ (reverse); the negative control lacking a p53 consensus was the interval -2925 to -2553, amplified with 5′- gccaggttgaactacagagtcag-3′ (forward) and 5′- aggctgaccttacagcctga-3′ (reverse). For the p21 promoter, the primers 5′-caccactgagccttcctcac-3′ and 5′-ctgactcccagcacacacactc-3′ were used to amplify the interval -2478 to -2029 containing an established p53 binding site (26).
Western blot analysis
Levels of hCLCA2 were measured by immunoprecipitation and western blotting with affinity-purified antibody TVE20, raised against the peptide TVEPETGDPVTLRLLDDGAG. Cleared lysates were prepared from confluent 10 cm dishes and immunoprecipitation and western analysis were performed as described (14). Primary antibody was detected with donkey anti-rabbit antibody tagged with IR680, then scanned and quantified on an Odyssey infrared detector (Licor). To detect other proteins, cleared lysates or total cell lysates were analyzed, loading 50 μg of protein per lane. Other antibodies were from Cell Signaling except anti-tubulin mAb 6G7 (W. Halfter via the Developmental Studies Hybridoma Bank, University of Iowa). Size marker, Dual color (Bio-Rad).
Adenoviral methods
hCLCA2-Ad-Easy was created by inserting a Not I fragment containing hCLCA2 into pAdTrack. Recombinant adenovirus was then generated according to established protocols, and viral supernatants were titrated on HEK293 cells and on each breast cell line. p53-AdEasy was obtained from Vector Labs. Adherent MCF10A, CA1D, and BT549 cells were typically infected overnight at an MOI of 20.
Luciferase assays
A BAC clone containing the hCLCA2 promoter region (#RK11-268K17, Oakland Research Institute) was used as template for PCR (Prime-Star, Takara). Error-free pGEM-T clones were inserted into pGL4 luciferase vector and co-transfected with Renilla control phRL-TK into 293T cells at a ratio of 25:1 using Lipofectamine2000 (Invitrogen). Plasmids encoding p53 fused to GFP (pC1-p53-GFP, gift of Y. Mo) or empty vector were included. Luciferase assays were performed on cell lysates using the Dual Luciferase kit (Promega) and quantified on a SIRIUS U3.1 luminometer. Data are reported as the mean of two experiments.
Growth curves
Cell proliferation was measured by seeding 12-well or 6-well plates with 100,000 cells per well, three wells per time point, and counting at daily intervals on a Vi-Cell Analyzer. Adenoviral infection was performed at the time of seeding.
Mitochondrial extraction
Cells were washed with cold PBS and lyzed in mitochondrial isolation buffer (MIB) (HEPES/KOH 20mM, KCl 10mM, MgCl2 1.5mM, EDTA 1mM, EGTA/EDTA 1mM, DTT 1mM, Sucrose 250mM) using a dounce homogenizer followed by passage through a 26 gauge needle. Homogenized samples were centrifuged at 1000g for 10 min at 4°C. Supernatants were spun at 10,000g for 20 min and pellets washed with MIB then resuspended in SDS-PAGE sample buffer, heated, and analyzed by SDS-PAGE. Supernatants were again spun at 14000g for 30 min to obtain the pure cytosolic fraction.
Cell synchronization and flow cytometry
For cell cycle analysis CA1d cells were infected with adenoviruses at an m.o.i. of 10. 24h after infection, cells were arrested with hydroxyurea (1 mM for 24h) followed by wash-out and nocodazole (0.2 μg/ml for 12h). Cells synchronized in G2/M were then released into fresh medium and samples harvested at 3h intervals. Cells were trypzinized, washed, and fixed in ethanol. Prior to FACS, cells were treated with RNase A, stained with 50 μg/ml propidium iodide and analyzed on a FACSCaliber instrument.
RNA interference
Expression of hCLCA2 was suppressed by infection with lentiviruses expressing shRNAs (GIPZ, Open Biosystems clones VLHS-181798, -183886 and nonsilencing control RHS4346). Packaging was performed by co-transfection into 293T cells with plasmids pCMV-dR8.74 and pCMV-VSV-G, purchased from Addgene. Cells were infected in the presence of 8 μg/ml polybrene and selected with 3μg/ml puromycin with frequent splitting until all cells were GFP-positive.
Measurement of pHi
BT549 cells were infected with Ad-GFP or Ad-hCLCA2 at an m.o.i. of 20. After 48h, cells were loaded with SNARF-AM (Invitrogen) and pHi was determined by flow cytometry as described by the manufacturer (http://probes.invitrogen.com/media/pis/mp01270.pdf). Details upon request.
Bioinformatics
Gene expression profiles were studied using Oncomine (oncomine.org) and Gene Expression Omnibus (ncbi.nlm.nih.gov/sites/entrez). The hCLCA2 promoter was analyzed using TFSearch (cbrc.jp/research/db/TFSEARCH.html) and TESS (cbil.upenn.edu/tess/).
Results
DNA damaging agents induce hCLCA2
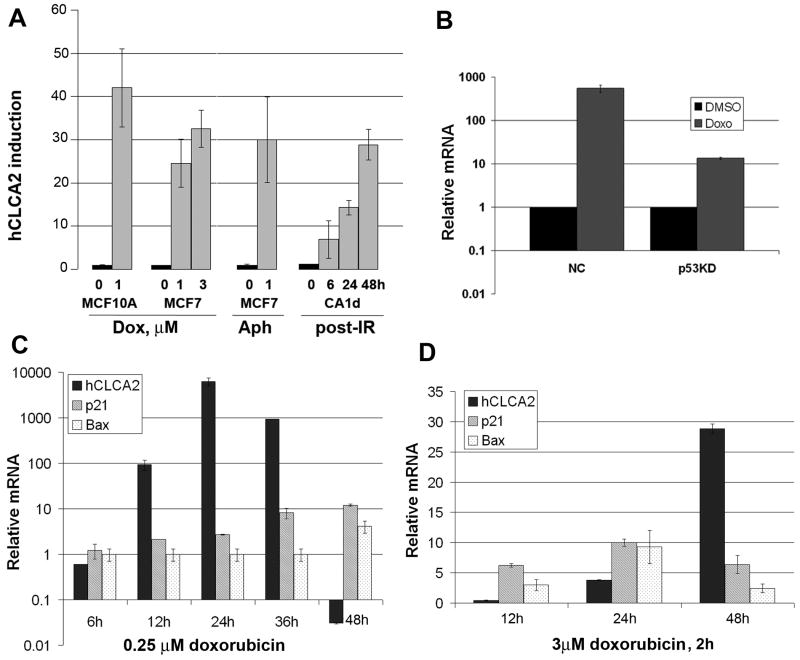
To determine whether hCLCA2 was induced in response to DNA damage, breast cell lines were treated with the topoisomerase inhibitor doxorubicin, the DNA polymerase inhibitor aphidicolin, or ionizing radiation (IR). All cause DNA damage, leading to either senescence, mitotic catastrophe, or apoptosis, depending on dosage and genetic background (4, 27, 28). An acute treatment of MCF10A immortalized cells with doxorubicin increased hCLCA2 mRNA expression by fortyfold (Figure 1A). Similar treatment of the p53-positive breast cancer cell line MCF7 with either doxorubicin or aphidicolin caused a 25-30 fold induction of hCLCA2 expression. Because MCF-7 cells are resistant to IR (4), we instead irradiated CA1d, a tumorigenic derivative of MCF10A, producing a thirtyfold induction of hCLCA2 by 48h post-irradiation (Figure 1A, right).
Figure 1.
Induction of hCLCA2 by chemotherapeutic agents in breast cancer cell lines. A, diverse forms of DNA damage induce hCLCA2. Cells were treated with doxorubicin or 1μg/ml aphidicolin and induction of hCLCA2 was measured by RT-qPCR. For IR, CA1d cells were treated with 8 Gy of ionizing radiation and allowed to recover for the indicated times, followed by RT-qPCR. Expression is relative to untreated cells (0h). B, inhibition of induction by p53 knockdown. MCF7 cells transduced with p53shRNA or non-silencing control were treated with 0.25μM doxorubicin for 24h, followed by RT-qPCR. C-D, kinetics and scale of induction vary with dosage. MCF7 cells were treated with different doses of drug and kinetics of induction were compared for hCLCA2, p21, and Bax by RT-qPCR. For drug treatments, expression is relative to that of the vehicle control, DMSO.
A survey of the cDNA microarray databases Oncomine and GEO using the key words “human CLCA2 and doxorubicin” revealed multiple studies that confirm and extend these observations. For example, hCLCA2 is induced by 45-fold in response to doxorubicin treatment of HT1080 fibrosarcoma cells (29) and 14-fold in Ewing's sarcoma A673 (30), placing it among the most highly induced genes in those studies (Supp. Figure S1). Thus, several forms of DNA damage induce hCLCA2 expression, and the phenomenon is not limited to breast.
Because of the well-established role of p53 in DNA damage response, we knocked down expression of p53 in MCF7 cells and measured induction of hCLCA2 in response to doxorubicin. Induction was reduced by 90% in the knockdown cells, implying that hCLCA2 is a p53 target gene (Figure 1B).
The kinetics of induction of p53 target genes can provide insight into their function. Genes responsible for arrest and repair tend to be induced earlier than those involved in apoptosis. This kinetic difference is mediated by auxiliary transcription factors and the severity of DNA damage (31). We measured the kinetics of hCLCA2 induction relative to p21 and Bax and found that the profile depended on the strength of the insult. At a low dose of doxorubicin, hCLCA2 induction peaked at 24h while p21 and Bax peaked at 48h, and hCLCA2 was no longer expressed (Figure 1C). However, at a higher dose, the kinetics were reversed, with hCLCA2 peaking at 48h and p21 and Bax peaking at 24h (Figure 1D). The scale of induction also differed. At the low dose, hCLCA2 induction was more than 1000-fold, compared to about thirtyfold at the higher dose. These results suggest that hCLCA2 may be involved in cell cycle arrest in response to moderate stress but apoptosis or senescence under extreme stress.
p53 binds to the hCLCA2 promoter and activates it
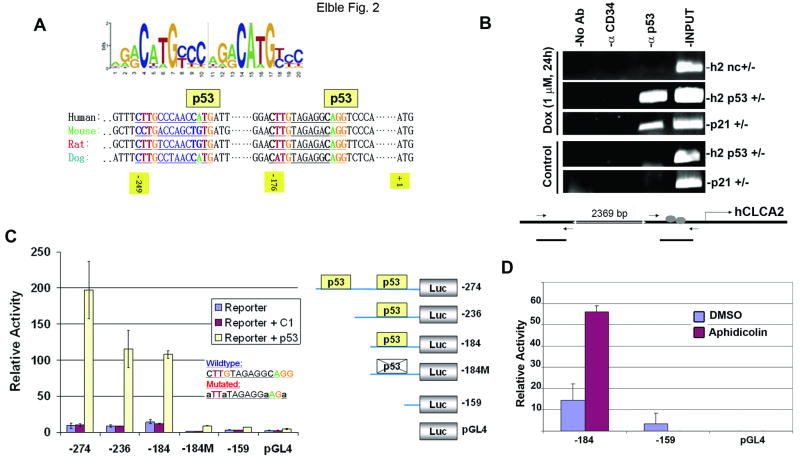
The p53 consensus binding site was recently defined (Figure 2A, top (9, 10)). We found two matches for this sequence within 250bp of the hCLCA2 initiation codon (Figure 2A, bottom). Comparison with the promoter regions of hCLCA2 orthologs from mouse, rat, and dog revealed that these elements are evolutionarily conserved (32). To establish that p53 bound to at least one of these elements in vivo, we performed chromatin immunoprecipitation, or ChIP, analysis. We found that a p53-specific antibody could immunoprecipitate the hCLCA2 promoter segment containing these consensus sites from cells transduced with p53-Ad (Figure S2) or from cells treated with doxorubicin to induce endogenous p53 (Figure 2B). A distal promoter segment that lacked such sites was not bound (Figure 2B). In addition, a high-affinity site from the p21 promoter was also precipitated by the p53 antibody, while an antibody against a cell surface protein, CD34, did not precipitate any of the fragments. Identical results were obtained in two cell lines. Thus, both ectopic and endogenous p53 bind to the hCLCA2 promoter.
Figure 2.
p53 binds to a consensus element in the hCLCA2 promoter. A, top, p53 binding consensus, reprinted from Wei et al. (9) with permission from Elsevier. Lower, matches for the consensus in the hCLCA2 promoter are conserved in orthologs from mouse, rat, and dog. Positions within the human promoter relative to the translation initiation codon are indicated. B, ChIP analysis with anti-p53 antibody 1C12 (Cell Signaling) showing p53 binding to the consensus elements in the hCLCA2 promoter (h2 p53) and to an established binding site from the p21 promoter (p21) but not to an adjacent region in the hCLCA2 promoter (h2 nc). An anti-CD34 mAb (Pharmingen) served as an additional negative control. Input, 10% of the lysate was subjected to PCR directly. Relative positions of the PCR products from the hCLCA2 promoter are depicted in the schematic. C, demonstration that the putative p53 binding sites from the hCLCA2 promoter are responsible for transcriptional activation by p53. Right, PCR products containing these sites or not were fused to a firefly luciferase reporter gene in pGL4. The constructs were transfected into 293T cells alone or along with a plasmid encoding p53 or no insert (C1). A plasmid expressing Renilla luciferase was co-transfected and readings were normalized to this control. Activities are expressed relative to the pGL4 vector control. Promoter segments include the interval from the indicated upstream nucleotide to the A of the translation initiation codon (+1). Inset, mutations introduced into the p53 binding site in the -184M construct are shown. Luc, luciferase. D, p53 role in induction by DNA damage. MCF7 cells were transfected for 8h with constructs bearing or lacking the p53 binding site, treated with aphidicolin for two hours, and lysed 24h later. Bars indicate standard error.
To establish whether p53 binding is functionally relevant, we tested segments of the hCLCA2 promoter for the ability to drive a luciferase reporter gene in response to co-transfection of p53 into 293T cells (Figure 2C). This analysis revealed that only segments containing one of the two p53 consensus sites could drive such expression. A segment containing both consensus sites (-274) produced a two-hundredfold response to p53, while loss of one site (-236) halved the activity. Mutation of the innermost site by deletion or C/G to A substitutions in the consensus abolished p53-responsive expression (-184M). Thus, p53 binding to these elements activates hCLCA2 gene expression. The reporter constructs were similarly p53-responsive in MCF7 cells except that the basal level of hCLCA2 expression was much higher than in 293T, suggesting that basal expression depends on breast-specific transcription factors absent from 293T (data not shown).
To confirm that p53 plays a role in the induction of hCLCA2 by DNA damage that was observed in p53+ MCF7 cells, these cells were transfected with the p53-responsive or -nonresponsive promoter constructs and treated with aphidicolin (Figure 2D). Only the construct bearing an intact p53-binding site showed induction, consistent with a major role for p53 in DNA damage-responsiveness of hCLCA2.
p53 transduction induces hCLCA2
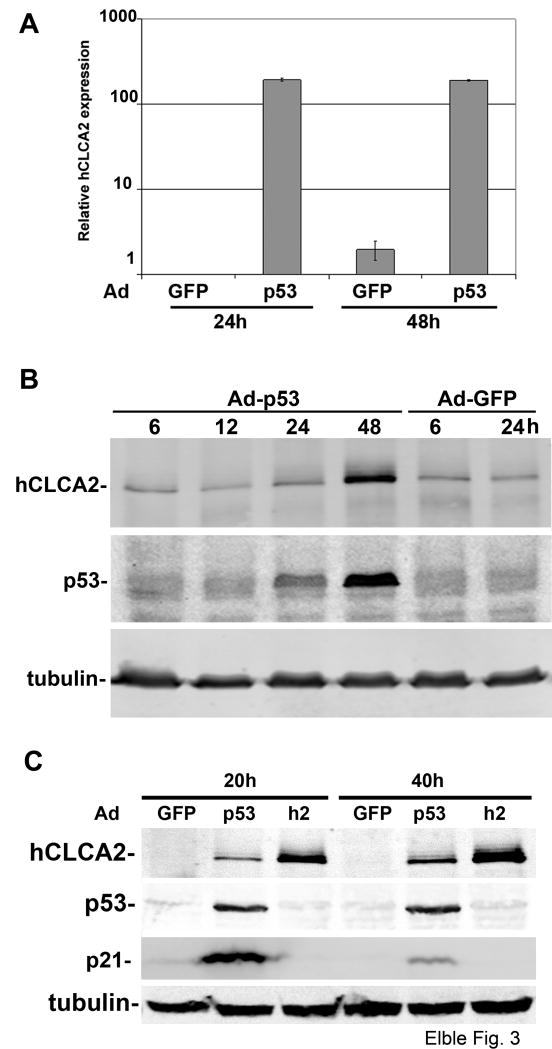
To further confirm that p53 drives expression of hCLCA2 from the natural promoter, we infected several breast cell lines with an adenovirus encoding p53, Ad-p53 (10). A prior survey of breast cancer cell lines established that MCF10A and BT549 were especially responsive to adenoviral expression of p53 (33). Accordingly, we found that transcription of hCLCA2 was induced more than a hundredfold by p53 in MCF10A cells (Figure 3A). Expression of hCLCA2 protein was induced 7.4-fold in parallel with a 4.3-fold induction of p53 (Figure 3B). In the metastatic p53-deficient cell line BT549, expression of p53 peaked around 40h post-infection, and hCLCA2 protein was again induced in parallel (Figure 3C). Interestingly, while the cell cycle arrest effector p21 had peaked by 20h and then subsided, hCLCA2 expression was robust at both times, consistent with roles in both cell cycle arrest and apoptosis.
Figure 3.
Induction of hCLCA2 by adenoviral transduction of p53. A, RT-qPCR shows 200-fold induction of hCLCA2 by 24h post-infection. Subconfluent MCF10A cells were infected with adenoviral supernatants at an m.o.i. of 30, and RNA was extracted and analysed. B, induction of hCLCA2 protein in parallel with p53. CA1d cells were infected with adenoviral supernatants and lysed at indicated intervals. Lysates were immunoblotted directly for p53 and tubulin or subjected to immunoprecipitation for hCLCA2 followed by immunoblotting. C, timing of hCLCA2 induction relative to p21. BT549 cells were infected and immunoblotted 20 or 40h later. Ad-GFP indicates Ad-Easy vector sans insert; p53, p53-Ad-Easy; h2, hCLCA2-Ad-Easy.
hCLCA2 inhibits growth of breast cancer cell lines
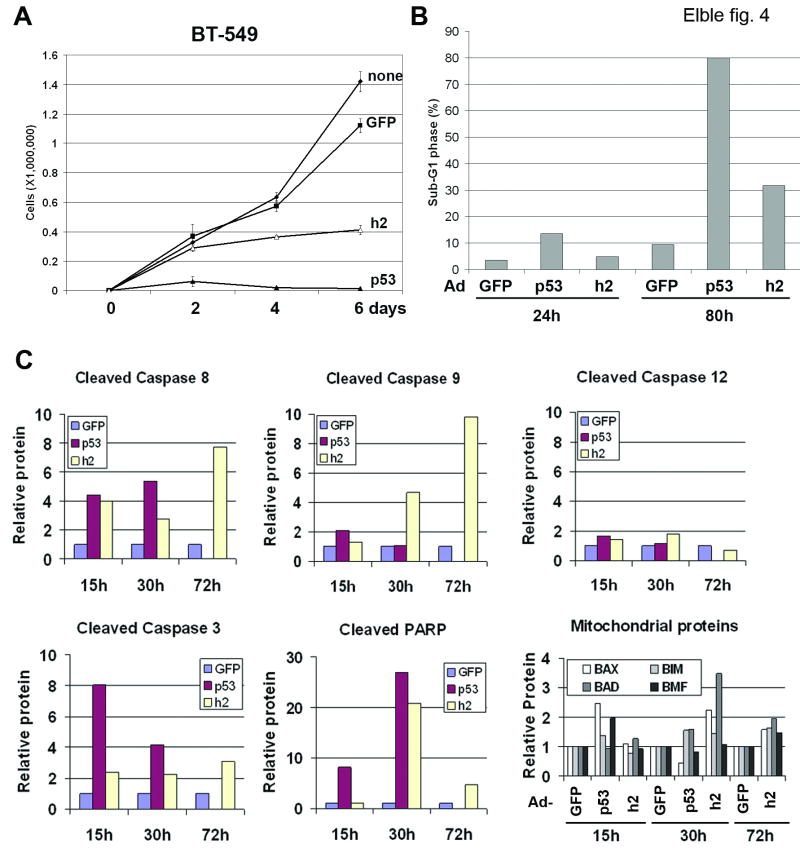
To determine whether hCLCA2 re-expression could inhibit proliferation of a malignant tumor cell line, BT549 cells were transduced with adenoviruses expressing hCLCA2 or a tumoricidal control, p53 (Figure 4A-C). This approach avoided artifacts associated with antibiotic selection, allowing short-term effects to be assessed more clearly, and enabled re-expression in transfection-refractory cell lines. In addition, the adenoviral backbone expressed GFP, allowing the virus to be easily titered and the extent of infection assessed (26). Infection with hCLCA2-Ad resulted in a steady increase in hCLCA2 protein expression to about three times the level induced by p53 at 40h post-infection (Figure 3C). Similar to p53-Ad but less potent, hCLCA2-Ad arrested proliferation of BT549 (Figure 4A). Quantification of apoptosis by flow cytometry of propidium iodide-stained cells revealed a fourfold increase in the sub-G1 fraction (Figure 4B). In a separate experiment, the frequencies of cell detachment and apoptotic nuclei were increased by 2.7- and five-fold, respectively (data not shown). Notably, hCLCA2-Ad did not induce expression of either p53 or the CDK inhibitors p21 (Figure 3C) or p27 (data not shown) in BT549. On the contrary, we observed induction of initiator caspases 8, 9, and 12, as well as executioner caspase 3 (Figure 4C). Because caspase 8 was strongly induced at 15h, well before 9 or 12, hCLCA2 appears to signal apoptosis via the extrinsic pathway. Cross talk between pathways then activates intrinsic pathways, manifested by the late induction of caspase 9 and translocation of Bax at 30h (Figure 4C).
Figure 4.
Restoration of hCLCA2 expression in breast cancer cells is growth-inhibitory. A-C, BT549 breast cancer cells were infected with adenoviruses bearing a GFP marker, p53, or hCLCA2 (h2) at an m.o.i. of 10. A, growth curves measured on a Vi-Cell instrument. B, flow cytometry of propidium iodide-stained cells 48h after infection showed a steep increase in the sub-G1 DNA peak. C, lysates were analyzed by immunoblotting for apoptotic markers. Signals were quantified by fluorometry (Licor) and normalized to actin. Units are relative to Ad-GFP control. Lower right, mitochondrial fractions were analyzed for translocation of Bax, Bad, Bim, and Bmf.
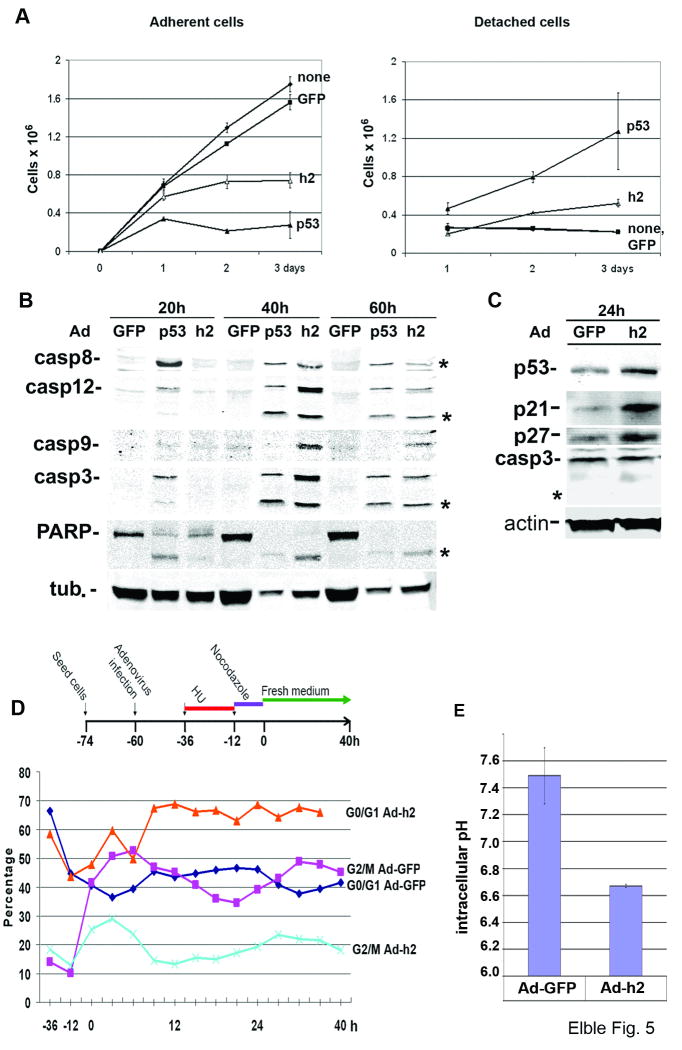
To determine the effect of hCLCA2 transduction on cells that normally express it, we infected MCF10A at the same m.o.i. as BT549. Growth curves showed a strong growth-inhibitory effect, arresting proliferation by 48h post-infection and causing about 40% cell detachment and death by 72h (Figure 5A). Detached cells showed blebbing typical of apoptosis. Biochemical analysis of apoptotic signaling 40h post-infection revealed activation of extrinsic, intrinsic, and ER initiator pathways as well as executioner caspase 3 in cells infected with either p53-Ad or hCLCA2-Ad (Figure 5B); notably, hCLCA2-Ad induced apoptosis about one day later than p53-Ad at similar m.o.i. Cleavage of PARP and cell death in response to p53 and hCLCA2 were also observed in MCF-7 and non-mammary cell lines as well (data not shown). The induction of apoptosis in MCF10A indicates that there is a threshold beyond which more hCLCA2 expression leads to cell death.
Figure 5.
Growth inhibition and apoptosis of MCF10A cells infected with hCLCA2 adenovirus. A, cells were infected as in Figure 4. Adherent (left) and detached (right) cells were counted daily. B, immunoblots of lysates processed at 20h intervals post-infection. Stars indicate proteolytic products diagnostic of apoptosis. C, immunoblots of subconfluent MCF10A cells 24h post-infection show induction of p53, p21, and p27 but not caspase 3. Star indicates absence of caspase 3 proteolytic product. This experiment was repeated four times with similar results. In B and C, equal amounts of total protein were loaded per lane (50 μg) based on the BCA assay. D, top, scheme for cell synchronization by sequential double block and release. Bottom, quantification of cells in G0/G1 versus G2/M after release from the double block as measured by flow cytometry. E, measurement of pHi in cells transduced with hCLCA2-Ad or control by the SNARF-AM method.
This presented a conundrum however, because confluent monolayers of MCF10A express hCLCA2 without displaying apoptosis, suggesting that its function in normal cells is to effect cell cycle arrest rather than apoptosis. To address this possibility, we infected MCF10A and analyzed cells 24h post-infection. Consistent with the preceding results, no cell detachment or apoptosis was observed at this time. Instead, immunoblots revealed induction of p53 and the CDKIs p21 and p27 (Figure 5C). Fluorometry showed that, after normalization to actin, p53 was induced 2.4-fold; p21, 7.2-fold; and p27, 3.5 fold. In contrast, no induction or cleavage of caspase 3 was detected at this time (Figure 5C). These results suggest that hCLCA2 causes cell cycle arrest in normal cells by activating p53 and CDKIs.
To determine whether hCLCA2 expression caused cell cycle arrest, we took advantage of CA1D cells, which we found exhibited an inhibition of proliferation but little apoptosis during the first 72h post-infection (data not shown). We synchronized cells by the double block technique with hydroxyurea followed by washout and nocodazole treatment (Figure 5D). Upon release, most of the hCLCA2-transduced population remained in G1 phase while Ad-GFP-infected cells resumed cycling.
To determine the genetic requirements for growth inhibition and apoptosis by hCLCA2, we obtained an isogenic set of HCT116 colon carcinoma cell lines bearing homozygous deletions of p53, p21, or Bax and infected them with hCLCA2-Ad or control. None of these deletions reduced growth-inhibition or cell death in response to hCLCA2 (Figure S3). However, inhibition of p53 by pifithrin in the p21 knockout line did reduce cell death compared to p21 knockout alone (Figure S3B). Knockdown of p27 also failed to reduce cell death (data not shown). These results indicate that hCLCA2 signals arrest and death via multiple pathways.
Knockdown of hCLCA2 enhances growth rate and resistance to doxorubicin
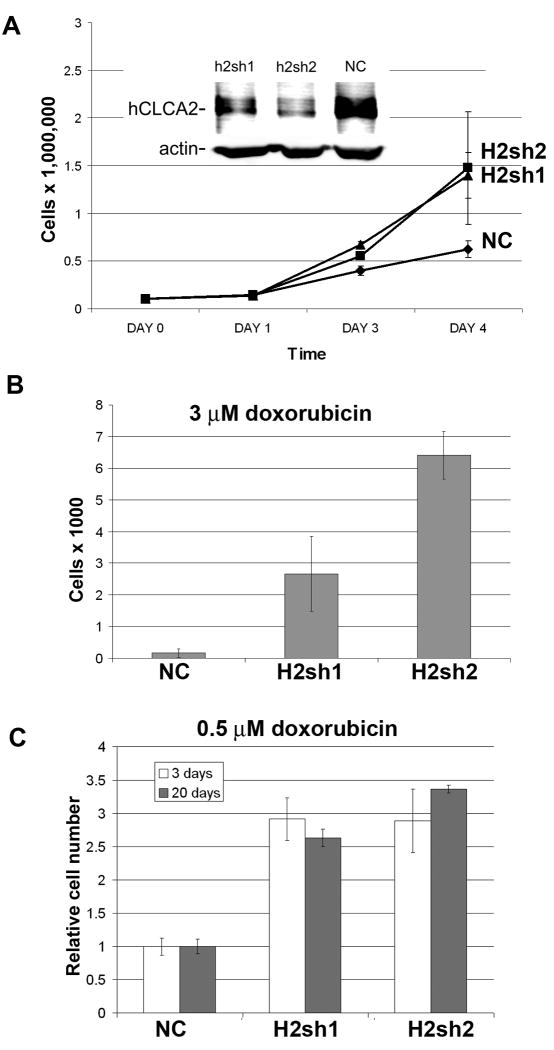
If hCLCA2 is a growth-inhibitor, then suppressing its expression in normal cells might enhance their growth rate or render them resistant to DNA damaging agents. To attenuate its expression in MCF10A, we used a lentiviral system encoding shRNAs embedded in a microRNA and bearing GFP and puromycin-resistance markers. This hybrid system has been reported to enhance knockdown efficiency and reduce off-target effects. Transduced populations were selected and knockdown was tested by immunoblot. While H2sh2 suppressed expression by about 90%, H2sh1 suppressed expression by only 50% compared to the nonsilencing control insert, NC (Figure 6A, inset). Nevertheless, both knockdown populations had significantly faster growth than the control over a four day time course (Figure 6A).
Figure 6.
Effect of hCLCA2 attenuation on cell proliferation and survival. Stable knockdown cell lines were generated by infection with lentiviruses expressing shRNAs (H2sh1; H2sh2; or NC, negative control), followed by puromycin selection. The extent of knockdown was measured by immunoblot (A, inset). To measure proliferation rate, identical numbers of cells were seeded in multiwell plates and counted at one day intervals on a Vi-cell instrument (A). B-C, surviving cell number after treatment with doxorubicin (B, 3 μM for 12h; C, 0.5 μM for 3 days) followed by medium change and recovery for 3 or 20 days.
To determine whether knockdown reduced sensitivity to DNA damage, cells were treated with varying doses of doxorubicin (0.5 – 3μM) and cell survival was measured. Both the knockdown cell lines were much more resistant to doxorubicin than was the control, especially at the highest dose tested (Figure 6B). The survival advantage was apparent whether drug exposure was acute or continuous and persisted over twenty days of exposure, suggesting that downregulation of hCLCA2 confers long-term drug-resistance to tumor cells (Figure 6C).
To further establish whether hCLCA2 expression is a determinant of sensitivity to chemotherapy, we searched public databases of gene expression profiles. One large scale study grouped 30 cancer cell lines according to resistance to the most common chemotherapeutics (34). Using the Oncomine Concepts Map tool to analyze these data, we found that hCLCA2 expression is predictive of sensitivity to doxorubicin, etoposide, mitomycin C, and vinblastine.
hCLCA2 reduces intracellular pH
Several studies link Cl- channel activation to reduction of intracellular pH and promotion of apoptosis (35-37). Intracellular acidification is known to activate p53, caspases, and a host of other responses (37-40). We tested whether cells transduced with hCLCA2 have lower intracellular pH. We found that hCLCA2 transduction reduced pHi from 7.49 to 6.67, a level shown previously to promote apoptosis (Fig. 5E and Fig. S4; (35-37)). We then confirmed that ectopic hCLCA2 enhanced transmembrane anion current in breast cancer cells and the current was further enhanced by calcium (Fig. S5).
Discussion
Considerable effort has been invested in the identification of new p53-client genes both for insight into the biology of stress response and for their therapeutic potential (9, 10). Many novel tumor suppressor genes have been identified by these studies. We have shown here that human CLCA2 is induced by several forms of DNA damage, that p53 binds directly to its promoter, and that mutation of the p53 binding site abolishes induction by both p53 and DNA damage. We further showed that re-expression of hCLCA2 is growth-inhibitory, provoking cell cycle arrest or apoptosis, depending on the cell line and timing. Notably, transduction of immortalized cells initially induced p53, p21, p27, and cell cycle arrest, but sustained expression produced apoptosis (Figure 5). Yet in BT549 and other malignant cell lines, which are characteristically incapable of G1 arrest, no induction of p53, p21 or p27 was detected and only apoptosis was observed, with more rapid kinetics than in MCF10A (15h vs. 40h). Based on these observations, we propose that the function of hCLCA2 in normal cells is to effect cell cycle arrest in response to stress but that in the absence of the capacity to G1-arrest, as in aggressive tumor cells, sustained expression leads instead to apoptosis. These results suggest that transient re-establishment of hCLCA2 expression in a clinical setting might be specifically lethal to tumor cells.
Consistent with this model, we found that attenuation of hCLCA2 expression in immortalized cells confers faster growth and resistance to DNA damage-induced cell death. Together, these results establish hCLCA2 as a multi-stress responder that may antagonize tumorigenesis at multiple stages. Thus, hCLCA2 satisfies many of the criteria for a tumor suppressor gene. It remains to be determined whether perturbation of its mouse orthologs predisposes to mammary tumorigenesis.
The finding that hCLCA2 is a growth inhibitor leads to the interesting question how immortalized cells are able to tolerate expression of a growth inhibitor. One possibility currently under study is that expression of hCLCA2 is conditional with respect to the cell cycle or other growth parameters as reported for the mouse ortholog mCLCA5 (23).
Growth arrest by hCLCA2 correlates with induction of the growth inhibitors p53, p21, and p27. However, the finding that arrest is not dependent on any of these genes individually suggests that a global change is involved. A number of recent studies link plasma-membrane ion channels to progress of the cell cycle and neoplasia (18, 41). For example, Cl- currents have been observed to vary with the cell cycle, and molecular expression of some Clc channels is cell cycle-regulated (18). Moreover, knockdown of Clc-3 induces p21, p27, and cell cycle arrest (42). In breast cancer, the intracellular putative Cl- channel CLIC4 has been found to be a p53-regulated gene that is downregulated in tumor cell lines (43). In addition, a subunit of the GABA receptor is proposed to be a breast tumor suppressor (44).
Cl- channels can also potentiate cell death by mediating intracellular acidification (18, 45). For example, transfection of wildtype, but not mutant, CFTR caused a drop in pHi to about 6.7 and potentiated apoptosis in mammary epithelial cells and lung fibroblasts (35-37). Both effects could be prevented by co-transfection with the NHE1 Na+/H+ exchanger (37). Changes in pH have profound effects on cell physiology, influencing the cell cycle, the cytoskeleton, and the activities of regulators such as Bcl-xL, caspases, annexins and p53 (38, 46-49). Thus, our finding that hCLCA2 acidifies the cytosol could explain its profound inhibitory effects on cell proliferation and survival. We further showed that hCLCA2 stimulates plasma membrane anion current in breast cancer cells. The channel or channels that are regulated by hCLCA2 have not been identified. However, it should be noted that a sister molecule, hCLCA1, is known to interact physically with CFTR (50). Future studies will determine whether hCLCA2 causes intracellular acidification by interacting with this or other channels.
In summary, we have identified a new player in stress response and demonstrated its potency as a growth inhibitor and its relevance to breast cancer progression. These results point to the need for more global studies of hCLCA2 dysregulation in cancer and more directed inquiry into its cellular function(s). Determining the precise mechanism and finding specific agonists or mimics of that action could lead to new therapeutic strategies for cancer treatment, as well as deepen our understanding of epithelial cell biology.
Supplementary Material
Acknowledgments
We thank the following for assistance: Judy Davie and Ko Watabe for ChIP protocols; Mike Flister and Sophia Ran for assistance with luciferase assays; Xu Peng for adenovirus construction tips; Deshou Cao, Xaoyi Lin, Shubam Gautam, and Michelle Lowy for technical assistance; B. Vogelstein for the HCT116 knockout cell lines; and Ko Watabe, Yin Mo, and Janet Factor for critical reading of the manuscript.
Financial support: United States Army Breast Cancer Research Fund Grant DAMD17-00-1-0219, a grant from the Philip Morris External Research Program, and SimmonsCooper Cancer Institute start-up funds (RCE); and NIH R01CA131445 (DN).
Footnotes
Conflicts of interest: none
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Vaziri H, West MD, Allsopp RC, et al. ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the post-translational activation of p53 protein involving poly(ADP-ribose) polymerase. Embo J. 1997;16:6018–33. doi: 10.1093/emboj/16.19.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garbe JC, Holst CR, Bassett E, Tlsty T, Stampfer MR. Inactivation of p53 function in cultured human mammary epithelial cells turns the telomere-length dependent senescence barrier from agonescence into crisis. Cell Cycle. 2007;6:1927–36. doi: 10.4161/cc.6.15.4519. [DOI] [PubMed] [Google Scholar]
- 4.Gewirtz DA. Growth arrest and cell death in the breast tumor cell in response to ionizing radiation and chemotherapeutic agents which induce DNA damage. Breast Cancer Res Treat. 2000;62:223–35. doi: 10.1023/a:1006414422919. [DOI] [PubMed] [Google Scholar]
- 5.El-Deiry WS. The role of p53 in chemosensitivity and radiosensitivity. Oncogene. 2003;22:7486–95. doi: 10.1038/sj.onc.1206949. [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 7.Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci Publ. 2004:247–70. [PubMed] [Google Scholar]
- 8.Langerod A, Zhao H, Borgan O, et al. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res. 2007;9:R30. doi: 10.1186/bcr1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei CL, Wu Q, Vega VB, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–19. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Zhang L, Hwang PM, Rago C, Kinzler KW, Vogelstein B. Identification and classification of p53-regulated genes. ProcNatlAcadSciUSA. 1999;96:14517–22. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pauli BU, Abdel-Ghany M, Cheng HC, Gruber AD, Archibald HA, Elble RC. Molecular characteristics and functional diversity of CLCA family members. ClinExpPharmacolPhysiol. 2000;27:901–5. doi: 10.1046/j.1440-1681.2000.03358.x. [DOI] [PubMed] [Google Scholar]
- 12.Elble RC, Widom J, Gruber AD, et al. Cloning and characterization of lung-endothelial cell adhesion molecule-1 suggest it is an endothelial chloride channel. JBiolChem. 1997;272:27853–61. doi: 10.1074/jbc.272.44.27853. [DOI] [PubMed] [Google Scholar]
- 13.Fuller CM, Ji HL, Tousson A, Elble RC, Pauli BU, Benos DJ. Ca(2+)-activated Cl(-) channels: a newly emerging anion transport family. Pflugers Arch. 2001;443 1:S107–S10. doi: 10.1007/s004240100655. [DOI] [PubMed] [Google Scholar]
- 14.Elble RC, Walia V, Cheng HC, et al. The putative chloride channel hCLCA2 has a single C-terminal transmembrane segment. JBiolChem. 2006;281:29448–54. doi: 10.1074/jbc.M605919200. [DOI] [PubMed] [Google Scholar]
- 15.Bothe MK, Braun J, Mundhenk L, Gruber AD. Murine mCLCA6 is an integral apical membrane protein of non-goblet cell enterocytes and co-localizes with the cystic fibrosis transmembrane conductance regulator. JHistochemCytochem. 2008;56:495–509. doi: 10.1369/jhc.2008.950592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson A, Lewis AP, Affleck K, Aitken AJ, Meldrum E, Thompson N. hCLCA1 and mCLCA3 are secreted non-integral membrane proteins and therefore are not ion channels. JBiolChem. 2005;280:27205–12. doi: 10.1074/jbc.M504654200. [DOI] [PubMed] [Google Scholar]
- 17.Huan C, Greene KS, Shui B, et al. mCLCA4 ER processing and secretion requires luminal sorting motifs. Am J Physiol Cell Physiol. 2008;295:279–87. doi: 10.1152/ajpcell.00060.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunzelmann K. Ion channels and cancer. J Membr Biol. 2005;205:159–73. doi: 10.1007/s00232-005-0781-4. [DOI] [PubMed] [Google Scholar]
- 19.Lastraioli E, Guasti L, Crociani O, et al. herg1 gene and HERG1 protein are overexpressed in colorectal cancers and regulate cell invasion of tumor cells. Cancer Res. 2004;64:606–11. doi: 10.1158/0008-5472.can-03-2360. [DOI] [PubMed] [Google Scholar]
- 20.Sontheimer H. An unexpected role for ion channels in brain tumor metastasis. Exp Biol Med (Maywood) 2008;233:779–91. doi: 10.3181/0711-MR-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh KS, Yuspa SH. Intracellular chloride channels: critical mediators of cell viability and potential targets for cancer therapy. CurrPharmDes. 2005;11:2753–64. doi: 10.2174/1381612054546806. [DOI] [PubMed] [Google Scholar]
- 22.Elble RC, Pauli BU. Tumor suppression by a proapoptotic calcium-activated chloride channel in mammary epithelium. JBiolChem. 2001;276:40510–7. doi: 10.1074/jbc.M104821200. [DOI] [PubMed] [Google Scholar]
- 23.Beckley JR, Pauli BU, Elble RC. Re-expression of detachment-inducible chloride channel mCLCA5 suppresses growth of metastatic breast cancer cells. JBiolChem. 2004;279:41634–41. doi: 10.1074/jbc.M408334200. [DOI] [PubMed] [Google Scholar]
- 24.Gruber AD, Pauli BU. Tumorigenicity of human breast cancer is associated with loss of the Ca2+-activated chloride channel CLCA2. Cancer Res. 1999;59:5488–91. [PubMed] [Google Scholar]
- 25.Li X, Cowell JK, Sossey-Alaoui K. CLCA2 tumour suppressor gene in 1p31 is epigenetically regulated in breast cancer. Oncogene. 2004;23:1474–80. doi: 10.1038/sj.onc.1207249. [DOI] [PubMed] [Google Scholar]
- 26.Saramaki A, Banwell CM, Campbell MJ, Carlberg C. Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Res. 2006;34:543–54. doi: 10.1093/nar/gkj460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang BD, Swift ME, Shen M, Fang J, Broude EV, Roninson IB. Molecular determinants of terminal growth arrest induced in tumor cells by a chemotherapeutic agent. Proc Natl Acad Sci U S A. 2002;99:389–94. doi: 10.1073/pnas.012602599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roninson IB. Tumor cell senescence in cancer treatment. Cancer Res. 2003;63:2705–15. [PubMed] [Google Scholar]
- 29.Lehnhardt M, Klein-Hitpass L, Kuhnen C, et al. Response rate of fibrosarcoma cells to cytotoxic drugs on the expression level correlates to the therapeutic response rate of fibrosarcomas and is mediated by regulation of apoptotic pathways. BMC Cancer. 2005;5:74. doi: 10.1186/1471-2407-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stegmaier K, Wong J, Ross K, et al. Signature-based small molecule screening identifies cytosine arabinoside as an EWS/FLI modulator in Ewing sarcoma. PLoS Med. 2007;4:e122. doi: 10.1371/journal.pmed.0040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aylon Y, Oren M. Living with p53, dying of p53. Cell. 2007;130:597–600. doi: 10.1016/j.cell.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Xie X, Lu J, Kulbokas E, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watts GS, Oshiro MM, Junk DJ, et al. The acetyltransferase p300/CBP-associated factor is a p53 target gene in breast tumor cells. Neoplasia. 2004;6:187–94. doi: 10.1593/neo.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Györffy B, Surowiak P, Kiesslich O, et al. Gene expression profiling of 30 cancer cell lines predicts resistance towards 11 anticancer drugs at clinically achieved concentrations. Int J Cancer. 2006;118:1699–712. doi: 10.1002/ijc.21570. [DOI] [PubMed] [Google Scholar]
- 35.Gottlieb RA, Dosanjh A. Mutant cystic fibrosis transmembrane conductance regulator inhibits acidification and apoptosis in C127 cells: possible relevance to cystic fibrosis. ProcNatlAcadSciUSA. 1996;93:3587–91. doi: 10.1073/pnas.93.8.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szabo I, Lepple-Wienhues A, Kaba KN, Zoratti M, Gulbins E, Lang F. Tyrosine kinase-dependent activation of a chloride channel in CD95-induced apoptosis in T lymphocytes. ProcNatlAcadSciUSA. 1998;95:6169–74. doi: 10.1073/pnas.95.11.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barriere H, Poujeol C, Tauc M, Blasi JM, Counillon L, Poujeol P. CFTR modulates programmed cell death by decreasing intracellular pH in Chinese hamster lung fibroblasts. AmJPhysiol Cell Physiol. 2001;281:C810–C24. doi: 10.1152/ajpcell.2001.281.3.C810. [DOI] [PubMed] [Google Scholar]
- 38.Xiao H, Li T, Yang J, Liu L. Acidic pH induces topoisomerase II-mediated DNA damage. Proc Natl Acad Sci U S A. 2003;100:5205–10. doi: 10.1073/pnas.0935978100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang F, Föller M, Lang K, et al. Cell volume regulatory ion channels in cell proliferation and cell death. Methods Enzymol. 2007;428:209–25. doi: 10.1016/S0076-6879(07)28011-5. [DOI] [PubMed] [Google Scholar]
- 40.Monastyrskaya K, Tschumi F, Babiychuk EB, Stroka D, Draeger A. Annexins sense changes in intracellular pH during hypoxia. BiochemJ. 2007 doi: 10.1042/BJ20071116. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Zhang Y, Cao L, et al. HERG K+ channel, a regulator of tumor cell apoptosis and proliferation. Cancer Res. 2002;62:4843–8. [PubMed] [Google Scholar]
- 42.Tang Y, Liu Y, Zhou J, Wang G, Qiu Q, Guan Y. Silence of ClC-3 chloride channel inhibits cell proliferation and the cell cycle via G/S phase arrest in rat basilar arterial smooth muscle cells. Cell Prolif. 2008;41:775–85. doi: 10.1111/j.1365-2184.2008.00551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernández-Salas E, Suh K, Speransky V, et al. mtCLIC/CLIC4, an organellular chloride channel protein, is increased by DNA damage and participates in the apoptotic response to p53. Mol Cell Biol. 2002;22:3610–20. doi: 10.1128/MCB.22.11.3610-3620.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klebig C, Seitz S, Arnold W, et al. Characterization of {gamma}-aminobutyric acid type A receptor-associated protein, a novel tumor suppressor, showing reduced expression in breast cancer. Cancer Res. 2005;65:394–400. [PubMed] [Google Scholar]
- 45.Lagadic-Gossmann D, Huc L, Lecureur V. Alterations of intracellular pH homeostasis in apoptosis: origins and roles. Cell DeathDiffer. 2004;11:953–61. doi: 10.1038/sj.cdd.4401466. [DOI] [PubMed] [Google Scholar]
- 46.Musgrove E, Seaman M, Hedley D. Relationship between cytoplasmic pH and proliferation during exponential growth and cellular quiescence. Exp Cell Res. 1987;172:65–75. doi: 10.1016/0014-4827(87)90093-0. [DOI] [PubMed] [Google Scholar]
- 47.Furlong I, Ascaso R, Lopez Rivas A, Collins M. Intracellular acidification induces apoptosis by stimulating ICE-like protease activity. J Cell Sci. 1997;110(Pt 5):653–61. doi: 10.1242/jcs.110.5.653. [DOI] [PubMed] [Google Scholar]
- 48.Zhao R, Oxley D, Smith TS, Follows GA, Green AR, Alexander DR. DNA damage-induced Bcl-xL deamidation is mediated by NHE-1 antiport regulated intracellular pH. PLoSBiol. 2007;5:e1. doi: 10.1371/journal.pbio.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monastyrskaya K, Tschumi F, Babiychuk E, Stroka D, Draeger A. Annexins sense changes in intracellular pH during hypoxia. Biochem J. 2008;409:65–75. doi: 10.1042/BJ20071116. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Venable J, LaPointe P, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–15. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.