Abstract
Enteroviruses elicit protective mucosal immune responses that could be harnessed as part of a strategy to prevent sexual transmission of the human immunodeficiency virus-1 (HIV-1). We report the construction of replication competent recombinant vectors of coxsackievirus B3 (CVB3) that express one or more portions of the HIV-1 Gag protein. Vectors containing the capsid domain of Gag were initially genetically unstable with protein expression lost after brief passage in tissue culture. Codon modification to increase the G/C content of the HIV-1 capsid sequence resulted in enhanced genetic stability of CVB3 vectors during in vitro passage. Cells infected with a vector expressing the matrix (MA) subunit of the HIV-1 Gag protein were susceptible to lysis by CD8 T cell clones specific for the SL9 epitope found within MA. These studies suggest that CVB3 vectors may be useful as vaccine vector candidates, if hurdles in class I antigen presentation and stability can be overcome.
Keywords: HIV-1, Vaccine, Coxsackievirus B3
1. INTRODUCTION
Approximately 36 million people worldwide are currently living with the Human Immunodeficiency Virus-type 1 (HIV-1) infection. Educational approaches, antiretroviral therapy, condom use, male circumcision, modifications of obstetrical care for infected women, and other strategies have been shown to diminish the risk of sexual and vertical transmission of HIV-1, but these strategies have not kept pace with the spread of the epidemic. Attempts to develop topical microbicides as a strategy to limit sexual transmission[1, 2] have been disappointing[3–5].
Given the historical success of vaccines for dealing with infectious disease epidemics, emphasis has been placed on development of an HIV-1 vaccine. The search continues for safe, effective, and practical vaccine agents that could prevent or modify the course of infection with HIV-1. Recently, a vaccine trial of HIV-1-uninfected persons using a recombinant adenovirus vector sought to prevent infection or reduce viral load after infection. The vaccine failed to accomplish either goal, and appeared to enhance the risk of infection in some study participants[6]. In order to create a successful prophylactic vaccine, new approaches to elicit both effective humoral and cellular immune responses will likely be necessary[7, 8]. Moreover, localization of these responses to the genital and gastrointestinal mucosa may also be crucial, as the vast majority of HIV-1 transmission occurs across mucosal barriers, and this is a major site of viral replication in established infection[9].
There has been interest in the potential use of poliovirus and other enteroviruses as potential vaccine vectors for HIV-1. Live poliovirus vectors have attracted interest since vaccines have nearly eliminated poliomyelitis in the world, produce long-lasting immunity, and produce potent mucosal immune responses following replication in the gastrointestinal tract[10]. In addition, recombinant live poliovirus vaccine vectors that express Simian Immunodeficiency Virus epitopes have been shown to elicit neutralizing mucosal IgA antibodies, SIV specific cytotoxic T cell responses, and appear to prevent and attenuate SIV infection in rhesus macaques following viral challenge[10–12].
Unfortunately, vaccine-associated paralytic poliomyelitis can occur as a complication of the live-attenuated poliovirus vaccine. This is rare, and usually involves infection of immunodeficient hosts and/or reversion to virulence during protracted in vivo passage. Despite its rarity, the potential for reversion to neurovirulence and plans to totally eliminate worldwide circulation of poliovirus [13] have led to reservations about using poliovirus as a vaccine vector for other diseases. Other enteroviruses, including the related coxsackieviruses, have therefore been considered as candidates. In one report, Halim et al inserted heterologous HIV-1 sequences into recombinant coxsackieviruses and demonstrated that these vectors could induce CD4+ T cell responses in mice[14]. However, these recombinants could accommodate only small sequences of HIV-1 CA since vectors containing more than 360 nucleotide insertions were genetically unstable[14].
To further explore the potential utility of enteroviruses as HIV-1 vaccine vectors, we constructed recombinant vectors based on coxsackievirus B3 that encode varying amounts of the Gag protein of HIV-1, and tested the genetic stability and ability of these vectors to delivery HIV-1 antigens.
2. MATERIALS AND METHODS
2.1 Cells and virus
HeLa (RW) cells were obtained from J. Lindsay Whitton (The Scripps Research Institute, San Diego, CA), and maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/ml of penicillin, and 100 µg/ml of streptomycin (complete DMEM). These cells were used for the titration of viral stocks by plaque assays. Virus stocks were produced by transfection of 293T cells using Fugene (Roche Diagnostics) with pPAR3126 (which expresses T7 polymerase) (a gift from F.W. Studier) along with either pCVB-EGFP [15], (kindly provided by J. Lindsay Whitton) or plasmids into which HIV-1 sequences were inserted in place of the coding sequence of green fluorescent protein. Cell passages were performed using A549 cells (kindly supplied by Timo Hyypia) and maintained with F-12 Nutrient Mixture (HAM) media (F-12; Gibco-BRL, Gaithersburg, Md.) supplemented with 10% FBS, 100 U/ml of penicillin, and 100 µg/ml of streptomycin. For cytotoxic T lymphocyte assays, A549 cells were stably transduced using a lentiviral vector driving the expression of the MHC class I HLA-A*0201 gene cloned from the Caco-2 cell line. The derived cell line (A549-A2.1JM) was maintained in the same culture conditions as the parental A549 cell line. Primary cytotoxic T lymphocytes isolated from patients were maintained in RPMI 1640 (Sigma-Aldrich) supplemented with 10% FBS and 50 U/ml IL-2.
2.2 Construction of Recombinant CVB3 vectors
To produce recombinant CVB3 vectors, HIV-1 sequences were inserted into the unique Sfi I restriction enzyme site of pCVB-EGFP, an infectious plasmid clone of CVB3 virus that expresses eGFP[15]. Removal of the EGFP open reading frame regenerated the parental vector pCVB3.1 described by Slifka et al [16]. Despite the fact that it replicates well in tissue culture, the CVB3.1 virus is less pathogenic than the myocarditic CVB3 strain (H3), from which it was derived[16]. DNA fragments encoding portions of the HIV-1 Gag gene were amplified by PCR from plasmid p83.2.1 vpr.CD24[17]. In this plasmid, the Gag gene from HIVNL4-3 was mutated to change the predicted amino acid sequence at Gag residues 77 to 85 to SLYNTVATL, the clade B consensus sequence for an immunodominant HLA-A*0201-restricted cytotoxic T cell (CTL) epitope. The forward primer CVB-MA6 (5’GGGGGCCGGAGGGGCCAGCGTATTAAGCGGG-3’) containing an Sfi I endonuclease restriction site followed by the codons 7 to 12 of the matrix domain was used for most constructs. Oligonucleotides CVB-MA-r (5’CCGGCCCCTCCGGCCCCGTAATTTTGGCTGAC-3’), CVB-CA-r (5’-CCGGCCCCTCCGGCCCCTATCATTATGGTAGCTGG-3’), CVB-NC-r (5’-CCGGCCCCTCCGGCCCCATTAGCCTGTCTCTCAGT-3’), and CVB-Gag-r (5’-CCGGCCCCTCCGGCCCCTTGTGACGAGGGGTCGCT-3’) were used to amplify fragments encompassing matrix (MA), matrix through P2 (MACA), matrix through nucleocapsid (MANC), or the entire Gag domain respectively, as shown in Figure 1. CVB-CA-r allowed amplification of the genuine CA domain of Gag, as well as the residues of the contiguous spacer peptide (SP1) interposed between the CA and NC domains of Gag. All of these oligonucleotides contained Sfi I restriction sites at their termini. The amplicons were first cloned into a TA-cloning vector (Invitrogen) prior to screening for correct length.
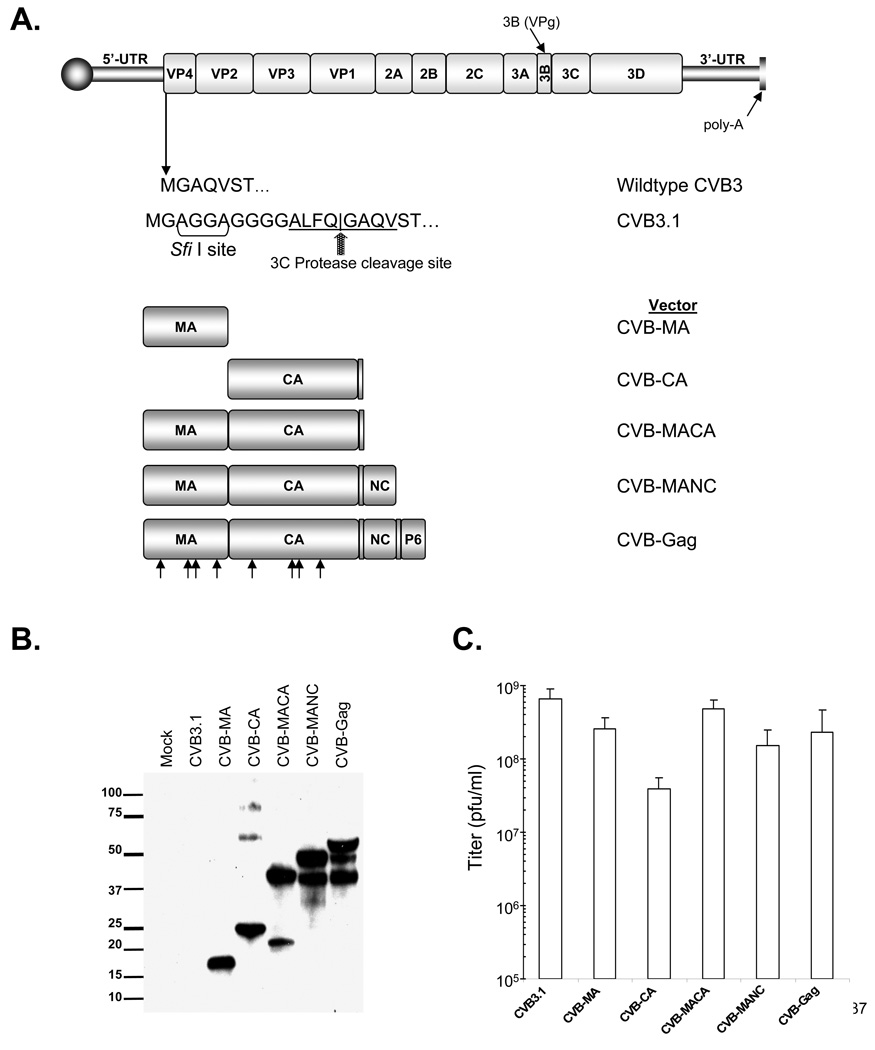
Figure 1. Construction and expression of HIV-1 protein by CVB3 vectors.
(A) Schematic diagram of CVB3 genome and constructs used to express HIV-1 sequences. In pCVB3.1 a Sfi I cloning site and a synthetic viral 3C protease cleavage site are present downstream of the initiation codon of the coxsackie polyprotein. HIV-1 sequences were PCR amplified with primers containing a Sfi I restriction site and ligated into CVB3.1. Arrowheads indicate regions of codon modification in vectors containing the M1–10 HIV-1 Gag mutations. (B) Expression of HIV-1 sequences as detected by immunoblot of lysates of transfected cells. Membrane probed with pooled serum from HIV-1 infected patients. (C) Titer of viruses produced by transfected cells. Values shown are in pfu/ml (mean ± standard deviation).
Inserts of the correct size were subjected to Sfi I digestion and transferred into the Sfi I site of pCVB-EGFP. The complete nucleotide sequences of the inserts were verified using primers at the insertion site. In a similar fashion, a recombinant coxsackievirus B3 containing only the HIV-1 capsid domain and spacer peptide (CA) was produced by amplification of HIV-1 sequences with the CVB-CA-r primer and the forward primer CVB-CA-f (5’-GGGGCCGGAGGGGCCATAGTGCAGAACCTCCAG −3’). Additional vectors were also constructed using pCMV55M1–10 (kindly provided by George Pavlakis) as the template[18]. In this plasmid, codon usage was changed by the introduction of silent mutations in 10 regions of the MA and CA coding sequence.
2.3 Cell Transfection, Virus Stock Preparation, and Virus Growth Assessment
293T cells were plated in 24-well plates after pre-treatment with poly-lysine. Cells were allowed to reach approximately 70% confluence before co-transfecting with plasmids encoding the CVB3 genome and pPAR3126. Transfections used the Fugene6 (Roche) reagent as per manufacturer’s instructions at a 3:1 ratio of reagent to DNA volume. The two-plasmid transfection was performed using a 2:1 ratio of viral genome and T7 plasmid, respectively. Cells were incubated with reagent for 72 hours or until cytopathic effect was observed. Cells were then scraped from the plates, pelleted by centrifugation, and half of the media volume was removed. Virus was liberated from the cells by performing three freeze/thaw cycles on dry ice. Viral titer was determined by plaque assay using Hela (RW) cells in triplicate wells, as described by others[15]. To assess viral growth kinetics, A549 cells were plated in a 24-well plate and allowed to reach near 80% confluence. Wells were simultaneously infected at a multiplicity of infection (MOI) of 3 infectious units per cell with each virus in 0.2 ml medium. At the time points indicated in the text, virus was harvested from replicate wells and titered as described above.
2.4 Immunoblot analysis
When cells showed cytopathic effects after transfection or infection, the cells were scraped, pelleted, and re-suspended in 75 µL of original medium. Protein gel loading buffer was added, and the proteins were separated by electrophoresis in 4–20% SDS-polyacrylamide gels. The proteins were transferred to PVD-F membranes and probed with pooled antiserum (diluted 1:10,000) from HIV-1-infected patients. After being washed with PBS/0.2% Tween-20, bound antibody was detected using horseradish peroxidase-conjugated sheep anti-human antiserum and chemiluminescence (Super Signal West Pico Substrate Chemiluminescence kit, Pierce).
2.5 ELISpot assay
CTL responses were assessed by standard IFN-γ ELISpot assay as previously described[19, 20], with minor modifications. Briefly, the primary HIV-1-specific CTL clone S1-SL9-3.23T recognizing the matrix epitope SLYNTVATL (SL9)[21, 22] was incubated with mock-infected A549-A2.1JM target cells or target cells infected with either CVB-MA or CVB-EGFP vectors (pre-infected at an MOI of 10 for 2 hours at 37°C). The target cells were incubated with CTLs, each at 10,000 cells per well in a 96- well filter plate. Positive controls for CTL activity included mock-infected target cells loaded with exogenous SL9 peptide, and stimulation with 2.5 µg/mL phytohaemagglutinin (PHA). Negative controls included A549 parental cells (not expressing HLA A*0201) infected with CVB-MA, A549-A2.1JM cells infected with CVB-EGFP, and wells with no target cells. Results were analyzed using Student’s T test.
2.6 Cytotoxic T cell co-culture/Chromium Release Assays
Either Caco-2 or A549-A2.1JM cells, which each express HLA-A*0201, were infected with CVB3 vectors at an MOI of 10 for two hours. The cells were then washed and cultured until 8, 10, or 12 hours post infection. These infected cells served as targets in standard chromium release assays for S1-SL9-3.23T, as previously described[21, 23]. Briefly, the target cells were labeled for one hour with 51Cr with or without the SL9 peptide at 10 µg/ml, followed by washing and plating in a 96-well U-bottom plate at 104 cells/well. The CTL clones were then added at 5 × 104 cells/well in a total volume of 200 µl during a four hour incubation. Spontaneous and maximal lysis wells contained medium without CTL and 2.5% Triton-X-100 (Sigma), respectively. Supernatants were then harvested to Lumaplates (Packard) and chromium counts were determined by microscintillation counting (Microbeta 1450, Wallac). Percent specific lysis was calculated as: 100 × (experimental release – spontaneous release)/(maximal release – spontaneous release).
3. RESULTS
3.1 Construction of recombinant CVB3 vectors
To explore the potential utility of recombinant coxsackievirus as expression vectors of HIV-1 sequences, varying lengths of the HIV-1 Gag protein coding sequence were inserted in place of the coding sequence for eGFP in pCVB-EGFP (Figure 1)[15]. These constructs contained genes for proteins ranging from the 17 kD HIV-1 MA protein up to the full length 55 kD Gag protein. One recombinant was also constructed to encode the HIV-1 CA protein alone since that is an immunodominant domain (Figure 1A)[24]. In all constructs containing the MA coding domain, the first five codons of the MA sequence, representing a myristoylation signal of Gag were omitted since the Sfi I cloning site is preceded by the myristoylation sequence for the CVB3 polyprotein. Cells transfected with these constructs expressed HIV-1 proteins of the expected sizes, as demonstrated immunoblot analysis using polyclonal HIV-1-specific human serum, as well as truncated Gag proteins for the CA-, MACA-, MANC-, and the full-length Gag-containing constructs (Figure 1B).
3.2 Recombinant CVB3 vectors are viable and express HIV-1 proteins in infected cells
Virus stocks harvested from the transfected cells were mostly similar in titer to the parental virus CVB3.1 (Figure 1C). The exception was the vector encoding the HIV-1 CA domain, which was approximately 6-fold lower. These viruses were used to infect HeLa (RW) cells. For the CVB-MA vector, cytopathic effects (CPE) were seen by 24 hours after infection, and immunoblot analysis revealed the expression of a protein of the predicted size for MA (Figure 2, lane 3). Similarly, cells infected with CVB-CA developed CPE by 24 hours post-infection and expressed the expected 24 kD protein (Figure 2, lane 4). In contrast, the CVB-MACA vector developed CPE more slowly (24 to 48 hours post infection). These cells expressed a minor amount of the expected 41 kD protein, but most of the HIV-1 protein detected appeared truncated (Figure 2, lane 5). Similarly, CPE was delayed up to 72 hours post infection with CVB-MANC and CVB-Gag vectors. The majority of the Gag protein expressed by CVB-MANC was of the expected size (about 48 kD), but the cells also contained smaller HIV-1 protein fragments (approximately 20 kD and 40 kD) (Figure 2, lane 6). CVB-Gag expressed HIV-1 products of several different sizes (Figure 2, lane 7), including a protein of the predicted size (55 kD), a protein with an apparent mass of 20 kD (similar to that observed with the CVB-MACA and CVB-MANC vectors), and several other small protein products. These fragments were possibly generated by cellular proteases, given the lack of HIV-1 protease in these constructs. Overall, these data showed that HIV-1 proteins of interest were expressed by the vectors.
Figure 2. Expression of HIV-1 proteins by recombinant CVB3 vector.
Membrane probed with pooled serum from chronically HIV-1-infected patients. A549 cells were infected with lysates from cells transfected with each of the CVB3 constructs. Lane 1: Mock infection. Lane 2: CVB3.1. Lane 3: CVB-MA. Lane 4: CVB-CA. Lane 5: CVB-MACA. Lane 6: CVB-NC. Lane 7: CVB-Gag. Markers to the right indicate proper size of 3C-processed insertions of HIV-1 protein. Lanes 3 and 4 show MA and CA as expected size proteins after cleavage from the coxsackievirus polyprotein. Lane 5 indicates MACA expression of the expected 41 kD protein as well as a predominant 20 kD truncated protein. Lane 6 shows that the CVB-MANC construct expresses protein of the expected size (48 kD), but also indicates a less intense 20 kD truncated protein. Lane 7 shows the full length HIV-1 Gag protein and multiple truncation products.
3.3 The genetic stability of HIV-1 genes in the CVB vectors is variable
Having verified that replication-competent viruses were produced by the recombinant CVB3 vectors, we next sought to examine the stability of expression of HIV-1 proteins. All of the recombinant viruses were assayed by serial passage except for the vector containing full-length Gag sequence, due to its limited expression of full-length protein in transfected cells.
Immunoblot revealed that the viral vector CVB-MA expressed the HIV-1 MA protein throughout ten passages in A549 cells (Figure 3A). Sequencing of viral RNA from cells at the tenth passage confirmed retention of the entire inserted sequence (data not shown).
Figure 3. Expression of HIV-1 proteins during serial passage.
Lysates from serial passages the CVB3 vectors were examined for stability of expression of the inserted HIV-1 protein coding regions. Diagrams show P1 region of coxsackievirus and, if applicable, where any deletions were found to occur during serial passage. Filled in arrowheads to right of gel signify expected size of proteins. If applicable, empty arrowheads indicate the majority deletion product. Data is representative of repeated experiments. (A) CVB-MA passages 6–10. The 17 kD matrix protein remained stable through the 10th passage. (B) Blot of passages 1–3 of CVB-CA construct lysates. First and second passage show expected 24 kD CA protein expression. By the third passage CA is deleted from virus. This experiment was repeated with identical results. (C) Early passage of CVB-MACA show expression of the 41 kD protein, but only truncated proteins are subsequently detected. (D) Serial passage of CVB-MANC 48 kD protein shows, similar to CVB-MACA, the HIV-1 sequence is truncated after the first passage.
In contrast, CVB-CA expressed HIV-1 CA protein for the first passage, but expression quickly dropped and was extinguished by the third passage (Figure 3B). Sequencing of viral RNA extracted from vector infected A549 cells revealed a deletion after the first 42 codons in the CA domain, extending beyond the downstream Sfi I cloning site and into the six following codons of VP4.
As noted above, the recombinant CVB-MACA construct directed the expression of the expected 41 kD protein as well as a truncated 20 kD protein. In contrast to the experiment shown in Figure 2, the 41 kD band was more prominent in the first of this series of passages of virus stock produced by transfection. However, during serial passaging, there was a progressive loss of the 41 kD species and by the third passage only the lower molecular weight protein was detectable (Figure 3C). RT-PCR, cloning, and sequencing of the viral RNA revealed one species that deleted nearly the entire coding sequence between the upstream and downstream Sfi I sites. In other clones, the coding sequence for MA and the first few codons of the HIV-1 CA domain were retained, but the remainder of CA was lost.
Similarly, CVB-MANC expressed two different sized proteins, including the expected 48 kD protein, and an apparently truncated form with a mass of about 38 kD. Upon passage, a band with apparent mass size of about 20 kD was also detected. By the third passage, all other detected protein forms disappeared except for this 20 kD species (Figure 3D). Viral RNA sequencing indicated a deletion after the MA coding region in the CA coding sequence. It spanned from 42 codons into the CA coding sequence through the fifth codon succeeding the second Sfi I site, indicating nearly complete deletion of the CA coding sequence.
Thus, the separate experiments shown in Figure 2 and Figure 3 together demonstrate the instability of the HIV-1 sequences in CVB-CA, CVB-MACA, and CVB-MANC vectors, resulting in truncation of inserted sequence within three passages of virus stock produced by plasmid transfection of producer cells.
3.4 Increased G/C content in the HIV-1 gene inserts modestly increases their stability in the CVB vectors
Others have shown that HIV-1 CA sequences are genetically unstable in viral vectors based on coxsackievirus B4, and that increasing the G/C content of HIV-1 sequence enhanced its stability in recombinant poliovirus vectors[14, 25]. Consequently, we tested whether codon-modified Gag RNA would have greater stability in our CVB3 vectors. Gag sequences were amplified from pCMV55M1–10, a vector containing multiple silent mutations to reduce the A/U content and enhance the nuclear export of unspliced HIV-1 mRNA in the absence of the viral HIV-1 Rev protein[18].
Vectors containing M1–10 sequences expressed proteins of the expected molecular weight, although some vectors co-expressed a truncated protein during initial passage (Figure 4). As for the codon-modified construct CVB-Gag M1–10, it continued to express multiple lower molecular weight proteins in addition to the expected 55 kD protein product (data not shown) and thus was not examined further.
Figure 4. Serial passage of sequence modified CVB3 vectors.
Expression of Gag M1–10 CVB3 vectors was examined using lysates from transfection and serial passage. Immunoblots revealed altered expression stability of the inserted HIV-1 protein coding regions. Filled in arrowheads on right of gel signify expected size of proteins. Where deletions were detected, empty arrowheads indicate the majority deletion product. (A) CVB-MA M1–10 passage 6 through passage 10. The 17 kD matrix protein again remained stable through 10 passages. (B) CVB-CA M1–10 immunoblots of passage 2 through passage 10, where deletion is nearly complete. Increased G/C content appeared to enhance the stability of CA expression. (C) Early serial passage of CVB-MACA M1–10 shows strong expression of the 41 kD protein. Expression still shifted however, favoring a truncated protein by the third passage. (D) Passage of CVB-MANC M1–10 vector still expressed both the expected 48 kD and approximately 38 kD protein as previously seen, but CVB-MACA M1–10 delayed complete truncation by one additional passage beyond the non-M1–10 vector.
The CVB-MA M1–10 vector maintained MA expression through the tenth serial passage, similar to the vector without codon modification (Figure 4A versus Figure 3A). As before, sequence analysis showed no evidence of mutations or deletions within the MA M1–10 coding sequence after these passages. Interestingly, a smaller band was reproducibly detected by immunoblot. Sequence analysis was performed using viral RNA extracted from the eighth passage; we found no evidence of truncation or mutation of the HIV-1 MA sequence (data not shown). We hypothesize that the smaller protein species may be the result of translation from a cryptic initiation site in the viral RNA.
The codon-modified CVB-CA M1–10 construct maintained expression of full-length inserted sequence until the tenth passage, when it faded nearly completely (Figure 4B), therefore persisting longer than the codon-unmodified version (Figure 3B). Loss of expression was associated with similar patterns of genetic deletion.
Similarly, the CVB-MACA M1–10 vector maintained expression for more passages than the codon-unmodified version, but also demonstrated a similar pattern of protein and gene truncation (Figure 4C versus Figure 3C).
The same pattern was observed for the CVB-MANC M1–10 vector, which expressed two large molecular weight species of the HIV-1 protein until the third passage when expression abruptly shifted to the 20 kD protein (Figure 4D versus Figure 3D). Again, sequence analysis showed multiple patterns of deletion including several species with the previously observed deletion in CA sequence noted for the non codon-modified insert vector.
3.5 CVB vectors containing HIV-1 CA have impaired replication kinetics while those with MA replicate normally
In view of the deletions that occurred in all constructs containing the HIV-1 CA sequence, and the apparent lower titer observed after transfection (Figure 1C), we compared the growth rates of CVB-CA with CVB-MA vectors and their M1–10 counterparts to the parental CVB3.1 virus (Figure 5, filled triangles). Virus was isolated at the time points indicated and titered by plaque assay.
Figure 5. One-step growth curve.
A549 cells were infected with recombinant CVB3 virus at an MOI of 3 using titered stocks produced by transfection. Squares (■) represent CVB-MA, open squares (□) represent CVB-MA M1–10, triangles (▲) represent CVB3.1, circles (●) represent CVB-CA, and open circles (○) represent CVB-CA M1–10. CVB-MA and the M1–10 version, in which the HIV-1 matrix protein is stable in both for at least 10 passages, had similar growth dynamics to the parental virus (CVB3.1). Titers of the three viruses peaked at 8 h post-infection. The CVB-CA vectors had slower growth rates than the parental virus, taking 12–24 hours to reach peak titer, but eventually achieve a similar titer by end of assay.
The CVB-CA vector appeared to have delayed kinetics compared to the empty vector CVB3.1 (Figure 5, filled circles), as it did not reach peak titer until at least 12 hours post-infection. However, its final titer was not significantly different than the parental vector (Figure 5, filled triangles). Curiously, despite enhanced stability over the non-optimized CVB-CA vector, the CVB-CA M1–10 vector had slower growth dynamics and did not reach peak titer until 24 hours post infection (Figure 5, open circles). The CVB-MA vector replicated well and reached peak titer at eight hours post-infection, similar to CVB3.1 (Figure 5, filled squares). The titer at 24 hours post-infection during a repeat experiment was observed to be higher than the CVB3.1 vector (data not shown), indicating that CVB-MA may not differ significantly in replication kinetics from CVB3.1. The CVB-MA M1–10 vector had similar growth rates to CVB-MA, but appeared to have slightly increased titers at 8 and 24 hours post-infection (Figure 5, open squares). These data and the immunoblot analysis indicated that insertion of HIV-1 MA sequences did not interfere greatly with CVB3 viral replication.
3.6 Antigen Presentation by Cells Expressing the HIV-1 MA Protein
To determine if the expressed HIV-1 proteins from recombinant CVB3 vectors are processed and presented via the MHC class I pathway, target cells were infected with CVB3 vectors expressing HIV-1 MA protein or the irrelevant EGFP protein as a negative control.
Chromium release assays were performed to assess the ability of virus-infected cells to trigger killing activity by an HIV-1-specific CTL clone recognizing an epitope in MA. In eight experiments, CaCo and A549-A2.1JM target cells infected with the CVB-MA vectors demonstrated significant but low levels of killing (typically <10% specific lysis) at an effector:target ratio of 5:1 (Figure 6A). CVB-MA (M1-M10) was also evaluated experiments, and yielded similar results to the CVB-MA vector (data not shown). Time course experiments comparing 8, 10, and 12 hours of infection demonstrated peak killing at 10 hours, which was similar for both the codon-unmodified and -modified versions (data not shown).
Figure 6. Antigen presentation by cells infected with CVB3 vectors.
(A) A549-A2.1JM cells were used as target cells for chromium release assays. Cells were either left uninfected (Mock) or infected at an MOI of 3 with CVB-MA, CVB-MA (M1–10), or CVB-EGFP vectors. At 10 hours post infection, cells were collected and incubated with a T cell clones specific to the SL9 epitope found in HIV-1 matrix protein. The graph shows percent specific lysis of uninfected cells, cells infected with vectors expressing either EGFP or MA, or cells infected with CVB-MA (M1–10) vector (Bars reflect mean and standard deviation from eight independent experiments). The specific lysis of control target cells labeled with SL9 peptide was 49% (standard deviation 9%)(not shown). (B) 104 cells of the same CTL clone were incubated 1 : 1 with target cells infected at an MOI of 10 using the above CVB3 vectors. A2.1JM + GFP represents HLA-A*0201 positive cells infected with the irrelevant CVB-GFP vector. A2.1JM + MA represents HLA-A*0201 cells infected with the CVB-MA vector. A549 + MA represents the parental cell line (which does not express HLA-A*0201) infected with the CVB-MA vector. All wells shown were done in quadruplicate. Numbers in graph represent mean number of spots per 100,000 cells; error bars indicate standard of deviation. This experiment is representative of three independent experiments. Positive controls using target cells labeled with SL9 peptide showed too many spots to count (not shown).
As a complementary approach, we utilized ELISpot to detect IFN-γ production by the same CTLs during co-culture with cells infected two hours previously with CVB vectors. We detected a small but statistically significant number of spot forming cells (80/100,000 cells) (Figure 6B). In related experiments, intracellular cytokine staining and flow cytometry also revealed activation and IFN-γ production by small numbers of CTLs during co-culture with target cells infected by MA containing vectors (data not shown).
Overall, these data indicated that presentation of the processed SL9 epitope from HIV-1 MA expressed by the CVB vectors was present but inefficient. The chromium release assay suggested that only a few percent of infected cells presented epitope at levels required for CTL recognition, and the ELISpot assay similarly demonstrated that the efficiency of recognition was as little as less than one percent of CTLs.
4. DISCUSSION
It is currently envisioned that a successful vaccine strategy for HIV-1 will require induction of both humoral and cellular immune responses[10–12, 14, 26]. Moreover, it may also be important to induce mucosal immune responses. The characteristic loss of CD4 T cells seen with HIV and Simian immunodeficiency virus (SIV) infection has been linked to replication of these viruses in the gastrointestinal tract associated lymphoid tissues (GALT), which is populated by a high fraction of the total body memory CD4 T cells. [9]. Consequently, we and others [9]have hypothesized that induction of cellular immune responses that home to GALT might potential prevent transmission or limit the replication of HIV acquired across the gastrointestinal and genital mucosa. Viruses may vary in their capacity to induce such responses; in one recent report, injection of a vaccinia vector encoding the HIV gag, pol, and env gene products failed to induce mucosal cellular immune response in most vaccinees[27], despite the fact that vaccinia vectors are highly effective at presenting HIV-1 epitopes in vitro [23, 28, 29].
In view of the replication of enteroviruses in the gastrointestinal tract, we sought to demonstrate the feasibility of using recombinant CVB3 as a possible vaccine vector to express varying regions of the HIV-1 Gag protein. CVB3 is a common enteroviral infection, but, in contrast to poliovirus, is rarely linked to cases of flaccid paralysis. We chose this specific strain in view of preceding studies by others showing that the CVB3 could stably carry foreign sequences for numerous passages[15],. Although CVB3 infection is common, this does not appear to rule out their potential utility: studies involving poliovirus vectors have shown that preexisting immunity does not prevent the generation of cellular immune responses to foreign sequences expressed by the vector [30].
Replication competent recombinant virus was produced by all of our constructs, and HIV-1 proteins of the expected sizes were expressed after transfection of cells with plasmids encoding for these vectors. Although packaging limits of the CVB3 virion have not yet been rigorously defined, we found that the CVB3 capsid could accommodate the insertion of the full coding sequence for Gag (Figure 2), approaching the theoretical maximum genome length for packaging by poliovirus and consistent with previously described results[25, 31].
Unfortunately, CVB3 vectors containing HIV-1 CA coding sequence were genetically unstable. Vectors with the CA sequence rapidly developed truncations leaving a short and relatively stable 20 kD leader-type sequence consisting mainly of the MA coding sequence. This truncated HIV-1 sequence remained stable, consistent with results previously observed using other picornavirus vectors (poliovirus and CVB4)[14, 31].
The overall A/U content in Gag could interfere with efficient replication of the vector, as has been described in poliovirus vectors[32]. The HIV-1 Gag M1–10 sequences inserted into CVB3 vectors contained about 5% greater G/C content than wildtype Gag sequence, and resulted in a modest increase in stability of HIV-1 sequences in all the vectors examined, consistent with other reports[18, 25].
The mechanism of deletions in inserted sequences is unclear, but studies of the deletion of foreign sequences from other picornaviruses have led to the hypothesis that slowing of the viral RNA polymerase while copying an inserted coding sequence potentiates jumps to a new area of template to circumvent the region[32]. Insertions of significant length or complex secondary structure could cause such a lag in RNA replication and thereby create deletions. This may be the case for CA coding sequence since stability of inserts containing complete CA sequence appeared problematic, even with changes in codon usage. As the CA region contains many useful CTL epitopes, individual epitopes could be inserted in tandem as a synthetic gene rather than the full-length coding sequence to circumvent stability issues and reduce insertion length.
In order to evaluate the ability of these vectors to deliver HIV-1 sequences in an antigenic manner, we tested their ability to activate HIV-1-specific CTLs recognizing an immunodominant epitope from MA. Unfortunately, induction of the activation of CTL clones and target cell lysis was inefficient compared to the maximum rate observed with peptide loading or whole HIV infection of target cells seen in other studies [21,23]. These data are in agreement with recent studies indicating CVB3 replication is associated with disruption of protein transport in the endoplasmic reticulum and disruption of the golgi apparatus, which interferes with MHC-I antigen display[33]. However, the low levels of observed CTL activity indicated that this mechanism of immune evasion is not complete, and therefore may be amenable to manipulation. Although similar growth kinetics were observed, the CVB-MA M1–10 vector appeared to have lower levels of specific lysis than wildtype CVB-MA, suggesting that replication rate, codon usage, and MHC-I presentation are not tightly linked.
Despite the low levels of HIV-1 antigen delivery to CTLs in our studies, there is evidence that cellular immune responses are involved in clearance of enterovirus infected cells in both non-human primate and mouse models[12, 16, 30, 34]. These data suggest that there are effective CTL responses that could be boosted by modification of the CVB3 3A and/or 2BC proteins which are believed to interfere with the MHC class I pathway. Alternatively, DNA priming with the epitope(s) encoded by the CVB3 vector prior to vaccination may elicit stronger CTL immune responses in vivo. Therefore, further development of CVB3 as a vaccine vector will therefore require directed approaches to cope with issues of insert stability and epitope presentation.
Further development of a coxsackievirus vector would likely necessitate additional attenuation of the virus. The parental vector employed in this study, CVB3.1, grows to lower titers in the organs of experimentally infected mice [16], and thus appears attenuated compared to the original molecular clone of the H3 strain of CVB3. However, replication was still seen in various organs in experimentally infected mice[16]. Additional attenuation could potentially be achieved by modifications of the IRES, or substitution of the IRES of less pathogenic CVB strains, as reviewed by others[35].
Finally, it will be essential to compare the magnitude, anatomic localization, and functional properties of immune responses induced by coxsackievirus vectors to those induced by other viral vectors (rhesus cytomegalovirus, picornavirus etc.) already shown to have an impact on SIV replication in simian AIDS models[11, 36].
Acknowledgements
This work was supported by grants from the NIH (AI060463) and the Center for AIDS Research to PK, a Universitywide AIDS Research Program predoctoral fellowship to JPM (CC02-LA-001), and a NIH grant (AI043203) to OY. PK also received support as an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Fletcher PS, et al. Candidate polyanion microbicides inhibit HIV-1 infection and dissemination pathways in human cervical explants. Retrovirology. 2006;3(1):46. doi: 10.1186/1742-4690-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shattock RJ, Moore JP. Inhibiting sexual transmission of HIV-1 infection. Nat Rev Microbiol. 2003;1(1):25–34. doi: 10.1038/nrmicro729. [DOI] [PubMed] [Google Scholar]
- 3.Tao W, Richards C, Hamer D. Enhancement of HIV infection by cellulose sulfate. AIDS Res Hum Retroviruses. 2008;24(7):925–929. doi: 10.1089/aid.2008.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Damme L, et al. Lack of effectiveness of cellulose sulfate gel for the prevention of vaginal HIV transmission. N Engl J Med. 2008;359(5):463–472. doi: 10.1056/NEJMoa0707957. [DOI] [PubMed] [Google Scholar]
- 5.Nonoxynol-9 does not protect against HIV. AIDS Patient Care STDS. 2008;22(1):89–93. doi: 10.1089/apc.2007.9963. [DOI] [PubMed] [Google Scholar]
- 6.HVTN Press Release. 2007 ( http://www.hvtn.org/media/pr/step1207.html).
- 7.McMichael AJ. HIV vaccines. Annu Rev Immunol. 2006;24:227–255. doi: 10.1146/annurev.immunol.24.021605.090605. [DOI] [PubMed] [Google Scholar]
- 8.Nathanson N, Mathieson BJ. Biological considerations in the development of a human immunodeficiency virus vaccine. J Infect Dis. 2000;182(2):579–589. doi: 10.1086/315707. [DOI] [PubMed] [Google Scholar]
- 9.Grossman Z, et al. Pathogenesis of HIV infection: what the virus spares is as important as what it destroys. Nat Med. 2006;12(3):289–295. doi: 10.1038/nm1380. [DOI] [PubMed] [Google Scholar]
- 10.Crotty S, Andino R. Poliovirus vaccine strains as mucosal vaccine vectors and their potential use to develop an AIDS vaccine. Adv Drug Deliv Rev. 2004;56(6):835–852. doi: 10.1016/j.addr.2003.10.042. [DOI] [PubMed] [Google Scholar]
- 11.Crotty S, et al. Mucosal immunization of cynomolgus macaques with two serotypes of live poliovirus vectors expressing simian immunodeficiency virus antigens: stimulation of humoral, mucosal, and cellular immunity. J Virol. 1999;73(11):9485–9495. doi: 10.1128/jvi.73.11.9485-9495.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crotty S, et al. Protection against simian immunodeficiency virus vaginal challenge by using Sabin poliovirus vectors. J Virol. 2001;75(16):7435–7452. doi: 10.1128/JVI.75.16.7435-7452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kew OM, et al. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu Rev Microbiol. 2005;59:587–635. doi: 10.1146/annurev.micro.58.030603.123625. [DOI] [PubMed] [Google Scholar]
- 14.Halim SS, Collins DN, Ramsingh AI. A therapeutic HIV vaccine using coxsackie-HIV recombinants: a possible new strategy. AIDS Res Hum Retroviruses. 2000;16(15):1551–1558. doi: 10.1089/088922200750006074. [DOI] [PubMed] [Google Scholar]
- 15.Feuer R, et al. Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J Virol. 2002;76(9):4430–4440. doi: 10.1128/JVI.76.9.4430-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slifka MK, et al. Using recombinant coxsackievirus B3 to evaluate the induction and protective efficacy of CD8+ T cells during picornavirus infection. J Virol. 2001;75(5):2377–2387. doi: 10.1128/JVI.75.5.2377-2387.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali A, Jamieson BD, Yang OO. Half-genome human immunodeficiency virus type 1 constructs for rapid production of reporter viruses. J Virol Methods. 2003;110(2):137–142. doi: 10.1016/s0166-0934(03)00110-1. [DOI] [PubMed] [Google Scholar]
- 18.Schneider R, et al. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71(7):4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang OO, et al. Genetic and stochastic influences on the interaction of human immunodeficiency virus type 1 and cytotoxic T lymphocytes in identical twins. J Virol. 2005;79(24):15368–15375. doi: 10.1128/JVI.79.24.15368-15375.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabado RL, et al. Detection of HIV-1-specific CTL responses in Clade B infection with Clade C Peptides and not Clade B consensus peptides. J Immunol Methods. 2005;296(1–2):1–10. doi: 10.1016/j.jim.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Bennett MS, et al. Epitope-dependent avidity thresholds for cytotoxic T-lymphocyte clearance of virus-infected cells. J Virol. 2007;81(10):4973–4980. doi: 10.1128/JVI.02362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adnan S, et al. Nef interference with HIV-1-specific CTL antiviral activity is epitope specific. Blood. 2006;108(10):3414–3419. doi: 10.1182/blood-2006-06-030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang OO, et al. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. J Virol. 1996;70(9):5799–5806. doi: 10.1128/jvi.70.9.5799-5806.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yusim K, et al. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J Virol. 2002;76(17):8757–8768. doi: 10.1128/JVI.76.17.8757-8768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SG, et al. Novel design architecture for genetic stability of recombinant poliovirus: the manipulation of G/C contents and their distribution patterns increases the genetic stability of inserts in a poliovirus-based RPS-Vax vector system. J Virol. 2002;76(4):1649–1662. doi: 10.1128/JVI.76.4.1649-1662.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porter DC, et al. Release of virus-like particles from cells infected with poliovirus replicons which express human immunodeficiency virus type 1 Gag. J Virol. 1996;70(4):2643–2649. doi: 10.1128/jvi.70.4.2643-2649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anton PA, et al. Differential immunogenicity of vaccinia and HIV-1 components of a human recombinant vaccine in mucosal and blood compartments. Vaccine. 2008;26(35):4617–4623. doi: 10.1016/j.vaccine.2008.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koup RA, et al. Detection of major histocompatibility complex class I-restricted, HIV-specific cytotoxic T lymphocytes in the blood of infected hemophiliacs. Blood. 1989;73(7):1909–1914. [PubMed] [Google Scholar]
- 29.Walker BD, et al. Long-term culture and fine specificity of human cytotoxic T-lymphocyte clones reactive with human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1989;86(23):9514–9518. doi: 10.1073/pnas.86.23.9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandl S, et al. Poliovirus vaccine vectors elicit antigen-specific cytotoxic T cells and protect mice against lethal challenge with malignant melanoma cells expressing a model antigen. Proc Natl Acad Sci U S A. 1998;95(14):8216–8221. doi: 10.1073/pnas.95.14.8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mueller S, Wimmer E. Expression of foreign proteins by poliovirus polyprotein fusion: analysis of genetic stability reveals rapid deletions and formation of cardioviruslike open reading frames. J Virol. 1998;72(1):20–31. doi: 10.1128/jvi.72.1.20-31.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pilipenko EV, Gmyl AP, Agol VI. A model for rearrangements in RNA genomes. Nucleic Acids Res. 1995;23(11):1870–1875. doi: 10.1093/nar/23.11.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitton JL, Cornell CT, Feuer R. Host and virus determinants of picornavirus pathogenesis and tropism. Nat Rev Microbiol. 2005;3(10):765–776. doi: 10.1038/nrmicro1284. [DOI] [PubMed] [Google Scholar]
- 34.Henke A, et al. The role of CD8+ T lymphocytes in coxsackievirus B3-induced myocarditis. J Virol. 1995;69(11):6720–6728. doi: 10.1128/jvi.69.11.6720-6728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tracy S, Gauntt C. Group B coxsackievirus virulence. Curr Top Microbiol Immunol. 2008;323:49–63. doi: 10.1007/978-3-540-75546-3_3. [DOI] [PubMed] [Google Scholar]
- 36.Hansen SG, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15(3):293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]