Abstract
Adenylosuccinate lyase (ASL), a catalyst of key reactions in purine biosynthesis, is normally a homotetramer in which three subunits contribute to each of four active sites. Human ASL deficiency is an inherited metabolic disease associated with autism and mental retardation. We have characterized five disease-associated ASL mutants: R194C and K246E are located at subunit interfaces, L311V is in the central helical region away from the active site, and R396C and R396H are at the entrance to the active site. The Vmax (at 25 °C) for R194C is comparable to that of WT; while those of L311V, R396C, R396H and K246E are considerably reduced and affinity for adenylosuccinate is retained. The mutant enzymes have decreased positive cooperativity as compared to WT. K246E exists mainly as dimer or monomer, accounting for its negligible activity; whereas the other mutant enzymes are similar to WT in the predominance of tetramer. At 37 °C, the specific activity of WT and these mutant enzymes slowly decreases 30-40% with time and reaches a limiting specific activity without changing significantly the amount of tetramer. Mutant R194C is unique in being rapidly inactivated at the harsher temperature of 60°C, indicating that it is the least stable enzyme in vitro. Conformational changes in the mutant enzymes are evident from protein fluorescence intensity at 25 °C and after incubation at 37 °C, which correlates with the loss of enzymatic activity. Thus, these disease-associated single mutations can yield enzyme with reduced activity either by affecting the active site or by perturbing the enzyme’s structure and/or native conformation which are required for catalytic function.
Adenylosuccinate lyase (EC 4.3.2.2) catalyzes two β-elimination reactions in the de novo synthesis of purines: the cleavage of adenylosuccinate (SAMP)1 to AMP and fumarate; and the conversion of 5-aminoimidazole-4-(N-succinylocarboxamide ribonucleotide) (SAICAR) to 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) and fumarate (1). Each reaction is considered as a “uni-bi” reaction, in which fumarate is released from the enzyme before AMP or AICAR (2). The rate limiting step of the reaction catalyzed by adenylosuccinate lyase and other members of the fumarase family is thought to be the cleavage of the C(γ)-N bond (3-5)
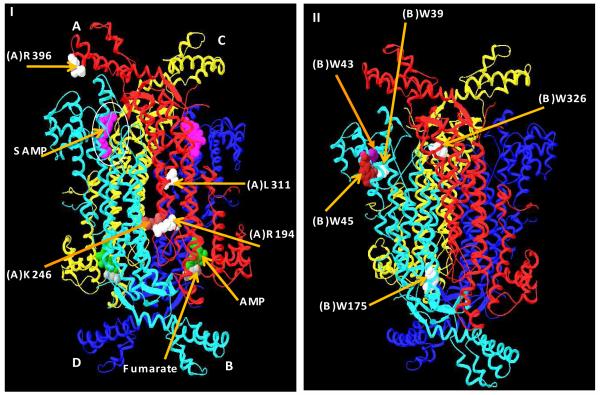
ASL is a homotetramer from all species examined (Figure 1 I); for human ASL, each subunit contains 484 amino acids and has a molecular mass of approximately 55 kDa. The enzyme has four active sites (Figure 1 I) and three subunits contribute amino acids to each active site (6). Recently, a new substrate analog, adenosine phosphonobutyric acid, 2′ (3′), 5′-diphosphate (APBADP) was synthesized (7). Kinetic studies showed that this substrate analog acts as a competitive inhibitor with respect to either SAMP or SAICAR (KI ∼ 0.2 μM), indicating that the two substrates occupy the same active site (7). Furthermore, binding studies showed that human ASL binds up to 4 mol of APBADP per mol of enzyme tetramer and exhibits positive cooperativity (7). The cooperative binding of the substrate in human ASL is supported by the recently deposited human ASL crystal structure (PDB # 2VD6) in which two active sites are occupied by the substrate SAMP and the other two active sites by the products AMP and fumarate, implying that half of the active sites are distinguishable from the other half of the sites (8).
FIGURE 1.
(I) The crystal structure of the human ASL which was crystallized with the products and the substrates (PDB # = 2vd6) (8). Each individual subunit is color coded and one active site of four is designated by a circle. The substrate, SAMP, is shown in pink and located in the two upper active sites and the products AMP (green) and fumarate (gray) are shown in the two lower active sites. The positions of the amino acids L311, R194, and R396, (in white) and K246 (in light brown) are shown in the A subunit. (II) The positions of Trp residues are shown in the B subunit: W39 (in white) W43 (in purple) W45 (in maroon) W175 and W326 (in white). The substrate and the products were removed from the active sites for clarity.
Adenylosuccinate lyase deficiency is an inherited error of metabolism which results in a profound effect on the central nervous system (9-11). Clinical symptoms of this deficiency are varying degrees of mental retardation accompanied by muscle wasting and behavioral changes such as autism, aggressiveness, and self-mutilation (9). It is common to see these symptoms developing within the first month after birth. Succinyladenosine (S-Ado) and SAICAriboside are dephosphorylated succinyl purines which are the derivatives of ASL substrates that accumulate as a result of a deficiency of this enzyme (10,11). ASL deficient patients have higher levels of S-Ado and SAICAriboside in cerebrospinal fluid (100 – 500 μM) than in plasma (2 – 12 μM) (10).
ASL deficiency occurs because of point mutations in the ASL gene. To date, more than 30 point mutations have been identified worldwide, predominantly from Europe (12,13). The majority of the point mutations are located relatively far from the active site, in the central helical region which seems to be important for the stability of the tetramer (13, 14).
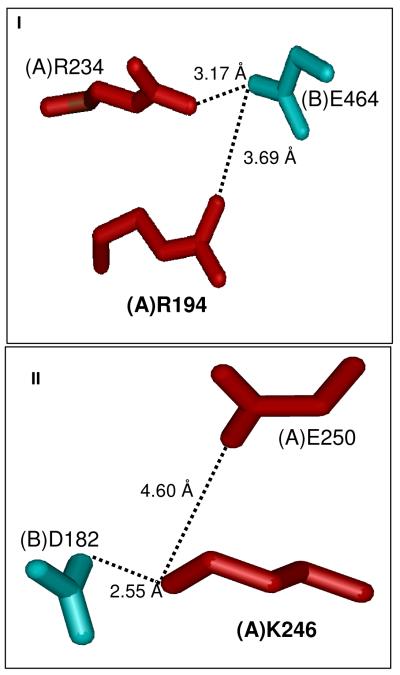
The present study focuses on five representative ASL-deficiency associated point mutations (R194C, K246E, R396C, R396H, and L311V) that are located at different regions of the enzyme (Figure 1I). The amino acids R194 and K246 are located in the A/B (or C/D) subunit interface, whereas amino acid R396 is found at the entrance to the active site (as designated by bound SAMP) and L311 is located in the central helical region (Figure1 I). In the WT enzyme, the subunit interface amino acids R194 and K246 each has its electrostatic counterpart. Thus, R194 of the A subunit interacts with E464 of the B subunit (3.69 Å apart) and K246 of the A subunit interacts with D182 of the B subunit (2.55 Å apart) (Figure 2). These mutant enzymes have been successfully expressed, purified, and characterized, as described in this paper.
FIGURE 2.
Electrostatic partners which are in close proximity to the subunit interface residues, R194C (I) and K246E (II). The color of each residue corresponds to the color scheme of the ASL tetramer shown in Figure 1.
Materials and Methods
Materials
SAMP, HEPES, dithiothreitol (DTT), RNase, DNase, and imidazole were purchased from the Sigma Chemical Company. Sephacryl S-200HR was obtained from Pharmacia. Bio-Rad dye concentrate was purchased from Bio-Rad Laboratories. All other reagents were from Fisher Scientific and were of reagent grade. DNA sequencing primers and oligonucleotides for site-directed mutagenesis were obtained from Biosynthesis, Inc. The QuikChange Site-Directed Mutagenesis kit was purchased from Stratagene, and the QIAprep Spin Miniprep kit was supplied by QIAGEN. E. coli Rosetta 2 (DE3) (pLysS) strain was purchased from EMD Bioscience.
Site-directed mutagenesis, enzyme expression, and purification
The Stratagene QuikChange site-directed mutagenesis kit was used to introduce single base mutations to the pETN25HASL plasmid which encodes human adenylosuccinate lyase (15). The oligonucleotides shown below and their complementary sequences were used to introduce the mutations into the cDNA: R194C, GAT GAC CTG TGC TTC CGG GGA GTA AAG GGT ACC; K246E, G CAG ACA TAT ACA CGA GAA GTG GAT ATT G; L311V, GCC CGC CAC GTG ATG ACC CTT GTC ATG GAC; R396H, GGA GGT AGC CAT CAG GAT TGC CAT GAG AAA ATC; and R396C, GGA GGT AGC TGC CAG GAT TGC CAT GAG AAA ATC. The cDNA was extracted and purified using QIAprep spin miniprep kit. The mutations were confirmed by DNA sequencing, which was performed at the University of Delaware Center for Agricultural Biotechnology using an ABI Prism model 377 DNA sequencer (PE Biosystems).
The WT and mutant enzymes were expressed as N-terminal His6-tagged proteins in E. coli Rosetta 2 (DE3) (pLysS) strain. The WT and mutant enzymes were purified to homogeneity using chromatography on Qiagen nickel nitrilotriacetic acid-agarose (15). The purity of the enzymes was assessed electrophoretically using 12 % polyacrylamide gels containing 0.1 % sodium dodecyl sulfate (16).The protein concentration was determined by absorbance at 280 nm using E2801% = 14.1 (7). After purification, the enzyme was aliquoted and stored at -80 °C in 50 mM potassium phosphate buffer, pH 7.0, containing 150 mM KCl, 1 mM EDTA, 1 mM DTT, and 10% glycerol (Enzyme Storage Buffer).
Kinetics
The activity of the preincubated WT or mutant enzymes was measured continuously by assaying (over a 1 min period) at 25 °C in 50 mM HEPES buffer, pH 7.4. The activity was measured from the decrease in absorbance at 282 nm as 60 μM SAMP is converted to AMP and fumarate (Standard Assay). The assay was linear over the 1 min period. The difference extinction coefficient of 10,000 M-1 cm-1 between SAMP and AMP was used to calculate the specific activity, which was expressed in μmol of substrate converted/min/mg of enzyme used (17).
The activities of the WT and mutant enzymes in fresh Enzyme Storage Buffer, pH 7.0, were measured by preincubating the enzymes (∼0.3 mg/mL) at 25 °C for 1 hr and then assaying for 1 min under Standard Assay conditions. Since the data did not obey simple Michaelis-Menten kinetics, they were analyzed using the Hill equation,
where n = Hill coefficient, and K0.5 = the substrate concentration yielding half the Vmax. The K0.5 values were determined by varying the SAMP concentrations (0.5 – 10 μM).The standard error (S.E.) estimates were obtained from the SigmaPlot software (SPSS Inc., Chicago, IL). Frozen enzyme samples were preincubated at 25 °C for ∼1 hr prior to any experiment since it has been shown both for the B. subtilis ASL (18) and for human ASL (15) that the restoration of full activity is a slow process.
Thermal stability
WT and mutant enzyme samples (0.35 mg/mL) in fresh Enzyme Storage Buffer, pH 7.0, were incubated at 37 °C for ∼75 hrs. Aliquots were withdrawn periodically and assayed for 1 min under Standard Assay conditions at 25 °C. All the enzyme samples were preincubated at 25 °C for ∼1 hr before being transferred to 37 °C for stability measurements. The kobs value for each reaction was determined from the slope of ln [(Et — E∞)/ (E0 — E∞)] versus time. For each case, E∞ = f E0, where E∞ is the limiting residual activity and f is the fraction of the limiting activity to the original activity. In order to find values for f, the data were fitted to Et/E0 = (1-f) e-kt + f, where Et, and E0 are the enzymatic velocities at a given time, t, and 0 time. At the end of the period of incubation at 37 °C, the concentrations of the enzyme samples were measured; there were no appreciable changes in the concentrations.
WT and mutant enzyme samples (0.35 mg/mL) in fresh Enzyme Storage Buffer, pH 7.0, were also incubated at the harsher temperature of 60 °C. Aliquots were removed periodically and assayed for 1 min under Standard Assay conditions at 25 °C. All the enzyme samples were preincubated at 25 °C for ∼1 hr before being transferred to 60 °C for stability measurements. The kobs value for each reaction was determined from the slope of ln (Et/E0) versus time.
Free SH analysis of WT, R396C, and R194C enzymes at 25 °C, 37 °C and 60 °C
WT and mutant enzyme samples (∼0.8 mg/mL) in fresh Enzyme Storage Buffer, pH 7.0, were incubated at 37 °C for ∼18 hr and separate enzyme samples of WT and R194C were incubated at 60 °C. Each sample was preincubated at 25 °C for ∼1 hr prior to transferring to the higher temperature water bath. Aliquots (500 μL) were withdrawn after each incubation and passed through a Sephadex G-25 column (5 mL), which was equilibrated with 50 mM sodium phosphate buffer, pH 7.0, containing 300 mM NaCl, to remove EDTA and DTT. Analysis of free SH groups was conducted at pH 7.0 and 25 °C as follows: an aliquot of the eluted enzyme sample was mixed with 10 % SDS in the same buffer as the enzyme (yielding a final concentration of 1 % SDS) and incubated at 25 °C for 2 min. Subsequently DTNB in the same buffer as the enzyme (final concentration of 1 mM), was added to the mixture and incubated at 25 °C for a further 2 min. The absorbance of the mixture was measured at 412 nm against a blank containing buffer, SDS, and DTNB. For each enzyme the absorbance at 280 nm was measured before and after the Sephadex G-25 column and aliquots were tested for activity under Standard Assay conditions. The number of free SH groups was calculated as follows:
where εDTNB is 14,150 M-1cm-1 (19), l is the path length, which is 1 cm, and C is the molar enzyme subunit concentration (subunit molecular weight is 57000).
Molecular Weight Determination using Analytical Ultracentrifugation (AUC)
Sedimentation equilibrium (SE) experiments of the WT and mutant enzymes were conducted using a Beckman Coulter ProteomLab™ XL-I analytical ultracentrifuge equipped either with an An-60Ti or an An-50Ti analytical rotor. Samples (∼0.35 mg/mL) were centrifuged at 11,000 rpm at either 25 °C or 37 °C and after equilibrium was reached (∼17 hr), stepwise radial scans were performed at 280 nm, using a step size of 0.001 cm (20). Initially, the equilibrium time was determined by scanning at 5 hr intervals for ∼20 hrs. Data were analyzed using the SEDPHAT program (21). In all cases the experimental data were globally fitted, assuming that there is a continuous distribution of non-interacting species, to all the possible theoretical models of an oligomer in equilibrium with species of different molecular weight. The models that gave best fits were chosen as representative of that sample. The density of the buffer at 25 °C and 37 °C was calculated using the SEDNTERP program (22). The partial specific volume of human ASL at 25 °C and 37 °C is 0.7369 and 0.7419, respectively. Prior to each SE experiment, the WT and mutant enzyme samples of 0.35 mg/mL were preincubated at 25 °C for ∼1-2 hrs until the maximum specific activity was reached. For the 37 °C experiments, the mutant and WT enzyme samples of 0.35 mg/mL in fresh Enzyme Storage Buffer, pH 7.0 were incubated at 37 °C for ∼60 hrs. Aliquots were withdrawn periodically (0 hr, 26 hrs, and 58 hrs) and SE experiments were performed at 37 °C.
Molecular Weight Determination using Gel Filtration Chromatography
A Sephacryl 200HR (1 × 88 cm) column was used and before loading each sample, the column was equilibrated with fresh Enzyme Storage Buffer, pH 7.0, at room temperature (∼22 °C). As soon as possible after each protein was purified, 500 μL of 3 mg/mL sample was loaded onto the column. Fractions (1 mL) were collected and the absorbance of the fractions was measured at 280 nm. Molecular weight standards from Amersham Biosciences were used to calibrate the column.
For WT and K246E enzymes, fractions designated in Results were pooled and concentrated. The absorbance of each pool was measured at 280 nm. The activities of the WT and mutant enzymes in fresh Enzyme Storage Buffer, pH 7.0, were measured by preincubating the enzymes at 25 °C and then assaying for 1 min under Standard Assay conditions. The K0.5 values were determined by varying the SAMP concentrations (0.5 – 10 μM).The standard error (S.E.) estimates were obtained from the SigmaPlot software (SPSS Inc., Chicago, IL).
Circular Dichroism Spectroscopy
The secondary structure of the WT and mutant enzymes was assessed using CD spectroscopy. Ellipticity was measured on an Aviv - 400 spectropolarimeter from 200 to 250 nm, in 1 nm increments using a 0.1 cm quartz cuvette. The samples were scanned three times, averaged, and the background from the buffer (50 mM KPO4 buffer, pH 7, containing 150 mM KCl, 1 mM EDTA, 1 mM DTT, and 10% glycerol) was subtracted. Final protein concentrations were measured using the Bio-Rad protein assay, which is based on the method of Bradford (23), with pure WT ASL as the protein standard. The mean molar residue ellipticity [θ] (deg cm2dmol-1) was calculated from the equation [θ] = θ/10 nCl, where θ is the measured ellipticity in millidegrees, C is the molar concentration of enzyme subunits, l is the path length in centimeters, and n is the number of residues per subunit (503, including the His6 tag and thrombin cleavage site). The WT and mutant enzyme samples of ∼0.34 mg/mL in Enzyme Storage Buffer, pH 7.0, were incubated at 25 °C for 30 min prior to acquiring CD data. The ellipticity of the samples was measured at 25 °C.
The WT enzyme sample (∼0.34 mg/mL) in Enzyme Storage Buffer, pH 7.0, were preincubated at 25 °C for 30 min and transferred to a 37 °C water bath. Aliquots were withdrawn periodically (0 hr, 25 hrs, and 48 hrs) and the ellipticity of the WT sample was measured at 37 °C.
Fluorescence Spectroscopy
The steady-state fluorescence spectra of WT and mutant enzymes in fresh Enzyme Storage Buffer, pH 7.0, were measured on a Perkin-Elmer MPF-3 fluorescence spectrometer either at 25 °C or at 37 °C. The enzyme solutions were excited at 290 nm, which is specific for tryptophan residues, and the emission spectra were scanned and recorded in the range of 300 – 390 nm. The spectra were corrected for the background contributed by the buffer. Prior to each fluorescent experiment, the WT and mutant enzyme samples were preincubated at 25 °C for ∼1-2 hrs until the maximum specific activity was reached. At the end of the period of incubation, either at 25 °C or at 37 °C, the concentrations of the enzyme samples were measured; there was no appreciable change in the concentration.
The WT and mutant enzyme samples of ∼0.34 mg/mL in Enzyme Storage Buffer, pH 7.0, were incubated in a cuvette that was thermostated either at 25 °C or at 37 °C for ∼70 hrs. Periodically, fluorescence spectra were obtained. The kFobs value for the reaction was determined from the slope of ln[(Ft — F∞)/ (F0 — F∞)] versus time. For each enzyme, F∞ = f F0, where F∞ is the limiting residual fluorescence intensity and f is the fraction of the limiting fluorescence intensity to the original fluorescence intensity. In order to find values for f, the data were fitted to Ft/F0 = (1-f) e-kt + f, where Ft and F0 are the fluorescence intensity of the enzyme at a given time, t, and 0 time, respectively.
The WT enzyme samples of ∼0.34 mg/mL in Enzyme Storage Buffer, pH 7.0, that has no glycerol were incubated separately at 25 °C and 37 °C for ∼1hr. Then a final concentration of 6 M guanidine hydrochloride (Gdn.HCl) was added to each sample and the fluorescence spectrum at 25 °C and 37 °C was scanned and recorded. The fluorescence spectra of the same concentration of free Trp (subunit concentration × 5) in Enzyme Storage Buffer, pH 7.0, at 25 °C and 37 °C were scanned and recorded.
Results
Expression and purity of the human ASL WT and mutants
WT and all the mutants were expressed and purified to homogeneity by the methods described previously (15) and the purity was assessed using SDS-PAGE (data not shown). Each His-tagged enzyme exhibited a single subunit band with the expected subunit molecular weight of ∼57,000.
Kinetics at 25 °C
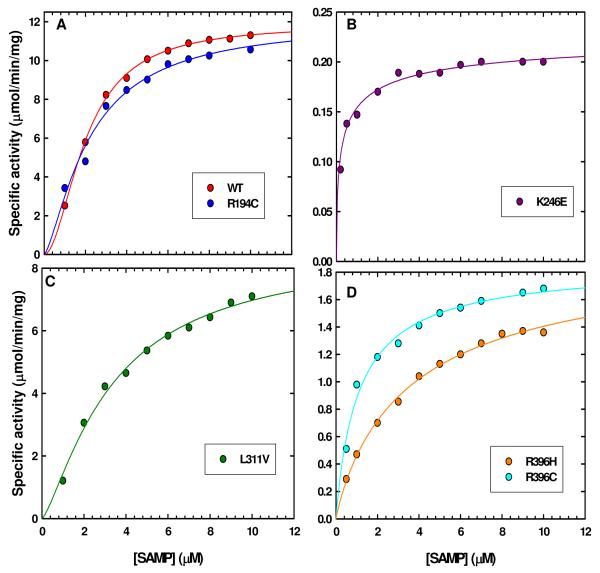
All of the disease-associated mutant enzymes had measurable catalytic activity and could be characterized kinetically. Kinetic data did not obey the simple Michaelis-Menten equation; therefore, they were analyzed using the Hill equation and the plots of velocity vs [SAMP] for WT and all the mutant enzymes are illustrated in Figure 3. The data at pH 7.4 in the direction of AMP formation for the WT and the mutant enzymes are summarized in Table 1.
FIGURE 3.
Specific activity (ν) vs [SAMP] plots of WT and the mutant enzymes. (A) Kinetic plots of WT and R194C mutant enzymes. (B) Kinetic plot of K246E. (C) Kinetic plot of L311V mutant enzyme. (D) Kinetic plots of R396 mutant enzymes.
Table 1.
Kinetic parameters of human ASL enzymes at 25 °C
| Enzyme |
Vmax ± S.Ea (μmol/min/mg) |
K0.5 ± S.Ea (μM) |
Hill coefficienta |
|---|---|---|---|
| WT | 11.9 ± 0.1 | 2.02 ± 0.04 | 1.9 ± 0.3 |
| R194C | 12.0 ± 0.9 | 2.2 ± 0.3 | 1.3 ± 0.2 |
| K246E | 0.23 ± 0.03 | 0.8 ± 0.2 | 0.5 ± 0.1 |
| L311V | 8.6 ± 0.7 | 3.4 ± 0.5 | 1.3 ± 0.1 |
| R396H | 1.9 ± 0.2 | 3.8 ± 0.8 | 0.9 ± 0.2 |
| R396C | 2.0 ± 0.2 | 1.1 ± 0.2 | 0.9 ± 0.1 |
The Vmax, K0.5, and Hill coefficient values were determined by varying the concentration of SAMP and fitting the data to the Hill equation using Sigma Plot. The values are shown along with their standard errors.
The kinetic data indicate that WT enzyme has a Hill coefficient of 1.9, suggesting positive cooperativity (Figure 3A). The Vmax values for the two mutant enzymes which are in the subunit interface (R194C and K246E) are very different from each other. The R194C mutant enzyme has a Vmax comparable to that of WT enzyme, whereas the Vmax for K246E mutant enzyme is only ∼2 % that of the WT enzyme. This result suggests that the charge reversal in the K246E mutant enzyme has greater repulsion than in the R194C mutant enzyme, leading to a less active enzyme. The K0.5 value for R194C is comparable to that of WT enzyme, while the K0.5 value for K246E is 50 % that of WT enzyme. Furthermore, the Hill coefficient for R194C mutant enzyme is 1.3 (Figure 3A), indicating that R194C mutant enzyme retains some positive cooperativity, albeit reduced. In contrast, the Hill coefficient of the charge reversal mutant enzyme K246E is 0.5 (Figure 3B) suggesting that it has completely lost its positive cooperativity; it may be that this enzyme is composed of various oligomeric species with different substrate affinities or alternatively, this mutant enzyme may consist of one species in which binding of substrate to the first site decreases the affinity of substrate for other enzyme sites.
The Vmax for L311V and R396H,C mutant enzymes are ∼72 % and ∼16 %, respectively, that of WT enzyme indicating that replacing position 396 either with Cys or His greatly affects the rate of the reaction. The K0.5 values for L311V and R396H mutant enzymes are comparable to that of WT enzyme, whereas the K0.5 value for R396C mutant enzyme is 50 % that of WT enzyme. The Hill coefficients indicate that the L311V mutant enzyme retains positive cooperativity although it is reduced (Figure 3C), while the two R396 mutant enzymes lose cooperativity (Figure 3D). These data suggest that even though L311V and the two R396 mutant enzymes are not in the subunit interface, they affect the rate of cleavage of the substrate SAMP.
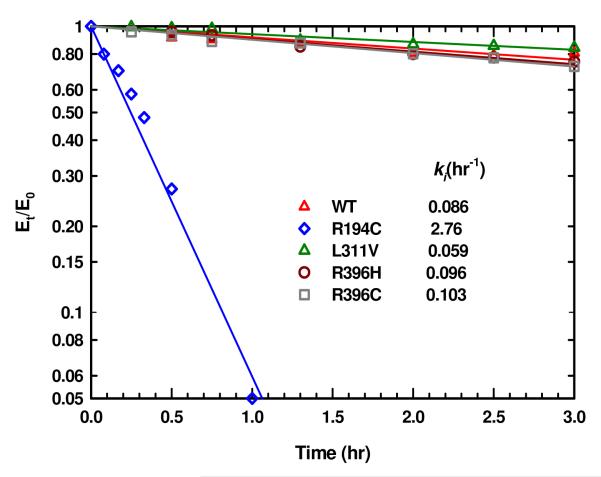
Thermal Stability at 37 °C
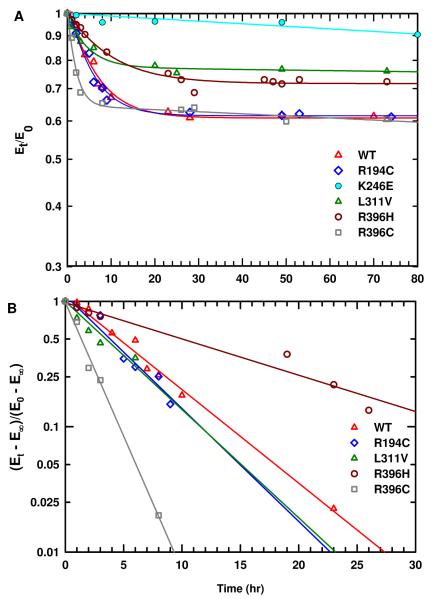
Mutant enzymes may be intrinsically defective in their kinetic parameters or may be particularly unstable at 37 °C while they are stable at 25 °C. Therefore, the stability of these ASL-deficiency associated point mutations as well as wild type enzyme was evaluated at 37 °C (to mimic the physiological conditions). The time-dependent inactivation plots are illustrated in Figure 4A. It is evident that within the first 15 hrs of preincubation at 37 °C there is a decrease in activity for each enzyme (except for the K246E mutant enzyme). Each of the enzymes reaches a limiting activity which does not change either by adding fresh DTT (final concentration of 1 mM) or by transferring the enzyme samples to a 25 °C water bath. These observations imply that the enzyme changes over time to another less active form, which is stable at 37 °C, and this process is irreversible. In contrast, K246E mutant enzyme had the lowest activity (2 % that of WT enzyme) and did not change appreciably at that temperature.
FIGURE 4.
(A) Time dependent thermal inactivation plots (Et/E0 vs time) of WT and the mutant enzymes at 37 °C. (B) The plots of ln [(Et — E∞) /(E0 — E∞)] vs time at 37 °C. The slope of each plot yields the kobs of each enzyme. The E0 and Et, are the enzymatic velocities at time 0 and time t, respectively. The E∞ is the limiting activity of each enzyme and E∞ = f E0 where f is the fraction of the limiting activity to the original activity of the enzyme.
The kobs value for the reaction at 37 °C was determined from the slope of ln [(Et — E∞)/ (E0 — E∞)] versus time as shown in Figure 4B. For each plot E∞ = f E0, where E∞ is the limiting residual activity, f is the fraction of the limiting activity to the original activity, and E0 and Et are, respectively, the enzymatic velocities at time 0 and the measured time, t. The f, E0, E∞, and kobs for each enzyme are summarized in Table 2. The kobs values for R194C and L311V mutant enzymes are comparable to that of WT enzyme, whereas the kobs value for R396H mutant enzyme is about half that of WT enzyme. In contrast, the kobs value for R396C mutant enzyme is ∼3 times that of WT enzyme. This result suggests that human ASL adapts different conformations when the amino acid at position 396 is changed either to Cys or His. The limiting activities, E∞, of R194C and L311V are comparable to that of WT. On the other hand, the E∞ of R396C and R396H mutant enzymes is only ∼15 % that of WT (Table 2).
Table 2.
Thermal inactivation and fluorescence data at 37 °C of human ASL enzymes
| Enzyme | Thermal inactivation studies a |
Fluorescence studies b |
|||
|---|---|---|---|---|---|
|
E0 (μmol/min/ mg) |
E∞ (μmol/min/mg) |
f |
kobs (hr-1) |
kFobs (hr-1) |
|
| WT | 11.3 | 6.8 | 0.61 | 0.16 ± 0.03 | 0.19 ± 0.02 |
| R194C | 10.8 | 6.5 | 0.61 | 0.20 ± 0.05 | 0.22 ± 0.03 |
| K246E | 0.45 ± 0.04 | ||||
| L311V | 8.5 | 6.6 | 0.77 | 0.22 ± 0.04 | 0.22 ± 0.03 |
| R396H | 1.37 | 1.0 | 0.72 | 0.10 ± 0.02 | 0.07 ± 0.03 |
| R396C | 1.47 | 0.9 | 0.64 | 0.55 ± 0.10 | 0.47 ± 0.02 |
The WT and mutant enzymes (∼0.34 mg/mL) in fresh Enzyme Storage Buffer, pH 7, were separately incubated at 37 °C for ∼73 hrs.
Periodically aliquots (20 μL) were withdrawn and assayed at 25 °C under Standard Assay conditions. E0 = enzymatic velocity at time 0, E∞ = limiting enzymatic velocity, and f = fraction of the limiting activityto the original velocity. In each case at 37 °C E∞ = f E0 and kobs = slope of ln [(Et — E∞)/ (E0 — E∞)] versus time plot.
Periodically, the fluorescence intensity at 37 °C was measured at emission λmax 337 nm. For each enzyme kFobs = slope of ln [(Ft — F∞)/ (F0 — F∞)] versus time plot, as shown in Figure 9B.
Since WT and the mutant enzymes are relatively stable at 37 °C we sought to evaluate the in vitro stability of these ASL-deficiency associated mutations under harsher conditions at 60 °C. WT, R396H, and R396C enzymes are relatively stable at 60 °C for about four hours (Figure 5); then they start to form insoluble aggregates. The L311V mutant enzyme is stable for a longer period of time (∼24 hrs) without forming any insoluble aggregates. In contrast, R194C mutant enzyme loses most of its activity within about one hour of incubation at 60 °C (Figure 5) suggesting that, although it is catalytically active and stable at 37 °C, R194C may be the most thermally unstable enzyme.
FIGURE 5.
Time dependent thermal inactivation plots (Et/E0 vs time) of WT and the mutant enzymes at 60 °C. It is assumed that Et/E0 goes to zero in calculating the rate constants for inactivation.
Free SH group analysis of WT, R194C and R396C enzymes
Human ASL has thirteen Cys residues in each subunit. According to the crystal structure, R396 is in close proximity to C399 of the same chain: the distance between the Cα of R396 and Cα of C399 is ∼5 Å, which is a common distance for disulfide bonds in protein (24). We considered that disulfide bond formation might account for the greater lability at 37 °C of R396C than the other mutant enzymes. The possibility of disulfide formation was investigated by analyzing the number of free SH groups in R396C mutant enzyme initially at 25 °C and after preincubating at 37 °C for ∼18 hr. At 25 °C, the number of free SH groups for WT and R396C was measured as 12.8 ± 0.2 and 13.7 ± 0.1, respectively, consistent with the extra cysteine present in the R396C mutant enzyme. Preincubation of either WT or R396C enzymes at 37 °C did not change the measured number of free SH groups (13.1 ± 0.3 and 14.3 ± 0.3, respectively) suggesting that there is no disulfide bond formation in R396C mutant enzyme during the 37 °C treatment.
Since the R194C mutant enzyme is the least stable enzyme at 60 °C, it seemed possible that oxidation of the Cys may be a cause for instability. This possibility was evaluated by measuring the number of free SH groups before and after incubation at 60 °C. For comparison, the R194C mutant enzyme was separately incubated at 25 °C and 37 °C for ∼18 hr and the number of free SH groups was measured. No change was found in the number of free SH groups either at 25 °C or 37 °C (13.9 ± 0.2 and 14.1 ± 0.4, respectively). However, when the enzyme was transferred to 60 °C and incubated for 10 min, the number of free SH groups for R194C mutant enzyme decreased to 8.9 ± 0.3, suggesting that disulfide bonds likely form. In contrast, incubation of WT enzyme at 60 °C for 20 min resulted in the smaller decrease of free SH groups to 11.4 ± 0.3 from its initial value of 13.0 ± 0.3, implying that there is less formation of disulfide bonds in WT enzyme. We conclude that the greater thermal instability of R194C at 60 °C is due to the more rapid oxidation of its cysteines under these conditions.
Molecular weights of the human ASL WT and mutants
At 25 °C
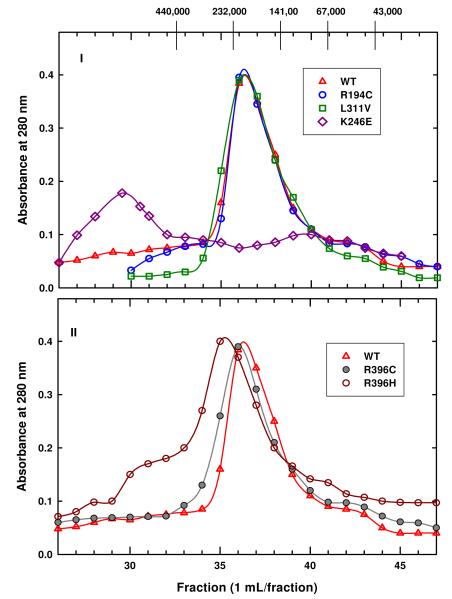
Since the kinetic parameters of the mutant enzymes at 25 °C are different from that of the WT enzyme, we sought to evaluate the effect of these disease-associated mutations on the oligomeric state of the enzyme at 25 °C using gel filtration. The elution profile of WT enzyme (Figure 6) indicates that this enzyme exists predominantly as tetramer (∼223 kDa) with some monomeric species.
FIGURE 6.
Gel filtration elution profiles of WT and mutant enzymes at 25 °C. These elution profiles were obtained by applying 500 μL of each protein (∼3 mg) to a Sephacryl 200HR column (1 × 88 cm) equilibrated with Enzyme Storage Buffer, pH 7.0. The molecular weight standards used are V0, blue dextran (30 mL); Ferritin-440 kDa; Catalase-232 kDa; Yeast alcohol dehydrogenase-141 kDa; Albumin- 67kDa; Ovalbumin-43 kDa. (I) The elution profiles of WT, R194C and L311V enzymes. (II) The elution profiles of WT, R396H and R396C enzymes.
Elution profiles of the R194C, L311V, and K246E mutant enzymes are illustrated in Figure 6,I and the elution profiles of R396C and R396H mutant enzymes are shown in Figure 6,II. The molecular weight results for these mutant enzymes are summarized in Table 3, column 5. It is evident that the elution profiles of R194C and L311V mutant enzymes are similar to that of WT (Figure 6), suggesting that they exist predominantly as tetramer with small amounts of monomer. In contrast, K246E mutant enzyme has a peak around the void volume, which is > 440 kDa and a broad peak at ∼100 kDa while R396C and R396H mutant enzymes have maxima at ∼232 kDa and ∼276 kDa, respectively. However, the elution profiles of K246E, R396C, and R396H mutant enzymes span a wide range of molecular masses (Figure 6), indicating that these enzymes exist as mixtures of species.
Table 3.
Molecular weight parameters of human ASL enzymes
| Enzyme | At 25 °C |
At 37 °C |
||||
|---|---|---|---|---|---|---|
| Molecular weight by AUC a [kDa (%)] |
Molecular weight by gel filtration c (kDa) |
Molecular weight by AUC d [k Da(%)] |
||||
| M—Tet | D—Tet | D—12mer | M—Tet | M—14mer | ||
| WT | 58(20) | 223 | 58(32) | |||
| 230(80) | 227(68) | |||||
| R194C | 56(37) | 223 | 57(33) | |||
| 233(63) | 225(67) | |||||
| K246E | 119(94)b | >440 | 59(91) | |||
| 668(6) | 100 | 802(9) | ||||
| L311V | 57(40) | 223 | 57(32) | |||
| 232(60) | 229(68) | |||||
| R396H | 112(28)b | 276 | 68(34)e | |||
| 230(72) | 238(66) | |||||
| R396C | 56(32)b | 232 | 56(40)e | |||
| 238(68) | 228(60) | |||||
The molecular weight by AUC was measured at 25 °C, using ∼0.34 mg/mL sample in fresh Enzyme Storage Buffer, pH 7.0, at 11,000 rpm. M = monomer, D = dimer, Tet = tetramer, and the dash (—) is used to indicate that different species are in equilibrium.
The experimental AUC data of R396C, R396H, and K246E mutant enzymes at 25 °C can also be fit well by a M-5mer (30 %-70 %), D-5mer (31 %-69 %), and M-9mer (90 %-10 %) models, respectively.
The molecular weight measurement by gel filtration was conducted at ∼22 °C, using a Sephacryl S-200 column (1 × 82 cm), equilibrated with fresh Enzyme Storage Buffer, pH 7.0.
Each sample (∼0.34 mg/mL) was incubated separately at 37 °C, in fresh Enzyme Storage Buffer, pH 7.0 and the molecular weight by AUC was determined at 37 °C using 11,000 rpm; measurements were made at 17, 43, and 73 hr.
The experimental AUC data for R396C and R396H mutant enzymes at 37 °C can also be fit well by M-5mer (36 %-64 %) and D—5mer (25 % - 75 %) model, respectively.
Since the K246E mutant shows an overall negative cooperativity, various fractions (Figure 6) of the mutant enzyme were characterized kinetically to ascertain whether the kinetics change with the various oligomeric species present. As a control experiment fractions of the WT enzyme were characterized kinetically. The data at pH 7.4 in the direction of AMP formation are summarized in the Supplementary Data Section (Table 1S). For WT enzyme only fractions 35 – 38 had appreciable activity and this enzyme exhibited positive cooperativity.
For K246E mutant enzyme, fractions 26 – 31 and fractions 32 – 36 had measurable catalytic activity whereas fractions 37 – 43 was inactive. The Vmax of the two pools is similar to the overall value. However, according to the Hill coefficients, fractions 26 – 31 exhibits negative cooperativity (n = 0.5) while fractions 32 – 36 shows no cooperativity (n = 1). The enzyme in fractions 32 – 36 exhibits a single Km value of 0.8 μM, suggesting that this pool contains a single species of enzyme. In contrast, the enzyme eluting in fractions 26 – 31 shows two distinct Km values: Km1 = 0.2 μM and Km2 = 0.8 μM (like the unfractionated K246E enzyme) suggesting that this pool is likely to be a mixture of species or a single species with different affinities. Since we can separate pools on the basis of molecular weight that have different kinetic properties, it is likely that the 0.5 Hill coefficient of the overall enzyme indicates that the K246E mutant enzyme is a mixture of species.
Because the elution profiles of the enzymes from gel filtration chromatography show that each enzyme is a mixture of species, we used analytical ultracentrifugation (AUC) in order to quantify the species present in each mutant enzyme sample. A constant concentration of ∼0.34 mg/mL was used for all the enzymes and the experimental AUC data at 25 °C were globally fitted to various theoretical models for an oligomer in equilibrium with species of different molecular masses. Representative residuals, which illustrate goodness of fit for various models [e.g. monomer-tetramer (M-tet) or dimer-tetramer (D-tet)], are shown in the Supporting Information section (Figure 1S and 2S). Results of the molecular weight analyses are summarized in Table 3, columns 2-4.
According to the best fit models, WT and the mutant enzymes (except the K246E mutant enzyme) have a predominant amount of tetramer. When the WT enzyme exhibits a high Vmax (>10 μmol/min/mg), it has a high amount of tetramer (∼80 %) at 25 °C; while some WT enzyme batches, which exhibited Vmax ∼8.5 μmol/min/mg at 25 °C, had only ∼63 % tetramer along with monomeric species. The only best fit model for R194C and L311V is the monomer-tetramer model whereas R396C and R396H mutant enzymes have multiple best fit models. The amount of tetramer for R194C and L311V enzymes is ∼63 % and ∼60 %, respectively, along with monomer. The best fit models for R396C indicate that it exists as ∼68 % tetramer along with monomer while R396H mutant enzyme also exists predominantly as tetramer (∼72 %), but in equilibrium with dimeric species. However, experimental AUC data of R396C and R396H mutant enzymes can also be fit with theoretical models of monomer-5mer and dimer-5mer, respectively, suggesting that the mutations at R396 position destabilize the tetramer. In contrast, the best fit models of K246E mutant enzyme indicate that the majority of the protein is present as a dimer (∼90 %) with small amounts of soluble aggregates. It is clear that the K246E mutation, which is in the subunit interface, greatly facilitates the dissociation of the tetramer, while the other subunit interface mutant enzyme R194C has little effect on the dissociation of the complete enzyme.
At 37 °C
Since 37 °C approximates the physiological temperature, the effect of these disease-associated mutations on the oligomeric state of the human enzyme at 37 °C was assessed using AUC. A constant concentration of ∼0.34 mg/mL of WT and mutant enzymes were preincubated at 37 °C and sample aliquots were removed at 17, 43, and 73 hr in order to conduct sedimentation equilibrium experiments at 37 °C. Since the AUC results at the three time periods were similar, the results were averaged and are shown in Table 3, columns 6 and 7.
It is interesting to note that at 37 °C the size distribution of the mutant enzymes is similar to that of 25 °C. The experimental AUC data for WT and the mutant enzymes, except K246E mutant enzyme, indicate that they have major amounts of tetramer along with some monomer. However, experimental AUC data of R396C and R396H mutant enzymes fit equally well with the monomer-5mer and dimer-5mer theoretical models, respectively. According to the best fit models, the WT and the mutant enzymes have ∼60 – ∼68 % tetramer. These values correspond well with the limiting Et/E0 values of 0.60 – 0.72 (Table 2), suggesting that the limiting activity is due to the residual tetramer present. At 37 °C the charge reversal subunit interface mutant enzyme, K246E, exists predominantly as monomer (∼90 %) with small amounts of soluble aggregates, consistent with the extremely low specific activity of this enzyme (Tables 1,2).
Circular Dichroism Spectroscopy
At 25 °C
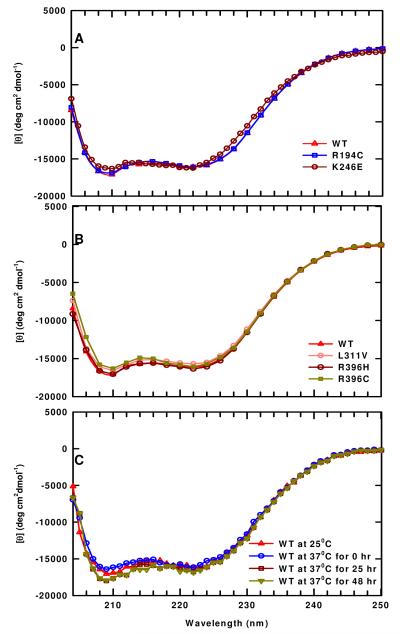
CD spectroscopy was used to evaluate any changes in the secondary structure of the mutant enzymes. A constant concentration of 0.34 mg/mL was used and the CD spectra of all the mutants and WT enzyme exhibit minima at 210 and 222 nm, which is typical of proteins containing appreciable amounts of α-helix. The CD spectra (Figure 7A,B) indicate that all of these mutants have appreciable amounts of α-helix with minimal changes in the secondary structure.
FIGURE 7.
CD spectra of WT and mutant enzymes. These spectra were determined with the protein (∼0.34 mg/mL) in Enzyme Storage Buffer, pH 7.0. (A) The CD spectra of WT enzyme and of R194C and K246E mutant enzymes at 25 °C. (B) The CD spectra of WT enzyme and of L311V, R396C, and R396H mutant enzymes at 25 °C. (C) The CD spectra of WT enzyme at 37 °C.
At 37 °C
Since, at 37 °C, the activity of WT enzyme decreases to a limiting value (Figure 4A, Table 2), we sought to evaluate any changes in its secondary structure using CD spectroscopy. The CD spectra obtained upon incubation of WT enzyme at 37 °C over 48 hrs are illustrated in Figure 7C. After 25 hrs incubation, a small change is observed at 209 nm, which does not change further (48 hr). These results show that when the WT enzyme reaches its limiting activity it has slightly more α-helical structure than at 25 °C, suggesting that there is a small change in the secondary structure under these conditions.
Fluorescence spectroscopy of the human ASL and mutant enzymes
Since the changes in the CD spectra are small, we considered that tryptophan fluorescence spectroscopy at 25 °C and 37 °C might provide a more sensitive method to detect conformational changes in human ASL enzyme caused by these disease-associated point mutations. Additionally, tryptophan fluorescence spectroscopy was used to assess the structural changes of WT enzyme in the presence of 6 M Guanidine hydrochloride (Gdn.HCl) at 25 °C and 37 °C.
At 25 °C
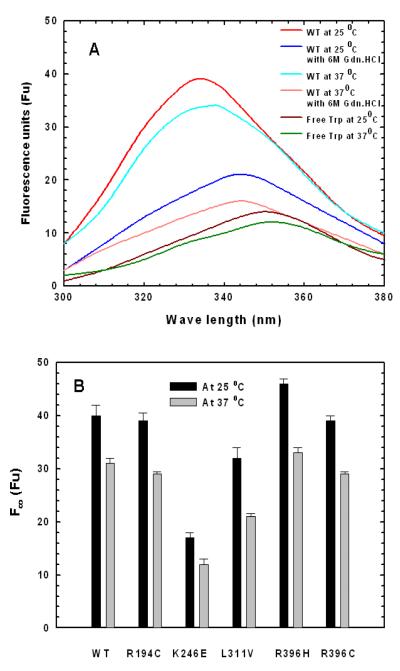
The emission λmax of free Trp is ∼350 nm and its fluorescence spectrum in the presence and absence of 6 M Gdn.HCl are very similar at 25 °C and 37 °C. Therefore, a representative of each temperature is shown in Figure 8A. Proteins with completely buried Trp show an emission λmax of 320 nm (25). In contrast, the fluorescence spectrum of native WT enzyme at 25 °C (Figure 8A) indicates a blue shift in emission λmax (with respect to free Trp) to 335 nm suggesting that most of the Trp residues in the enzyme are internalized. However, in the presence of 6 M Gdn.HCl the λmax shifts to 345 nm indicating that the enzyme is unfolded and some of the Trp residues are solvent exposed. It is evident that there is a large enhancement in fluorescence intensity of native WT enzyme relative to free tryptophan and unfolded WT enzyme. These observations suggest that the local environment of Trp residues in the native WT enzyme is predominantly non-polar (which agrees with the crystal structure). Similar fluorescence enhancement is observed in the folded Transmembrane Protein OmpA (26).
FIGURE 8.
The fluorescence spectra of WT and mutant enzymes at 25 °C and 37 °C were determined with the protein (∼0.33 mg/mL) in fresh Enzyme Storage Buffer, pH 7.0. (A) The fluorescence spectra of WT enzyme at 25 °C and 37 °C in the absence and presence of 6 M Gdn.HCl and the fluorescence spectra of free Trp in the Enzyme Storage Buffer, pH 7.0. (B) Limiting fluorescence intensities of WT and mutant enzymes at 25 °C (measured at 335 nm) and at 37 °C (measured at 337 nm).
At 25 °C, the fluorescence intensity of native WT and mutant enzymes did not change appreciably during the ∼70 hr incubation time. The results were averaged and are shown in Figure 8B. All the mutant enzymes also exhibit an emission λmax of 335 nm. The fluorescence intensities of R194C and R396C mutant enzymes are comparable to that of native WT. However, the fluorescence intensities of K246E and L311V mutant enzymes are decreased, while there is an enhancement in the fluorescence intensity of R396H mutant enzyme (∼42 %, ∼75 %, and ∼115 %, respectively) as compared with that of native WT enzyme. The results imply that the overall structures of the R194C and R396C mutant enzymes are similar to that of native WT enzyme, while the overall conformations of the L311V, R396H and K246E mutant enzymes are altered.
At 37 °C
There is a 2 nm red shift in the emission λmax of the native WT enzyme as the temperature is increased from 25 °C to 37 °C (Figure 8A) indicating that there is slight increase in solvent exposure of the Trp residues at 37 °C. Furthermore, there is a large enhancement of the fluorescence intensity in native WT enzyme at 37 °C as compared to that of free Trp (∼258 % of free Trp) and unfolded WT enzyme but less than that of native WT enzyme at 25 °C. The results imply that the highly non-polar local environment of Trp in ASL at 25 °C decreases slightly as the temperature is increased to 37 °C.
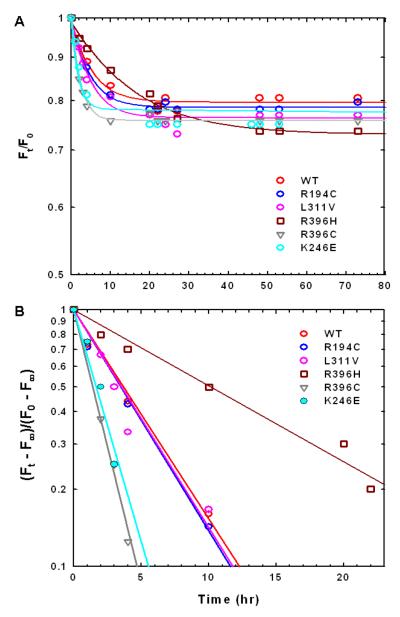
Time dependent fluorescence intensity changes in native WT and the mutant enzymes at 37 °C are shown in Figure 9A. The fluorescence intensities of all the enzymes, including WT, decrease initially and reach a limiting fluorescence intensity as illustrated in Figure 9A. The kFobs values for the fluorescence changes were determined from the slope of ln [(Ft — F∞)/ (F0 — F∞)] versus time (Figure 9B) and are reported in the last column of Table 2. The kFobs values are similar to the rate constants obtained from the thermal stability studies at 37 °C (Table 2, column 5), indicating that the change in fluorescence measures a subtle conformational change, which is responsible for the partial inactivation at 37 °C. The limiting fluorescence intensity of R194C, R396C, and R396H is similar to that of native WT enzyme at 37 °C (as shown in Figure 8B) suggesting that the overall conformation of those mutant enzymes is similar to that of WT enzyme. In contrast, the limiting fluorescence intensity of K246E and L311V mutant enzymes decreases to ∼39 % and ∼71 %, respectively, that of WT enzyme at 37 °C, implying that their overall structures are different from that of WT enzyme.
FIGURE 9.
(A) Time dependent fluorescence change at 37 °C (Ft/F0 vs time) measured at 337 nm of WT and mutant enzymes. (B) The plots of ln [(Ft — F∞) /(F0 — F∞)] vs time and the slope of each plot yields the kFobs of each enzyme. The F0 and Ft, are the fluorescence intensities at time 0 and time t, respectively. The F∞ is the limiting fluorescence intensity of each enzyme.
DISCUSSION
Human adenylosuccinate lyase deficiency is an autosomal recessive disorder which results in autistic features, muscle wasting, and various magnitudes of mental retardation. Most of these ASL-deficiency-associated mutations occur in the central helical region that seems to be important for the stability of the tetramer (12). In the present study we evaluated five disease-associated point mutations (R194C, K246E, R396C, R396H, and L311V) from various parts of the enzyme structure (Figure 1 I).
For R194C mutant enzyme, the V at 25 °C, the rate constant for inactivation at 37 ° max C as well as the limiting activity and amount of tetramer at 37 °C are comparable to that of WT enzyme (Table 2,3). Furthermore, at 25 °C, the fluorescence intensity of R194C is comparable to native WT suggesting that overall conformation of R194C is comparable to native WT. When (A)R194 is mutated to Cys, the electrostatic attraction to (B)E464 is lost (Figure 2-I). The crystal structure indicates that there is another Arg residue, (A)R234, which is even closer to (B)E464 (3.17 Å apart) that could be stabilizing the A/B and C/D subunit interfaces (Figure 2-I). This (A)R234-(B)E464 interaction may be the reason why the R194C mutant enzyme is similar to WT enzyme at 25 °C and 37 °C. On the other hand, under harsh conditions such as 60 °C, the R194C mutant enzyme is the least stable enzyme. Preliminary data suggest that inclusion of the non-cleavable substrate analog, APBADP, in the incubation solution of R194C mutant enzyme at 60 °C provides appreciable stabilization of the enzyme. Thus, an appropriate nucleotide analog, by overcoming the instability of a disease-associated ASL mutant, may be useful in treating some types of ASL deficiency.
In contrast to R194C, the K246E mutant enzyme exhibits an extremely low Vmax (only 2 % that of WT enzyme) and exists as a mixture of monomer (90 %) and aggregates at 37 °C. The K246E mutant enzyme, which is a charge reversal, exerts severe repulsion between (A)K246E and its partner, (B)D182 (Figure 2-II). Furthermore, there are no other positively charged residues around (B)D182 to compensate for the absence of K246; hence a drastic destabilization of the human ASL tetramer is observed, yielding dimers or monomers. Since each catalytic site requires contributions of amino acid side chains from 3 subunits (6), it is clear why the K246E enzyme is so low in activity.
Residue L311 is in the central helical region towards the middle (Figure 1 I). At 25 °C, its Vmax is ∼76 % that of WT enzyme and the amount of tetramer is ∼60 %, which is lower than that of WT enzyme (∼80 %), while at 37 °C the amount of tetramer is similar to that of WT enzyme (∼68 %). Since Val is less hydrophobic than Leu (27), it might be expected that there would be weakening of the long range hydrophobic interactions in the L311V enzyme (27-29 ). Replacing Leu by Val (with its lower molar volume) increases the flexibility of the hydration shell (28, 29) which leads to some destabilization of the tetramer, perhaps decreasing somewhat the subunit interactions as reflected in the reduced Hill coefficient (n = 1.3). In addition, its decreased fluorescence intensity indicates that the L311V enzyme has an altered conformation which can account for its diminished catalytic activity.
Our experimental results show that R396C and R396H mutant enzymes exhibit substantially decreased residual activity (∼17 % that of WT enzyme) at 25 °C and 37 °C while maintaining similar amounts of tetramer as WT enzyme. Residue R396 is in close proximity to the entrance to the active site (Figure 1 I), where its side chain points towards the solvent. Many proteins have positively charged Arg residues around the active site entrance while the side chain faces toward bulk water (30). Water forms more ordered hydration shells around polar groups than the non-polar groups, leading to longer residence times of water (30). This ordering of water extends the proteins’ electrostatic surfaces well away from their physical surfaces (made up of amino acid side chains) and increases the capturing of visiting ligands (30), in this case the negatively charged SAMP or SAICAR. Any substitution for these positively charged Arg residues on the surface decreases the extended electrostatic surface, leading to diminished capturing of the substrate molecules and a decrease in activity, as in the R396H and R396C enzymes.
For wild type enzyme, the dependence of initial velocity on adenylosuccinate concentration provides the first evidence for kinetic cooperativity in this enzyme, probably because the kinetics were examined over a wider range of substrate concentration than previously. Also, we recently reported that direct binding of the non-cleavable substrate analog, APBADP, by human ASL shows positive cooperativity (7); we now conclude that human ASL shows positive cooperativity in kinetics as well as in substrate binding. The K0.5 data of these disease-associated point mutations of ASL indicate that there are only small changes in the affinity of the enzyme for its substrate whereas the greatest change is in the extent of cooperativity. These changes in the extent of cooperativity might be due to the conformational changes caused by the disease-associated point mutations that lead to a change in the degree of communication among the subunits.
Such conformational changes were investigated using tryptophan fluorescence spectroscopy, which is extremely sensitive to any changes in the molecular environment and polarity (25,31-33). In native human ASL there are five Trp in each subunit: (B)W45 is surface exposed; (B) W39, (B) W43 and (B) W175 are buried; and (B) W326 is shared between B/A and B/C subunit interfaces (Figure 1 II). However, the fluorescence emission intensity of native WT enzyme at 25 °C is much greater than that of free Trp and WT enzyme in the presence of 6 M Gdn.HCl at 25 °C, implying that the local environment (within 6 A) of Trp is predominantly non-polar.
The fluorescence intensity of R194C mutant enzyme at 25 °C is similar to that of WT enzyme indicating that the overall conformation of R194C mutant enzyme is comparable to that of WT enzyme. The two human ASL crystal structures indicate that there are subtle changes near Arg194 during catalysis; therefore, changing Arg at position 194 to a Cys leads to a slight destabilization of the subunit interface without changing either Vmax or the overall conformation, which reduces positive cooperativity as indicated by the Hill coefficient (n=1.3). A similar scenario has been documented for phenylalanine ammonia-lyase (34).
In contrast, K246E mutant enzyme (which exists as a mixture of monomer-dimer along with some aggregates) has the lowest fluorescence intensity and is similar to that of the unfolded WT enzyme at 25 °C. These observations suggest overall conformational changes in the mutant protein greatly influence the communication among the subunits as indicated by the plot of initial velocity vs [SAMP] of K246E mutant enzyme, which exhibits apparent negative cooperativity (i.e., n is <1). It is possible that one site of K246E mutant enzyme binds substrate with high affinity and decreases the binding affinity for the enzyme’s other sites (35). Alternatively, the apparent negative cooperativity can be attributed to the presence in the preparation of several species of active enzyme with different affinities for substrate (35, 36). For K246E mutant enzyme, the gel filtration data demonstrate that there are several species of the enzyme with different affinities for its substrate, so in this case that is probably the preferred explanation for the apparent negative cooperativity.
At 25 °C, as indicated by the fluorescence data the overall conformation of R396H mutant enzyme is different from native WT, whereas the overall conformation of R396C mutant enzyme is similar to that of native WT enzyme. According to the crystal structure, (A)R396 belongs to helix-19, where several residues in that helix have direct electrostatic interactions with the residues in subunits B and C and there is indirect stabilization of some of these inter-subunit interactions by (A)R396. Furthermore, a comparison between the two human ASL crystal structures (PDB # 2J91 and 2VD6) indicates that there are significant conformational changes in helix-19 after the catalysis of SAMP, which might be important for maintaining positive cooperativity. Changes in helix-19 may therefore cause destabilization of the tetramer and loss of inter-subunit communication, as indicated by the Hill coefficient (n = 1).
Most of the ASL deficient patients identified to date are compound heterozygotes (12). The mutations that we investigated in the present study are found paired in patients as follows: R396H/L311V (37) and R396C/R194C (9). These two patients have been described as having severe mental retardation while the two reported patients with the K246E mutation exhibit moderate to severe mental retardation (38). Although, the individual R194C and L311V mutant enzymes have comparable activity to WT enzyme, the genetic combination of R194C/R396C and L311V/R396H result in marked ASL deficiency. One or more of the following may explain the symptoms of these patients: 1. the protein might be a collection of hybrids (i.e., each tetramer is composed of some subunits of L311V and some of R396H) with kinetic and physicochemical characteristics different from those of either mutant alone or of the average of the two mutants; or 2. the expression of the mutant enzymes may be low. These possibilities may lead to a drastic decrease in catalytic activity, with resultant accumulation of S-Ado and SAICAriboside (the dephosphorylated substrates of SAMP and SAICAR) in high concentrations in the cerebrospinal fluid. These possibilities are currently being evaluated in this laboratory. It has been suggested that the accumulating S-Ado and/or SAICAriboside are toxic and may interact with the adenosine receptor in neurological tissues; however, experimental studies have not yet supported this possibility (13). Although the biochemical mechanism of central nervous system damage has not been established, there is a clear association of adenylosuccinate lyase deficiency with autistic features and mental retardation. It is anticipated that elucidation of molecular basis of ASL deficiency may lead to effective pharmacological treatment of the disease.
Concluding Remarks
This paper describes the study of five ASL-deficiency associated point mutations: K246E, R194C, L311V, R396C and R396H. We conclude that the most detrimental point mutation is the interfacial K246E mutant enzyme, which is observed as a monomer or dimer, but no tetramer, and has a residual activity of only 2% that of WT enzyme with no positive cooperativity. In contrast, the interfacial R194C mutant enzyme is very similar to WT enzyme in kinetic properties, and state of oligomerization at 25 °C and 37 °C; however, it exhibits marked thermal instability, which is best observed at 60 °C. The L311V mutation (located in the central helical region) results in an enzyme with a Vmax of 72% that of WT enzyme; however its state of oligomerization at 25 °C and 37 °C is similar to WT and shows conformational changes along with decreased positive cooperativity. Finally, the two replacements for Arg396, R396C and R396H, yield enzyme with the relatively low activity of ∼17% that of WT, which exhibit conformational changes and loss of positive kinetic cooperativity; these effects are probably due to the location of Arg396 at the entrance to the active site. Thus, these disease-associated single mutations can yield enzymes with reduced activity either by affecting the catalytic reaction or by perturbing the enzyme’s multimeric structure and/or native conformation, which are required for catalytic function.
Supplementary Material
Footnotes
This work was supported by NIH grant R01-DK60504 and by a grant from Autism Speaks. The Beckman Optima XL-I analytical ultracentrifuge used in this study was obtained and supported by NIH 2P20 RR016472.
- ASL
- adenylosuccinate lyase
- SAMP
- adenylosuccinate
- SAICAR
- 5-aminoimidazole-4-(N-succinylocarboxamide ribonucleotide)
- AICAR
- 5-aminoimidazole-4-carboxamide ribonucleotide
- AMP
- adenosine 5′-monophosphate
- HEPES
- N-(2-hydroxyethyl)piperazine-N’-ethanesulfonic acid
- AUC
- Analytical ultracentrifugation
- CD
- circular dichroism
- SDS-PAGE
- sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- WT
- wild type
- SE
- sedimentation equilibrium
- DTT
- dithiothreitol
- EDTA
- ethylenediamine tetraacetic acid
- KCl
- potassium chloride
- APBADP
- adenosine phosphonobutyric acid
- 2′ (3′)
- 5′-diphosphate
- S-Ado
- succinyladenosine
- Gdn.HCl
- guanidine hydrochloride
- DTNB
- 5,5′-dithiobis-2-nitrobenzoic acid.
Supporting Information Available The pools of WT and K246E mutant enzymes obtained from gel filtration chromatography at 25 °C were characterized kinetically and the kinetic parameters are shown in Supporting Table 1S. Experimental AUC data of the WT and mutant enzymes were globally fitted to various theoretical models for an oligomer in equilibrium with species of different molecular masses. Representative residuals at 25 °C and 37 °C, which illustrate goodness of fit for various models, are shown in Supporting Figure 1S and Figure 2S, respectively. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Ratner S. Argininosuccinases and adenylosuccinases. In: Boyer PD, editor. The Enzymes. 3rd Ed Vol. 7. Academic Press, Inc.; New York: 1972. pp. 167–197. [Google Scholar]
- 2.Bridger WA, Cohen LH. The kinetics of adenylosuccinate lyase. J. Bio. Chem. 1968;243:644–650. [PubMed] [Google Scholar]
- 3.Bulusu V, Srinivasan B, Bopanna MP, Balaram H. Elucidation of the substrate specificity, kinetic and catalytic mechanism of adenylosuccinate lyase from Plasmodium falciparum. Biochim. et Biophys Acta. 2009;1794:642–654. doi: 10.1016/j.bbapap.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 4.Wu C-Y, Lee H-J, Wu S-H, Chen S-T, Chiou S-H, Chang G-G. Chemical mechanism of the endogenous arginosuccinate lyase activity of duck lens δ2-crystallin. Biochem. J. 1998;333:327–334. doi: 10.1042/bj3330327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nuiry I, Hermes JD, Weiss PM, Chen C-Y, Cook PF. Kinetic mechanism and location of rate-determining steps for aspartase from Hafnia alvei. Biochemistry. 1984;23:5168–5175. doi: 10.1021/bi00317a013. [DOI] [PubMed] [Google Scholar]
- 6.Brosius JL, Colman RF. Three subunits contribute amino acids to the active site of tetrameric adenylosuccinate lyase: Lys268 and Glu275 are required. Biochemistry. 2002;41:2217–2226. doi: 10.1021/bi011998t. [DOI] [PubMed] [Google Scholar]
- 7.Sivendran S, Colman RF. Effect of a new non-cleavable substrate analog on wild-type and serine mutants in the signature sequence of adenylosuccinate lyase of Bacillus subtilis and Homo sapiens. Protein science. 2008;17:1162–1174. doi: 10.1110/ps.034777.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stenmark P, Moche M, Arrowsmith C, Berglund H, Busam R, Collines R, Dahlgren LG, Edwards A, Flodin S, Flores A, et al. Human adenylosuccinate lyase in complex with its substrate N6-(1,2-dicarboxy-ethyl)-AMP, and its products AMP and fumarate. RCSB protein Data Bank. :2VD6. [Google Scholar]
- 9.Mouchegh K, Zikanova M, Hoffmann GF, Kretzschmar B, Kuhn T, Mildenberger E, Stoltenburg-Didinger G, Krijt J, Dvorakova L, Honzik T, Zeman J, Kmoch S, Rossi R. Lethal fetal and early neonatal presentation of adenylosuccinate lyase deficiency: Observation of 6 Patients in 4 Families. J. Pediatr. 2007;150:57–61. doi: 10.1016/j.jpeds.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 10.Jaeken J, Van den Berghe G. An infantile autistic syndrome characterized by the presence of succinylpurines in body fluids. Lancet. 1984;II:1058–1061. [PubMed] [Google Scholar]
- 11.Jaeken J, Wadman SK, Duran M, Van Sprang FJ, Beemer FA, Holl RA, Theunissen PM, de Cock P, Van den Bergh F, Vincent MF, Van den Berghe G. Adenylosuccinase deficiency: an inborn error of purine nucleotide synthesis. Eur. J. Pediatr. 1988;148:126–131. doi: 10.1007/BF00445919. [DOI] [PubMed] [Google Scholar]
- 12. Adenylosuccinate Lyase Mutations Database Home Page ( http://www.icp.ucl.ac.be/adsldb/mutations.html)
- 13.Speigel EK, Colman RF, Patterson D. Adenylosuccinate lyase deficiency. Mol. Genet. Metab. 2006;89:19–31. doi: 10.1016/j.ymgme.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 14.Van den Berghe G, Jaeken J. Adenylosuccinate lyase deficiency. In: Scriver CR, Beaudt AL, Valle D, Sly WS, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic and Molecular Basis of Inherited Diseases. 8th ed Vol.II. McGraw-Hill; New York: 2001. pp. 2653–2662. [Google Scholar]
- 15.Lee P, Colman RF. Expression, purification, and characterization of stable, recombinant human adenylosuccinate lyase. Protein Expr. Purif. 2007;51:227–234. doi: 10.1016/j.pep.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli UK. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Tornheim K, Lowenstein JM. The purine nucleotide cycle: The production of ammonia from aspartate by extracts of rat skeletal muscle. J. Biol. Chem. 1972;247:162–169. [PubMed] [Google Scholar]
- 18.Palenchar JB, Colman RF. Characterization of a mutant Bacillus subtilis adenylosuccinate lyase equivalent to a mutant enzyme found in human adenylosuccinate lyase deficiency: Asparagine 276 plays an important structural role. Biochemistry. 2003;42:1831–1841. doi: 10.1021/bi020640+. [DOI] [PubMed] [Google Scholar]
- 19.Riddles PW, Blakeley RL, Zerner B. Ellman’s reagent: 5,5′-dithiobis(2-nitrobenzoic acid): A reexamination. Anal. Biochemistry. 1979;94:75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- 20.Hearne JL, Colman RF. Catalytically active monomer of class Mu glutathione transferase from rat. Biochemistry. 2006;45:5974–5984. doi: 10.1021/bi060249e. [DOI] [PubMed] [Google Scholar]
- 21. SEDPHAT program Home Page ( http://www.analyticalultracentrifugation.com/sedphat/sedphat.htm)
- 22.Laue TM, Shah BD, Ridgeway TM, Pelletier SL, Harding SE, Rowe A, Horton JC. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, U.K.: 1992. Computer-aided interpretation of analytical sedimentation data for proteins; pp. 90–125. [Google Scholar]
- 23.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt B, Ho L, Hogg PJ. Allosteric disulfide bonds. Biochemistry. 2006;45:7429–7433. doi: 10.1021/bi0603064. [DOI] [PubMed] [Google Scholar]
- 25.Vivian JT, Callis PR. Mechanisms of tryptophan fluorescence shifts in proteins. Biophy. J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez KM, Gable JE, Schlmadinger DE, Kim JE. Effects of tryptophan microenvironment, soluble domain, and vesicle size on the thermodynamics of membrane protein folding: lessons from the Transmembrane Protein OmpA. Biochemistry. 2008;47:12844–12857. doi: 10.1021/bi800860k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lienqueo ME, Mahn A, Asenjo JA. Mathematical correlations for predicting protein retention times in hydrophobic interaction chromatography. J. Chromatogr. A. 2002;978:71–79. doi: 10.1016/s0021-9673(02)01358-4. [DOI] [PubMed] [Google Scholar]
- 28.Martorana V, Bulone D, San Biagio PL, Palma-Vittorelli MB, Palma MU. Collective properties of hydration: long range and specificity of hydrophobic interactions. Biophy J. 1997;73:31–37. doi: 10.1016/S0006-3495(97)78044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H. Viscosity B-coefficients and standard partial molar volumes of amino acids, and their roles in interpreting the protein (enzyme) stabilization. Biophys. Chem. 2006;122:157–183. doi: 10.1016/j.bpc.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Hildebrandt A, Blossey R, Rjasanow S, Kohlbacher O, Lenhof H-P. Electrostatic potentials of proteins in water: a structured continuum approach. Bioinformatics. 2006;23:e99–e103. doi: 10.1093/bioinformatics/btl312. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Barkley MD. Toward understanding tryptophan fluorescence in proteins. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 32.Hernández-Alcántara G, Rodríguez-Romero A, Reyes-Vivas H, Peon J, Cabrera N, Ortiz C, Enríquez-Flores S, De la Mora-De la Mora I, López-Velázquez G. Unraveling the mechanism of tryptophan fluorescence quenching in the triosephosphate isomerase from Giardia lamblia. Biochim. Biophys. Acta. 2008;1784:1493–1500. doi: 10.1016/j.bbapap.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 33.Burley SK, Petsko GA. Aromatic-aromatic interaction: A mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 34.Hanson KR. Phenylalanine ammonia-lyase: A model for the cooperativity kinetics induced by D-and L-phenylalanine. Arch. Biochem. Biophys. 1981;211:564–574. doi: 10.1016/0003-9861(81)90491-4. [DOI] [PubMed] [Google Scholar]
- 35.Segel IH. Enzyme kinetics: Behavior and analysis of rapid equilibrium and stedy-state enzyme systems. John Wiley and Sons, Inc.; 1975. [Google Scholar]
- 36.Budde RJA. Evidence for kinetically distinct forms of pp60c-src with different Km values for their protein substrate. J. Biol. Chem. 1993;268:24868–24872. [PubMed] [Google Scholar]
- 37.Castro M, Perez-Cerda C, Merinero B, Garcia MJ, Bernar J, Gil Nagel A, Torres J, Bermudez M, Garavito P, Marie S, Vincent F, Van den Berghe G, Ugarte M. Screening for adenylosuccinate lyase deficiency: clinical, biochemical and molecular findings in four patients. Neuropediatrics. 2002;33:186–189. doi: 10.1055/s-2002-34493. [DOI] [PubMed] [Google Scholar]
- 38.Marie S, Cuppens H, Heuterspreute M, Jaspers M, Zambrano TE, Gu XX, Legius E, Vincent MF, Jaeken J, Cassiman JJ, Van den Berghe G. Mutation analysis in adenylosuccinate lyase deficiency: eight novel mutations in the re-evaluated full ADSL coding sequence. Hum Mutat. 1999;13:197–202. doi: 10.1002/(SICI)1098-1004(1999)13:3<197::AID-HUMU3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.