Abstract
The binding of cAMP to the Escherichia coli catabolite gene activator protein (CAP) produces a conformational change that enables it to bind specific DNA sequences and regulate transcription, which it cannot do in the absence of the nucleotide. The crystal structures of the unliganded CAP containing a D138L mutation and the unliganded WT CAP were determined at 2.3 and 3.6 Å resolution, respectively, and reveal that the two DNA binding domains have dimerized into one rigid body and their two DNA recognition helices become buried. The WT structure shows multiple orientations of this rigid body relative to the nucleotide binding domain supporting earlier biochemical data suggesting that the inactive form exists in an equilibrium among different conformations. Comparison of the structures of the liganded and unliganded CAP suggests that cAMP stabilizes the active DNA binding conformation of CAP through the interactions that the N6 of the adenosine makes with the C-helices. These interactions are associated with the reorientation and elongation of the C-helices that precludes the formation of the inactive structure.
Keywords: allostery, cAMP, helix-turn-helix, transcription activator
A reduction in the levels of glucose in Escherichia coli results in an increase in the concentration of the secondary messenger 3′-5′ cAMP, which then binds to the catabolite gene activator protein (CAP). This binding of cAMP produces a conformational change that activates the binding of CAP to specific DNA sequences and stimulates the transcription of >100 genes. The subunit of the homodimeric CAP consists of an N-terminal domain that binds the cAMP, a C-terminal domain that binds DNA and a long α-helix between them called the C-helix that serves as the hydrophobic core for the dimerization interface. Although the structure of CAP complexed with cAMP and that of CAP with cAMP and DNA have been determined (1–3), to our knowledge, there is no structure of apo-CAP to explain its lack of affinity for specific DNA sequences and the mechanism by which the binding of cAMP activates it.
Biochemical data imply that the mutation of aspartic acid 138, located in the hinge that lies between the C-helix of the cAMP binding domain and the DNA binding domain, into a hydrophobic residue stabilizes the inactive conformation of CAP (4). Equilibrium sedimentation analysis of these mutant proteins under mild denaturing conditions indicated a tighter subunit association, whereas circular dichroism spectroscopy showed that the mutant CAP did not exhibit any change in its net secondary and tertiary structure. However, the D138L mutation remained an anomaly, because it showed a reduced affinity for DNA yet exhibited a subunit association similar to that of WT CAP (5). We chose to crystallize the D138L mutant CAP, because the mutation stabilized the inactive form of the protein.
The crystal structures of CAP in the absence of cAMP have been obtained for both the D138L mutant and WT proteins at 2.3 and 3.6 Å resolutions, respectively. Importantly, these structures reveal that the DNA recognition helix (F-helix) is buried within the core of the apo-CAP structure, which explains its low affinity for DNA in the absence of cAMP. This structure, as well as that of the unliganded WT protein, shows that the two DNA binding domains interact to form one rigid body that can assume various orientations relative to the cAMP binding domain. Last, a comparison of these unliganded structures with that of the liganded CAP explains how the binding of cAMP and certain mutations in the nucleotide binding domains, the DNA binding domains, and the C-helix that connects them shift the equilibrium between the inactive and active forms of CAP.
Results and Discussion
Overall Structure of the Apo-CAP.
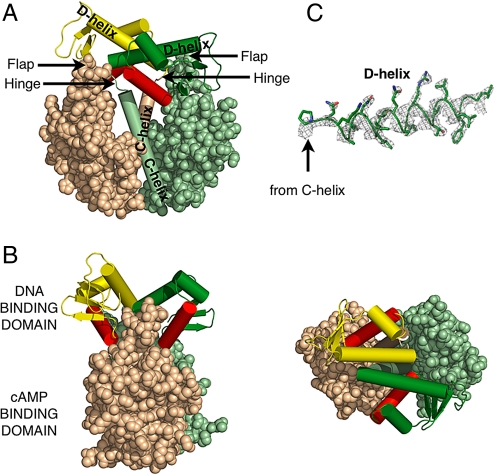
The D138L mutant CAP could be crystallized after the cAMP liganded CAP impurity was removed by separating the DNA complex formed only by the liganded CAP. Crystals of the mutant protein had one dimer in the asymmetric unit, and its structure shows the key features of the inactive form (Fig. 1 and Table 1). The most striking feature is an antiparallel coiled-coil formed between the D-helices of the two individual DNA binding domains, which welds them together as one rigid body. This conformation positions the F-helix of each monomer between the DNA binding domain and the nucleotide binding domain where it cannot interact with the DNA.
Fig. 1.
Structure of the unliganded D138L CAP mutant. The DNA binding domain of one monomer is colored forest, whereas the other is yellow. The DNA recognition helices are colored red. The cAMP binding domain of one monomer is colored pale green, whereas the other is colored wheat. This color scheme is used in all other figures unless noted. (A) The nucleotide binding domains are represented as spheres, whereas the C-helices and the helices of the DNA binding domains are represented as cylinders. (B) Alternative views of the mutant are shown. Rotation of the molecule by 90° along its central vertical and horizontal axis, respectively, produces the “side view” (Left) and “top view” (Right). (C) Electron density corresponding to the D-helix, contoured at 2σ, that was calculated using experimental phases and observed amplitudes to 2.4-Å resolution.
Table 1.
Data collection and refinement statistics
| Molecule | D138L mutant | WT |
|---|---|---|
| Space group | P43212 | P3212 |
| Unit cell dimensions | ||
| a, Å | 68.0 | 125.3 |
| b, Å | 68.0 | 125.3 |
| c, Å | 230.1 | 224.7 |
| Resolution, Å | 50–2.3 (2.4–2.3) | 50–3.6 (3.7–3.6) |
| Rmerge, % | 9.0 (71.3) | 8.6 (100) |
| I/σ | 31.2 (2.1) | 23.5 (1.1) |
| Completeness, % | 98.9 (90.9) | 99.9 (100) |
| Redundancy | 13 (9) | 11.3 (10.8) |
| Rcryst,% | 23.9 | 29.5 |
| Rfree,% | 27.4 | 31.8 |
| rmsd bond length, Å | 0.007 | 0.004 |
| rmsd bond angle, ° | 1.106 | 0.893 |
| Protein statistics from Ramachandran plot | ||
| Residues in favored regions | 382 (95.3%) | 958 (82.1%) |
| Residues in allowed regions | 13 (3.2%) | 154 (13.2%) |
| Outliers | 6 (1.5%) | 55 (4.7%) |
The values in parentheses are for the highest-resolution shell. Rmerge is Σ|Ij−〈I〉|/ΣI, where Ij is the intensity of an individual reflection, and 〈I〉 is the mean intensity for multiple recorded reflections. Rcryst is Σ |Fo − Fc|/ ΣFo, where Fo is an observed amplitude and Fc a calculated amplitude; Rfree is the same statistic calculated over a subset, 5%, of the data that have not been used for refinement.
The crystal form of the WT CAP has three dimers/asymmetric unit, each of whose structures reveal different orientations of the two domains relative to the cAMP binding domains (Fig. S1; Table 1). Overall, the structures of the cAMP binding domains of the mutant and the WT molecules were fairly similar, and the Cα atoms could be superimposed with an average rmsd of 1.61 Å (Fig. S1). Most of the differences between the structures of the nucleotide binding domains were slightly alternative orientations of the flap and the loops near the cAMP binding site. However, despite the low resolution of WT CAP data, the electron density was continuous and clear for the tracing of the main chain except for the hinge residues 131 and 132 (Fig. 1C). A superposition of the pairs of D-helices of the DNA binding domains from the three WT dimer molecules and the pair in the mutant protein reveals that these helices exhibit the same relative orientations with respect to each other. Neither their structures nor their lengths vary, which suggests that the mutation of D138 to leucine does not change the structure of the inactive form, but rather stabilizes it relative to the active form. However, the relative orientations of these pairs of helices with respect to the nucleotide binding domains do differ in each molecule indicating that the pair of DNA binding domains are mobile as one rigid body about the hinge (Movie S1). Because the majority of the differences between the mutant and the WT structures occur only as alternate orientations of the DNA binding domains, the higher resolution mutant structure was chosen to make generalizations about the inactive form of CAP in its apo state.
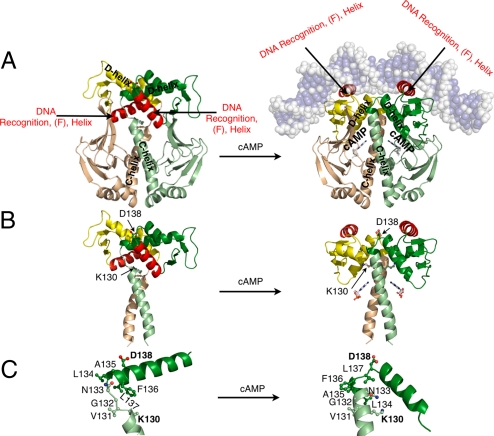
Comparison of the Unliganded and the Liganded Structures.
Comparison of the structures of the cAMP-CAP and apo-CAP show that the apo-CAP cannot bind DNA site specifically, because the two DNA recognition helices are buried in the inactive form in contrast to their being solvent exposed on the surface of the DNA binding domains of CAP in the active form. Also, although the conformation of the nucleotide binding domain is largely unaltered, the orientations of the DNA binding domains and the structure of the portion of the C-helices that lies near the nucleotide binding site differ dramatically between the two forms (Fig. 2). In the inactive form, the D-helix becomes four residues longer and spans residues 135–152, whereas, in the active form, only residues 139–152 form the α-helix. However, the length of the C-helix becomes six residues shorter (residues 110–130) in the inactive structure compared with its active structure length (residues 110–136). Therefore, the C-helix of the inactive form is ≈1.5 helical turns shorter than the C-helix of the active form, whereas its D-helix is longer by the same amount (Fig. 2 B and C). Analyses of NMR NOE's and amide proton exchange rates of apo-CAP had shown that residues 127–133 have little or no secondary structure, whereas residues 134–139 become helical, which is in agreement with these crystal structures (6). Although, the C-helix in these structures is slightly longer (approximately three residues) than concluded from the NMR studies, the helical residues 127–130 of at least one of the two subunits show a larger bend than the rest of the C-helix, indicating the inherent flexibility of this region. That the C terminus of the C-helix is flexible is also supported by the analysis of CAP with a C-helix containing cysteine substitution (7). Associated with the differences in the lengths of the C- and D-helices is a difference in the position of the hinge, which spans residues ≈130–134 in the inactive form and ≈136–139 in the active form (Fig. 2C).
Fig. 2.
Comparison of the inactive (Left) and the active forms of CAP (Right). (A) Schematic representation of CAP shows β-strands as arrows and α-helices as coils. The cAMP is shown in ball and stick representation. DNA is shown as transparent spheres and its bases are colored slate blue. (B) The cAMP binding domains and DNA have been excluded to emphasize the orientations of the DNA binding domains and the C-helices. Residues capping the C- and D-helices and cAMP are shown as ball and stick. In the inactive form, K130 is located at the C termini of the C-helices, whereas in the active form, D138 is located at the N termini of the D-helices. (C) The C terminus of the C-helix, the N terminus of the D-helix, and the hinge residues of one protomer of the inactive and the active form.
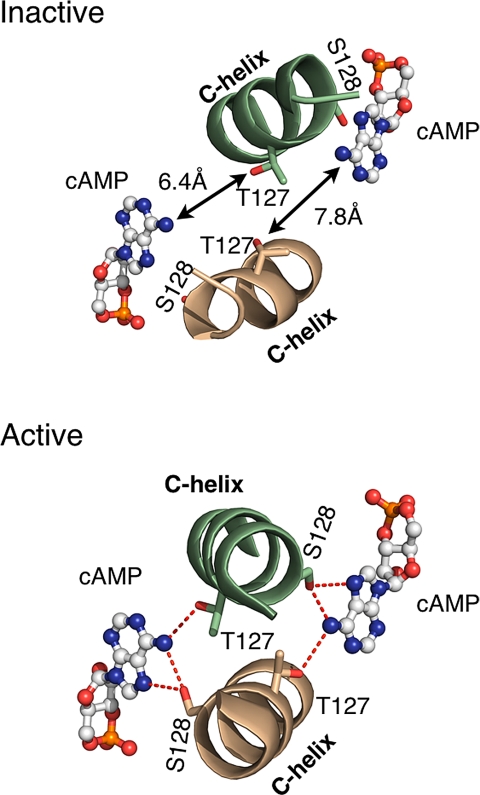
It has been previously established that residues of the C-helix that lie near the cAMP binding pocket, such as T127 and S128, as well as the hinge, have an important role in transmitting the signal from the nucleotide binding site to the DNA binding domains (8). Comparisons of the structures of the inactive and the active forms reveal that T127 and S128 from opposite helices lie closer to cAMP in the active form and both hydrogen bond to the N6 of the nucleotide (Fig. 3). Indeed, these interactions of T127 and S128 with cAMP may have the critical role in shifting the equilibrium between the active, cAMP complexed structure and that of the apo-CAP.
Fig. 3.
The nucleotide binding pocket of the inactive and the active forms. The cAMP is modeled in the nucleotide binding pocket of the inactive form (Upper). The distances between N6 of adenosine and the hydroxyl group of residues, S128 and T127, are too large to allow hydrogen bonding between cAMP and these two residues in the inactive form. However, N6 of cAMP forms key hydrogen bonds with the S128 and T127 in the active form (Lower).
Structural Interpretation of Existing Biochemical and Biophysical Data.
In the inactive form, the two DNA binding domains interact through their D-helices to form a rigid body that buries their DNA recognition helices, which explains the inability of CAP to recognize DNA site specifically, and is also consistent with proteolytic digestion data that indicated that it has a different quaternary structure in the absence of the nucleotide (9, 10). The proteolysis of CAP in the presence or absence of cAMP yields different sized protein fragments, because the hinge region is protected from the solvent by the rigid body shell of DNA binding domain in the apo-CAP. In the active form, the hinge is exposed and is accessible to proteolytic digestion. For example, chymotrypsin cleaves CAP with bound cAMP at position F136; however, no proteolytic fragments are observed in the absence of the nucleotide. In the active form of CAP, this residue is located in the hinge; however, in the inactive form, it is located in the D-helix and, thus, protected from the protease (Fig. 2). The formation of the coiled-coil between the D-helices that is observed in the inactive form also suggests why mutations in this helix would affect the DNA binding affinity of CAP (11). Mutations that destabilize the coiled-coil between the two D-helices would destabilize the inactive form; thereby, leading to formation of the active conformation.
Although proteolytic digestion data are consistent with CAP existing in alternative conformations in the presence and absence of cAMP, small-angle neutron scattering (SANS) experiments have been used to determine the overall shape of CAP in solution on binding cAMP. Although the orientations of the DNA binding domains are different in the two structures, the calculated radius of gyration (Rg) of CAP is essentially the same (< 1%) in the two forms. The Rgs experimentally determined by SANS were 23.0 Å for the apo form and 22.3 Å for the cAMP complex (12), which are nearly identical to the 23.1 Å Rg that we calculated for all of the apo structures and the 22.7 Å Rg calculated for the liganded form, consistent with the crystal structures presented here being the same as the structures in solution.
The unique arrangement of the DNA binding domains locked together as one rigid body through the interactions between the D-helices is consistent with earlier biochemical data showing that mutations in these helices result in the F-helix being solvent exposed (13). Measurements of FRET between the donor 1,5-I-AEDANS (N-iodoacetylaminoethyl-1-naphthylamine-5-sulfonate) attached to cysteine 178 and the acceptor tryptophan 85 (14) are consistent with the interactions between the D-helices presented here being the same as existing in solution. The cysteine is located in the DNA binding domain and the tryptophan is located in the nucleotide binding domain. The binding of cAMP causes an increase in FRET efficiency, which corresponds to an 8-Å change in the distance between these two residues: 26.6 ± 3.9 Å to 18.7 ± 0.8 Å. In the apo crystal structures, the average distance between Cα of C178 and W85 is 26.3 Å, whereas the distance is 19.3 Å in the previously determined liganded structures. Therefore, the agreement of the results from the FRET experiments with those obtained from the crystal structures further support the crystal and solution structures of the inactive form being the same.
The structures of the liganded and the unliganded CAP provide a structural basis for understanding how other mutations also alter the equilibrium between the inactive and the active conformations. These mutations include mutations that lie in the nucleotide binding site, the flap formed by the β4-, β5-strands and loop between them, the C-helix, the hinge between the C- and D-helices (Fig. 1). Because the G141K, A144T, and L148K mutations (15) all convert hydrophobic residues located at the contact interface between the coiled-coil formed by the D-helices in the inactive form into charged or polar ones, they presumably destabilize this interaction and the inactive form; thus, explaining why these mutant proteins have a lower dimer association constant and a higher DNA binding affinity in the absence of the nucleotide (Fig. 2). For example, the G141Q mutation shows weakened intersubunit interactions, whereas interdomain interactions remain similar to WT CAP. Sedimentation profiles under denaturing conditions indicated that the mutant exhibited that the dimer association constant is 7.1 × 106 M−1 compared with the 109 − 1010 M−1 for the WT (16). Providing further support for our observation, NMR and biochemical data suggest that the F-helix is buried, is involved in interdomain interactions, and becomes solvent accessible on cAMP binding (6). A mutation D138, which is located at the positive N terminus of the helix, to leucine could both effectively destabilize the active form by removing the helix capping property of D138 and stabilize the formation of the antiparallel coiled-coil found in the inactive form through additional hydrophobic interactions. The D138L mutant CAP may have facilitated our crystallization of apo-CAP by its stabilization of the inactive form (Fig. 2B).
Energetic Contributions to the Stabilization of the Inactive Form.
The shift in the equilibrium between the inactive apo and the active cAMP bound form of CAP can also be understood by examining the factors that stabilize each state. The inactive form is stabilized in part by the total surface area that is buried on dimer formation (≈3,950 Å2) being ≈800 Å2 greater than that of the active form (≈3,120 Å2). The increase in total buried surface area of the inactive form is due, in part, to the hydrophobic interface created by the dimerization of the elongated D-helices, in contrast to the active form where the DNA binding domain of each protomer exhibits little or no interactions with the other. Presumably, hydrophobic interactions arising from additional buried surface area are responsible for stabilizing the inactive conformation of CAP and exceeds the entropic cost of immobilizing the DNA binding domains into one rigid body.
The cAMP Induced Conformational Changes That Lead to the Activation of CAP.
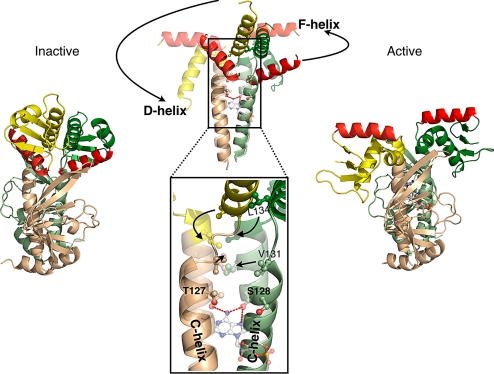
The mechanism by which the binding of cAMP activates CAP can be explained by the differences in the lengths of the C- and D-helices in the inactive and the active forms, because they correlate with the reorientations of the DNA binding domains and their DNA recognition helices (Fig. 2). Most significantly, the binding of cAMP to CAP shifts its equilibrium toward the active conformation through interactions of the N6 of its adenosine with S128 and T127, which is associated with the reorientation and the lengthening of the C-helices. Although a shortened C-helix is a notable feature of the unliganded CAP structures, the dimerization of the DNA binding domains by an extended D-helix is perhaps the most functionally significant (Fig. 2 B and C). The D-helix not only provides a key dimerization interface, but its increased length is directly correlated with the decreased length of the C-helices.
Comparison of the unliganded and the liganded structures of CAP suggests that the primary way that cAMP stabilizes the active form is through its exocyclic NH2 group interacting with S128 and T127 on the trans and cis C-helices. S128 from the opposite subunit acts as a hydrogen bond donor and acceptor with the N6 of adenine moiety of cAMP, and T127 from the same subunit, to which cAMP is bound also, contributes a hydrogen bond (Fig. 3). The hydroxyl groups of these two residues are at the optimal distance and orientation to interact with cAMP in the active form, but not in the inactive form. Therefore, it is likely that the hydrogen bonding between these two residues of CAP and cAMP is necessary to stabilize the active form.
The side chains of S128 and T127 appear to have the principle role in the cAMP induced conformational change in CAP. Their interactions with cAMP in the binding pocket require that the C-helices are positioned closer to the nucleotide and to each other than they are in the inactive form. Also, this reorientation of the C-helices allows hydrophobic residues on the C-helices to interact with each other, which thereby, stabilizes an extended C-helix. The crystal structures suggest that associated with an extension of the C-helices is a relocation of the hinge and a corresponding reduction in the length of the D-helices by one helical turn that destabilizes the coiled-coil maintained by the pair (Fig. 4). The destabilization of the coiled-coil interactions made by the D-helix removes the restraint this interaction imposes on the DNA binding domains and allows the F-helices to adopt the orientation from where they are buried to a position that is necessary for DNA site-specific recognition (Movie S1). This mechanism agrees with previous protein footprinting experiments that had indicated changes in the C termini of the C-helices and the hinge on cAMP binding may lead to rigid body movements of the DNA binding domains and its D-, E-, and F-helices, and indicated that the F-helix and the C terminus of the C-helix become more solvent accessible in the presence of cAMP (13).
Fig. 4.
The cAMP induced conformational changes of CAP. The side view of CAP is shown. Structure of the unliganded CAP (Left) and of the liganded CAP (Right). A superposition of these two structures along their C-helices is shown in the middle of the figure. The unliganded structure is solid, whereas the liganded structure is transparent. To emphasize the conformational changes on cAMP binding, only the C-, D-, and F-helices are shown.
The crystal structures presented here show that the DNA binding domains have dimerized and the F-helices are buried, which is consistent with a majority of biochemical and biophysical data. These structures and previous NMR data (6) only partially agree with a recent NMR structure of apo-CAP (17). Although all of the structures show that C-helix is shortened, the recent NMR structure did not show a lengthened D-helix, nor a buried F-helix. Because the earlier NMR data did not reveal the relative orientations of domains, the recent NMR structure measured longer distances by modifying cys-mutants, including a K57C mutation located in the flap of the nucleotide binding domain, with nitroxide spin labels to make Paramagnetic Relaxation Enhancement measurements. However, the crystal structures show that K57 and other flap residues are involved in asymmetric interdomain interactions extending to the hinge of domain rotation. Spin labeling of mutations in the flap may be sterically preventing the formation of the inactive form seen here, and account for the difference between the NMR and X-ray structures.
Comparisons with Proteins That Bind cAMP and Those That Belong in the CAP/FNR Family.
Because bound cAMP induces a conformational change in CAP through critical trans interactions with the other subunit, its mechanism of ligand-induced conformational change appears to be significantly different from that used by a homologous but different family of cAMP regulating proteins. The conformational change caused by the binding of cAMP to members of this family is stabilized only by residues of the subunit to which it binds, and thus, cAMP exerts its effects in cis rather than in trans as it does in CAP. The proteins in the cis-regulated family include protein kinase A, Epac proteins, and cyclic nucleotide regulated gated channels (18). The conformational change induced by nucleotide binding involves L75 and F102 in the nucleotide binding domain (using CAP numbering) forming a hydrophobic pair in which the position of one affects the position of the other (Fig. S3). The position of L75 changes on cAMP binding, which results in F102 moving and occupying the previous position of leucine, as well as other conformational changes that are associated with activation. In contrast, L75 and F102 remain in essentially the same positions before and after the binding of cAMP to CAP.
Although domains homologous to the cAMP binding domains are used by proteins that regulate various biological activities including protein phosphorylation and the electrostatic potential across membranes, the helix-turn-helix motif of the DNA binding domains is used to recognize a specific DNA sequence and regulate transcription (19). The orientation of the helix-turn-helix of the unliganded CAP, especially its recognition helix, suggests that other members of the cAMP receptor protein (CRP) subgroup of the CAP/FNR superfamily of transcriptions factors would exhibit a similar inactive conformation and may use the same underlying principle of burying their DNA recognition helix. For example, the apo crystal structures of the inactive forms of CooA and CprK, two transcription factors that belong in this family, indicate that their DNA recognition helices are shielded from solvent (20, 21). However, because these transcription factors contain additional structural features at their N and C termini, they would most likely use an alternative mechanism of activation.
Materials and Methods
Expression and Purification.
The D138L mutant, the selenomethionyl-labeled D138L mutant, and the WT CAP were overexpressed in BL21 cells. The cells were induced with 1 mM IPTG after growth for 4 h in Luria Broth supplemented by kanamycin at 37 °C. Cells were harvested ≈2 h after induction. Cells were resuspended in buffer A (50 mM Tris·HCl pH 7.8/1 mM EDTA/100 mM KCl/1 mM DTT/0.2 mM PMSF/5% Glycerol) and then lysed. The lysate was clarified by centrifugation and was added to ≈35 g BioRex70 that had been preequilibrated at pH 7.5 with buffer A. The slurry was mixed, and the supernatant was then removed. Fresh buffer A was added, and the slurry was allowed to settle again. This procedure was repeated five times until the OD278/OD260 ratio was >1.5, and then, the matrix was packed into a column and washed with ≈100 mL of buffer A. CAP was gradient-eluted with buffer A containing 1.0 M KCl and dialyzed overnight against buffer A. The sample was then centrifuged to remove the precipitate. Approximately 15g of hydroxyapatite (HTP) was resuspended with double distilled (dd) H2O (4 °C) and packed into a column, which was then loaded with the centrifuged CAP sample. The column was then washed with ≈50 mL of buffer B (50 mM K2HPO4-KH2PO4, pH 7.5/1 mM EDTA/1 mM DTT/10% Glycerol) and gradient-eluted with buffer B containing 0.50 M K2HPO4, pH 7.5. The sample was then slowly precipitated with ammonium sulfate and was loaded onto a Phenyl Sepharose column that was prewashed with ≈1 L of ddH2O and then preequilibrated with ≈300 mL of buffer C [1.0 M (NH4)2SO4/50 mM Tris·HCl, pH 7.8/100 mM KCl/1 mM EDTA/1 mM DTT]. The column was then washed with ≈50 mL of buffer C and gradient-eluted with ddH2O and equilibrated with storage buffer (50 mM Tris·HCl, pH 7.8/1 mM EDTA/100 mM KCl).
Crystallization.
A 2 mL of solution containing the D138L mutant protein that had been concentrated to ≈10 mg/mL was crystallized by dialysis against a 1 L solution containing 8% PEG 4K, 200 mM Proline, 10 mM Tris·HCl, pH 7.8, and storage buffer. Before the crystallization, the dialysis solution containing the precipitant was slowly added to the protein solution and mixed until only a slight amount of precipitate was unable to go back into solution. This tiny amount of precipitate was left in the solution. This procedure allowed crystals to grow within 2–3 days. These crystals were then used as seeds to grow selenomethionyl-labeled D138L mutant CAP crystals.
Crystals of the selenomethionyl-labeled D138L mutant CAP (Se-CAP) were obtained by vapor diffusion when the protein in storage buffer was mixed with 10% PEG 4K, 200 mM Proline, 10 mM Tris·HCl, pH 7.8 solution in a 1:1 ratio. Native D138L crystals were crushed and used as seeds in a 1:10 ratio to the concentrated solution of Se-CAP before adding the crystallization solution. The crystals grew within 2–3 days and were stabilized in a solution containing an increased amount of PEG 4K. They were then cryoprotected in two steps with stabilizing solutions containing 15 and 30% glucose, and flash-frozen in liquid propane.
WT CAP was concentrated to ≈20–25 mg/mL in storage buffer and mixed with various amounts of native D138L CAP seeds. Crystals were obtained by vapor diffusion against storage buffer. Storage solution without KCl served as the stabilizing solution. The crystals were then cryoprotected stepwise with this solution containing 10, 20, and 30% glycerol. Last, they were flash-frozen in liquid propane.
Structure Determination.
Diffraction data for the unliganded D138L mutant and the unliganded WT CAP were collected at the beam line station, ID24 at the Argonne Advanced Photon Source (Argonne, IL). Initial processing and scaling of the raw data were carried out by using HKL2000 (22). General handling of scaled data were done in CCP4 (23). Seleno-methionine sites were found by using ShelxD and ShelxE (24), and the initial phases were calculated by using SOLVE (25). The maps were solvent flattened by using the program RESOLVE. The model of the D138L mutant was built by using the programs O (26) and COOT (27). Individual nucleotide binding domains and DNA binding domains from the original liganded structure (1) were used as rigid bodies and placed in the initial electron density. Refinement was carried out by using REFMAC with TLS (translation, libration, and screw rotation parameters) and restrained refinements. TLS groups were determined by using a Difference Distance matrix. Difference distance matrix plots were produce using the Differences Distance Matrix Program (DDMP) from the Center for Structural Biology at Yale University, New Haven, CT. The final Rfree was 27.4%, and the Rfactor was 23.9% (Table 1). Figures were generated using the program Pymol (28). The initial electron density map of the unliganded WT CAP was obtained using measured amplitudes phased by molecular replacement using the program PHASER (29). The structure of the mutant D138L was used as a search model. Z-scores were low; therefore, the mutant model without the DNA binding domains was used. Initial electron density of the DNA binding domains was poor and precluded model building. The program DMMULTI was used to carry out seven cycles of 6-fold averaging of the density from the six independent cAMP binding domains in the asymmetric unit, which resulted in improved density. Masks were made by using the program NCSMASK to protect the DNA binding domains from solvent flattening. The improved electron density maps showed the location of the pair of D-helices in each of the three dimers in the asymmetric unit. Models of the DNA binding domains were built into this density, and the overall structure was refined using the same procedures as the mutant protein. At this point, the Rfree and the Rfactor stabilized to 39.3 and 33.8%. Subsequently, each strand and helix of the nucleotide binding domains was defined as a TLS group. The model of the WT refined and stabilized to an Rfree of 31.8% and Rfactor of 29.5% (Table 1).
Supplementary Material
Acknowledgments.
We thank the staff at the beam line station 24ID at the Advanced Photon Source for help during data collection. This work is supported by the National Institutes of Health Grant GM57510 and in part by the National Basic Research Program of China Grants 2007CB914304 and 2007CB714500, the National Natural Science Foundation of China Grants 30970631 and 30770423, the Pujiang Program of Shanghai City Grant 09PJD007, and the Shanghai Leading Academic Discipline Project B109.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3FWE for D138L and 3HIF for WT CAP).
This article contains supporting information online at www.pnas.org/cgi/content/full/0908380106/DCSupplemental.
References
- 1.McKay DB, Steitz TA. Structure of catabolite gene activator protein at 2.9 Å resolution suggests binding to left-handed B-DNA. Nature. 1981;290:744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- 2.Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: The DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 3.Passner JM, Steitz TA. The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer. Proc Natl Acad Sci USA. 1997;94:2843–2847. doi: 10.1073/pnas.94.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryu S, Kim J, Adhya S, Garges S. Pivotal role of amino acid at position 138 in the allosteric hinge reorientation of cAMP receptor protein. Proc Natl Acad Sci USA. 1993;90:75–79. doi: 10.1073/pnas.90.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu S, Lee JC. Role of residue 138 in the interdomain hinge region in transmitting allosteric signals for DNA binding in Escherichia coli cAMP receptor protein. Biochemistry. 2004;43:4662–4669. doi: 10.1021/bi0362166. [DOI] [PubMed] [Google Scholar]
- 6.Won HS, et al. Structural understanding of the allosteric conformational change of cyclic AMP receptor protein by cyclic AMP binding. Biochemistry. 2000;39:13953–13962. doi: 10.1021/bi000012x. [DOI] [PubMed] [Google Scholar]
- 7.Tomlinson SR, Tutar Y, Harman JG. CRP subunit association and hinge conformation changes in response to cAMP binding: Analysis of C-helix cysteine-substituted CRP. Biochemistry. 2006;45:13438–13446. doi: 10.1021/bi0610855. [DOI] [PubMed] [Google Scholar]
- 8.Youn H, Kerby RL, Conrad M, Roberts GP. Study of highly constitutively active mutants suggests how cAMP activates cAMP receptor protein. J Biol Chem. 2006;281:1119–1127. doi: 10.1074/jbc.M509421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Bobin S, Krakow JS. Characterization of the CRPCY core formed after treatment with carboxypeptidase Y. Nucleic Acids Res. 1991;19:4253–4257. doi: 10.1093/nar/19.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heyduk E, Heyduk T, Lee JC. Intersubunit communications in Escherichia coli cyclic AMP receptor protein: Studies of the ligand binding domain. Biochemistry. 1992;31:3682–3688. doi: 10.1021/bi00129a017. [DOI] [PubMed] [Google Scholar]
- 11.Garges S, Adhya S. Sites of allosteric shift in the structure of the cyclic AMP receptor protein. Cell. 1985;41:745–751. doi: 10.1016/s0092-8674(85)80055-6. [DOI] [PubMed] [Google Scholar]
- 12.Krueger S, et al. Determination of the Conformations of cAMP Receptor Protein and Its T127L, S128A Mutant with and without cAMP from Small Angle Neutron Scattering. J Biol Chem. 1998;273:20001–20006. doi: 10.1074/jbc.273.32.20001. [DOI] [PubMed] [Google Scholar]
- 13.Baichoo N, Heyduk T. Mapping conformational changes in a protein: Application of a protein footprinting technique to cAMP-induced conformational changes in cAMP receptor protein. Biochemistry. 1997;36:10830–10836. doi: 10.1021/bi970714v. [DOI] [PubMed] [Google Scholar]
- 14.Polit A, Blaszczyk U, Wasylewski Z. Steady-state and time-resolved fluorescence studies of conformational changes induced by cAMP and DNA binding to cyclic AMP receptor protien from Escherichia coli. Eur J Biochem. 2003;270:1413–1423. doi: 10.1046/j.1432-1033.2003.03497.x. [DOI] [PubMed] [Google Scholar]
- 15.Youn H, Koh J, Roberts GP. Two-state allosteric modeling suggests protein equilibrium as an integral component for cAMP specificity in the cAMP receptor protein of Escherichia coli. J Bacteriol. 2008;190:3336–3343. doi: 10.1128/JB.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng X, Lee JC. Differential perturbation of intersubunit and interdomain communications by glycine 141 mutation in Escherichia coli CRP. Biochemistry. 1998;37:51–60. doi: 10.1021/bi9719455. [DOI] [PubMed] [Google Scholar]
- 17.Popovych N, et al. Structural basis for cAMP-mediated allosteric control of the catabolite gene activator protein. Proc Natl Acad Sci USA. 2009;106:6927–6932. doi: 10.1073/pnas.0900595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehmann H, Wittinghofer A, Bos JL. Capturing cyclic nucleotides in action: Snapshots from crystallographic studies. Nat Rev Mol Cell Biol. 2007;8:63–73. doi: 10.1038/nrm2082. [DOI] [PubMed] [Google Scholar]
- 19.Steitz TA, Weber IT, Matthew JB. Catabolite gene activator protein: Structure, homology with other proteins, and cyclic AMP and DNA binding. Cold Spring Harb Symp Quant Biol. 1983;47:419–426. doi: 10.1101/sqb.1983.047.01.049. [DOI] [PubMed] [Google Scholar]
- 20.Lanzilotta WN, et al. Structure of the CO sensing transcription activator CooA. Nat Struct Biol. 2000;7:876–880. doi: 10.1038/82820. [DOI] [PubMed] [Google Scholar]
- 21.Komori H, Inagaki S, Yoshioka S, Aono S, Higuchi Y. Crystal Structure of CO-sensing Transcription Activator CooA Bound to Exogenous Ligand Imidazole. J Mol Biol. 2007;367:864–871. doi: 10.1016/j.jmb.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 22.Levy C, et al. Molecular basis for halorespiration control by CprK, a CRP-FNR type transcriptional regulator. Mol Microbiol. 2008;70:151–167. doi: 10.1111/j.1365-2958.2008.06399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode Methods. Enzymol Macromol Crystallogr. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.DeLano WL. The PyMol Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 30.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.