Abstract
The biological function of regulatory factor X1 (RFX1), the prototype member of the transcription factor RFX family, is not clear. We have used gene trap technique to disrupt the expression of RFX1 in mice. Although heterozygous RFX1WT/GT mice appear normal and fertile, homozygous RFX1GT/GT embryos died at an early stage (most likely before embryonic day 2.5). Our results indicate that RFX1 regulates expression of genes that are essential for early embryonic development/survival and that RFX1 function can not be compensated by other RFX1 family members.
Keywords: Embryonic lethality, gene trap, regulatory factor X1
1. Introduction
The regulatory factor X (RFX) proteins are within the winged-helix subfamily of helix-turn-helix transcription factors [1]. These proteins have a highly conserved 76-amin acid DNA binding domain that can bind X-box consensus sequences. Seven RFX proteins (named RFX 1 – 7) have been identified so far in mammals [2] and each of them may have distinct functions. RFX5 has been shown to play an important role in regulating the expression of the major histocompatibility complex class II genes and knockout of Rfx5 gene results in bare lymphocyte syndrome in mice [3]. Knockout of Rfx3 gene causes severe ciliaophathies that can result in diabetes and left-right asymmetry specification [4,5].
The function of RFX1, the prototype of the RFX family, is not clear. Knockout of RFX homologue in Caenorhabditis elegans leads to severe sensory defects [6]. The Drosophila RFX homologue is necessary for ciliated sensory neuron differentiation [7]. RFX1 has been shown to be expressed in the highest amount in the mammalian brain, compared with many other tissues and organs [2]. We have shown that RFX1 proteins are expressed in the neurons of rat brain and contribute to the regulation of the expression of the neuronally expressed glutamate transporter type 3 [8]. These results suggest a role of RFX1 in the nervous system. In addition, RFX1 has also been shown to regulate the expression of proteins, such as interleukin-5 receptor α chain[9], whose expression may be broader than just in the nervous system. To further study the biological functions of mammalian RFX1, we decided to generate mutant mice by knocking out the Rfx1 gene.
2. Materials and Methods
2.1. Generation of RFX1 mutant mice
The embryonic stem (ES) cell clone RRO347 containing pGT2Lxf gene trap vector integrated into the intron sequence between the exon 2 and exon 3 was obtained from BayGenomics/Mutant Mouse Regional Resource Center (San Francisco, CA, USA) and was a product of the International Gene Trap Consortium [10]. This gene trap clone has only one insertion site with the gene trap vector. The ES cells were from mouse strain 129 and were used to generate chimeras by blastocyst injection with C57BL/6J wild-type embryos. The chimeras were then crossed with wild-type C57BL/6J to generate heterozygous RFX1WT/GT (C57Bl/6J × 129) F1 offspring. The heterozygous mice were intercrossed to obtain F2 offspring that was genotyped by PCR. F2 heterozygous mice were intercrossed again to generate F3 offspring that was also genotyped.
2.2. Genotyping of the RFX1 gene trap mutant mice
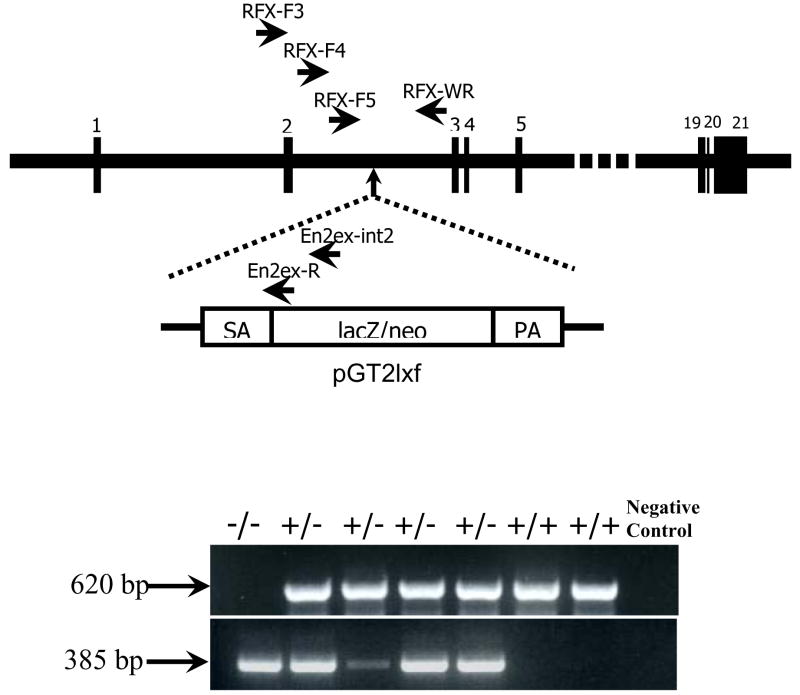
To identify the site of integrated RFX1 gene trap vector, following primers were used: Rfx-F3, 5′-ctcacacctctggttgggcagt-3′ and En2ex-int2, 5′-gggacctgggacctggttgtcatgga-3′ (Fig. 1). The PCR program was at 94°C for 5 min, followed by 40 cycles at 94 °C for 30 s, 68 °C for 20 s, and 72 °C for 30 s, with the final step at 72 °C for 10 min. The PCR product was purified for sequencing analysis. All isolated embryos and viable mice were genotyped by PCR amplification. Genomic DNA of embryos or mouse tails was isolated by QIAamp DNA Micro Kit (QIAGEN, Valencia, CA, USA). Primers Rfx-F4, 5′-cactggagaggatgaacagggaggta-3′, and En2ex-R, 5′-ttgggttagaggggtctcaaagtcag-3′ (Fig. 1), were used to amplify the products from the mutant allele. Primers Rfx-F5, 5′-ctagagatgatggcagggagatcaggtt-3′, and Rfx-WR, 5′-gggaggaaaggaggggaaagtagagtc-3′ (Fig. 1), were used to amplify the products of the wild-type allele. The PCR program was at 94°C for 5 min, followed by 40 cycles at 94°C for 30 s, 62°C for 30 s, and 72 °C for 30 s, with the final step at 72°C for 10 min.
Fig. 1. The gene trap mutation in the mouse RFX1 gene.
The pGT2Lxf vector was integrated into the second intron of the RFX1 gene that consists of 21exons. The locations of RFX1-specific and vector-specific primers used in PCR-based genotyping of embryos and mice are shown in the top panel. SA: splice acceptor sequence; PA: polyadenylation signal. A representative gel image of the PCR products is shown in the bottom panel. The wild-type and mutant alleles produce products of 620 and 385 nucleotides, respectively.
2.3. Isolation and in vitro culture of mouse embryos
Female heterozygous mice at 4 weeks old were injected with pregnant mare serum gonadotropin (PMSG) (NHPP, Torrance, CA, USA) followed 48 hour later by injection of human chorionic gonadotropin (HCG) (ICN, Costa Mesa, CA, USA). The female mice were then mated with sexually mature male heterozygous mice. Fertilized eggs were collected in FHM medium (Millipore, Billerica, MA, USA), treated with hyaluronidase (Sigma, St Louis, MO, USA) and rinsed with FHM. The embryos were then cultured in KSOM medium (Millipore) overlaid with mineral oil in a humidified air containing 5% CO2 at 37°C. After culture, each embryo was lyzed in a lysis buffer consisting of 50 mM Tris-HCl (pH 8.0), 0.5% triton X-100 and 0.2% proteinase K. The lysis was performed at 55°C for 5 h, followed by 95°C for 10 min. The lysate was then used for PCR.
3. Results
The ES cell clone RRO347 contains the gene trap vector inserted into the second intron of the RFX1 gene (Fig. 1). Total 154 mice from heterozygous parents were genotyped when they were ~25-days-old, no homozygous RFX1GT/GT mice were detected, suggesting that homozygous RFX1 mutant mice were not viable (Table 1).
Table 1.
Genotypes of progeny from heterozygous RFX1 parents
| Age | N | Genotype | |||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | ND | ||
| P25 | 154 | 47 | 107 | 0 | 0 |
| E15.5 | 7 | 2 | 5 | 0 | 0 |
| E10.5 | 15 | 3 | 12 | 0 | 0 |
| E3.5 | 22 | 3 | 19 | 0 | 0 |
| E2.5 | 20 | 5 | 6 | 1 | 8 |
ND: not determined
We then isolated embryos at E15.5 and E10.5 for genotyping and did not find homozygous RFX1GT/GT embryos either, indicating that RFX1 disruption caused early embryonic lethality. To identify when embryos with RFX1 disruption might die, 67 embryos at stage of E0.5 were isolated from four superovulated heterozygous mice and cultured in vitro. Ten embryos showed no obvious nuclei when embryos were isolated. Fifteen embryos still remained in two-cell stage at E1.5; while most embryos entered into four-cell stage. These twenty-five embryos that were abnormal or developmentally delayed were collected and genotyped. Unfortunately, none of their genotypes can be identified by PCR, possibly due to insufficient DNA samples. Four embryos that remained in four-cell stage at E2.5 were collected and genotyped. One homozygous RFX1GT/GT embryo was detected among these four embryos. We also genotyped 16 embryos that developed into morula at E2.5 and we did not detect any homozygous RFX1GT/GT embryos from them. All of the rest embryos at E3.5 developed into blastocysts. Three of them were wild type and the other nineteen were heterozygous.
4. Discussion
RFX1 has been implicated to play an important role in the nervous system because knockout of RFX homologue in Caenorhabditis elegans and Drosophila results in severe sensory defects [6,7]. A few studies have shown that RFX1 can regulate expression of various gene products, such as glutamate transporter type 3 and interleukin-5 receptor, in mammalian cells [8,9]. However, the biological functions of mammalian RFX1 are not clear. Our current study showed that targeted knockout of RFX1 expression leads to embryonic lethality, suggesting critical functions of RFX1.
We have examined a large number of postnatal mice and have not found any homozygous RFX1 knockout mouse. This situation is very different from the knockout of other types of mammalian RFX genes. Knockout of two other RFX genes has been performed in mice. RFX5−/− mice can survive but had a severe immunodeficiency due to the lack of major histocompatibility complex II gene expression in various lymph cells and macrophages [3]. Although most RFX3−/− mice died before birth, some of them did survive to adulthood [4,5]. These results suggest that various RFX1 proteins have very different functions and that the lack of one RFX protein is not compensated by the other RFX proteins.
We did not find any RFX1−/− postnatal mice or embryos at a stage later than E2.5. The only RFX1−/− embryo was found at E2.5 and this embryo was still at 4 cell stage. These results suggest that RFX1 knockout leads to development delay/lethality at very early stage.
We used a gene trap technique to produce targeted gene knockout in mice. As described before [11,12], this technique involves inserting a trap vector in the targeted gene. The vector contains a polyA sequence that will prematurely terminate the transcription of the gene. In our study, the trap vector was inserted in the second intron of the RFX1 gene. This mutated gene will produce a truncated mRNA that contains the sequence transcribed from the first two exons out of the 21 exons for RFX1 gene. This process will result in disruption of normal RFX1 protein expression.
In summary, our results show that disruption of RFX1 expression in mouse leads to early embryonic lethality. These results suggest that RFX1 regulates expression of genes that are essential for early embryonic development/survival and that RFX1 function can not be compensated by other RFX1 family members.
Acknowledgments
This study was supported by grants (R01 GM065211 and R01 NS045983 to Z Zuo; 5P30CA044579-19901609 to W Xu) from the National Institute of Health, Bethesda, MD, by a grant from the International Anesthesia Research Society (2007 Frontiers in Anesthesia Research Award to Z Zuo), Cleveland, OH, by a Grant-in-Aid from the American Heart Association Mid-Atlantic Affiliate (0755450U to Z Zuo), Baltimore, MD, and the Department of Anesthesiology, University of Virginia.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gajiwala KS, Chen H, Cornille F, Roques BP, Reith W, Mach B, Burley SK. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 2.Aftab S, Semenec L, Chu JS, Chen N. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol Biol. 2008;8:226. doi: 10.1186/1471-2148-8-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clausen BE, Waldburger JM, Schwenk F, Barras E, Mach B, Rajewsky K, Forster I, Reith W. Residual MHC class II expression on mature dendritic cells and activated B cells in RFX5-deficient mice. Immunity. 1998;8:143–155. doi: 10.1016/s1074-7613(00)80467-7. [DOI] [PubMed] [Google Scholar]
- 4.Bonnafe E, Touka M, AitLounis A, Baas D, Barras E, Ucla C, Moreau A, Flamant F, Dubruille R, Couble P, Collignon J, Durand B, Reith W. The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol Cell Biol. 2004;24:4417–4427. doi: 10.1128/MCB.24.10.4417-4427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ait-Lounis A, Baas D, Barras E, Benadiba C, Charollais A, Nlend Nlend R, Liegeois D, Meda P, Durand B, Reith W. Novel function of the ciliogenic transcription factor RFX3 in development of the endocrine pancreas. Diabetes. 2007;56:950–959. doi: 10.2337/db06-1187. [DOI] [PubMed] [Google Scholar]
- 6.Swoboda P, Adler HT, Thomas JH. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- 7.Dubruille R, Laurencon A, Vandaele C, Shishido E, Coulon-Bublex M, Swoboda P, Couble P, Kernan M, Durand B. Drosophila regulatory factor X is necessary for ciliated sensory neuron differentiation. Development. 2002;129:5487–5498. doi: 10.1242/dev.00148. [DOI] [PubMed] [Google Scholar]
- 8.Ma K, Zheng S, Zuo Z. The transcription factor regulatory factor X1 increases the expression of neuronal glutamate transporter type 3. J Biol Chem. 2006;281:21250–21255. doi: 10.1074/jbc.M600521200. [DOI] [PubMed] [Google Scholar]
- 9.Iwama A, Pan J, Zhang P, Reith W, Mach B, Tenen DG, Sun Z. Dimeric RFX proteins contribute to the activity and lineage specificity of the interleukin-5 receptor alpha promoter through activation and repression domains. Mol Cell Biol. 1999;19:3940–3950. doi: 10.1128/mcb.19.6.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stryke D, Kawamoto M, Huang CC, Johns SJ, King LA, Harper CA, Meng EC, Lee RE, Yee A, L’Italien L, Chuang PT, Young SG, Skarnes WC, Babbitt PC, Ferrin TE. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiles MV, Vauti F, Otte J, Fuchtbauer EM, Ruiz P, Fuchtbauer A, Arnold HH, Lehrach H, Metz T, von Melchner H, Wurst W. Establishment of a gene-trap sequence tag library to generate mutant mice from embryonic stem cells. Nat Genet. 2000;24:13–14. doi: 10.1038/71622. [DOI] [PubMed] [Google Scholar]
- 12.Hansen J, Floss T, Van Sloun P, Fuchtbauer EM, Vauti F, Arnold HH, Schnutgen F, Wurst W, von Melchner H, Ruiz P. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc Natl Acad Sci U S A. 2003;100:9918–9922. doi: 10.1073/pnas.1633296100. [DOI] [PMC free article] [PubMed] [Google Scholar]