Summary
Protein synthesis is a regulated cellular process that links nutrients in the environment to organismal growth and development. Here we examine the role of genes that regulate mRNA translation in determining growth, reproduction, stress resistance and lifespan. Translational control of protein synthesis by regulators such as the cap-binding complex and S6 kinase play an important role during growth. We observe that inhibition of various genes in the translation initiation complex including ifg-1, the worm homologue of eIF4G, which is a scaffold protein in the cap-binding complex; and rsks-1, the worm homologue of S6 kinase, results in lifespan extension in Caenorhabditis elegans. Inhibition of ifg-1 or rsks-1 also slows development, reduces fecundity and increases resistance to starvation. A reduction in ifg-1 expression in dauers was also observed, suggesting an inhibition of protein translation during the dauer state. Thus, mRNA translation exerts pleiotropic effects on growth, reproduction, stress resistance and lifespan in C. elegans.
Keywords: C. elegans, eIF4G, lifespan, mRNA translation, protein synthesis, S6 kinase
Introduction
The genetics of lifespan in model invertebrate organisms has yielded considerable information on the possible physiological determinants of aging rate. Many of the major genetic modulators of lifespan, including those in the insulin/IGF-1 and target of rapamycin (TOR) pathways, influence aging in organisms ranging from single-celled yeast, fruit flies, nematodes and, in the case of insulin/IGF-1 signalling, rodents (Kapahi & Zid, 2004; Kenyon, 2005). Protein synthesis is a key regulated cellular process that links nutrients to organismal growth, and is also modulated by the insulin and TOR pathways (Sonenberg et al., 2000; Shamji et al., 2003). The eukaryotic initiation factor 4F (eIF4F) initiation complex mediates mRNA translation (Sonenberg et al., 2000; Harris & Lawrence, 2003; Shamji et al., 2003). This is accomplished by regulating the association of the mRNA cap-binding protein eIF4E to the scaffold protein eIF4G, both components of the eIF4F complex (Supplementary Fig. S1). Eukaryotic initiation factor 4G helps assemble the eIF4F complex by bridging the poly(A)-binding proteins (PABPs) with eIF4E (Sonenberg et al., 2000). This leads to the circularization of mRNAs, which has a synergistic effect on the rate of translation (Sonenberg et al., 2000). Ribosomal S6 kinase (S6K) phosphorylates and thereby activates the ribosomal protein S6 on multiple serine residues, under the control of TOR, and is a key regulator of mRNA translation and growth (Thomas, 2002; Shamji et al., 2003; Ruvinsky & Meyuhas, 2006). S6K was also recently found to be associated with the eIF3 pre-initiation complex, which coordinates protein synthesis (Holz et al., 2005). In this study we examine the effect of modulating mRNA translation on various life history traits including lifespan, reproduction and stress resistance.
Results
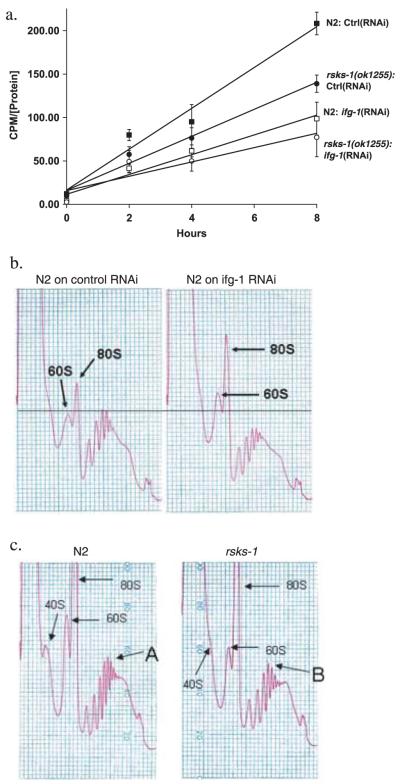
mRNA translation was inhibited using RNA interference (RNAi) of ifg-1 (an eIF4G homologue in Caenorhabditis elegans) and the deletion mutant rsks-1(ok1255) (C. elegans ribosomal S6K, homologue of mammalian p70S6K). A 1.6-kb deletion in exon 4 of rsks-1 was confirmed using nested polymerase chain reaction (PCR) (data not shown). The rate of protein synthesis was measured by incorporation of 35S-methionine from media. Radioactivity in worm protein extracts was measured as a function of exposure time to 35S-methionine-labelled OP50 Escherichia coli (Fig. 1a). The slope of this function indicates the rate of protein synthesis (see Supplementary Fig. S2 for feeding rates). These results demonstrate a significant decrease in the rate of incorporation of 35S-methionine upon inhibition of either ifg-1 by RNAi or in the rsks-1(ok1255) mutant (Fig. 1a). Moreover, inhibition of ifg-1 in the rsks-1(ok1255) mutant background resulted in an additive decrease in protein translation (Fig. 1a). Translation state was also analyzed by measuring polysomal distribution. Inhibition of ifg-1 led to a reduction in total polysomal translation compared with controls (Fig. 1b). Mutation in rsks-1 resulted in a smaller decrease in translation compared to that observed upon ifg-1 inhibition. The most abundant polysome levels shifted from the sixth peak (A) in wild type (N2) to the fifth peak (B) in rsks-1(ok1255) animals (Fig. 1c), consistent with an overall reduction in translation. The data from both the methionine incorporation assay and the polysome profiles suggest a decrease in mRNA translation upon inhibition of ifg-1 and rsks-1 compared to controls.
Fig. 1.
Inhibition of either rsks-1 or ifg-1 reduces the rate of protein translation. (a) Reduction in rates of protein synthesis measured by 35S-methionine incorporation upon inhibition of ifg-1and rsks-1. Age-matched adults were placed in 35S-methionine-labelled OP50 for 0, 2, 4, and 8 h. Radioactivity was measured in the TCA precipitated protein extract and standardized for protein concentration. Protein synthesis rate was evaluated by measuring the slope of the curves during incorporation. The following slopes were observed: N2 on L4440 (control RNAi), 23.5 (cpms (counts per minute) μg-1) h; rsks-1(ok1255) on L4440 (control RNAi), 15.5 (cpms μg-1) h; N2 on ifg-1(RNAi), 11.4 (cpms μg-1) h; rsks-1(ok1255) on ifg-1(RNAi), 8.3 (cpms μg-1) h. Each data point represents the mean of three biological replicates. Similar results were obtained in two separate experiments. (b,c) Polysomal profiles were generated by measuring absorbance profiles at 260 nm of worm extracts separated on 10-50% sucrose gradients. (b) The N2 strain on control RNAi and ifg-1(RNAi) and from (c) N2 and rsks-1(ok1255). 40S, 60S, and 80S labels show the position of ribosomal subunits and whole ribosomes. There was an increase in the 60S and the 80S peaks upon inhibition of ifg-1 accompanied by a decrease in the polysomal peaks suggesting a significant reduction in ribosomes that were involved in mRNA translation but an increase in free ribosomes (b). Maximum absorbance in N2 was detected in the sixth ribosome peak (A) vs. the fifth (B) in the rsks-1(ok1255) (c). Results are representative of three experiments performed.
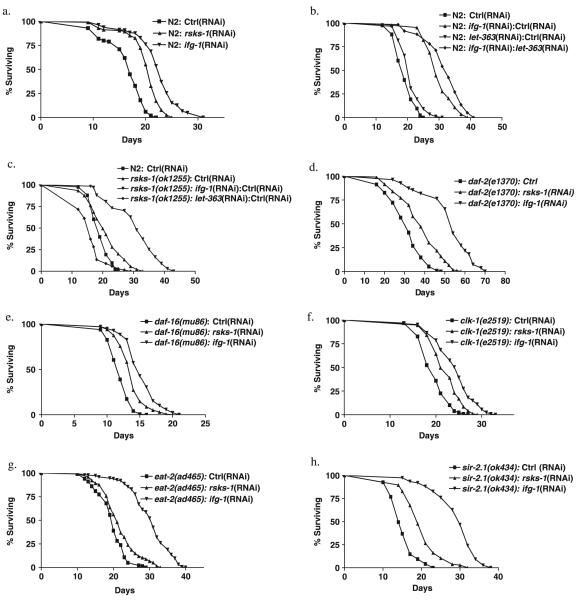
Then we examined the effect on lifespan upon inhibition of genes that regulate protein translation. Inhibition of ifg-1 during adulthood led to a 36% increase in mean lifespan (Fig. 2a). We also examined the effect on lifespan of inhibition of other genes that are part of the cap-binding complex. Inhibition of at least one eIF4E isoform and two of the three PABPs also has a significant effect on lifespan extension (Supplementary Fig. S3). Inhibition of rsks-1 using RNAi during adulthood led to a 22% increase in mean lifespan (Fig. 2a). Mean lifespan was also extended in a deletion mutant of rsks-1(ok1255) compared to the parental control N2 strain (Fig. 2c). Together, these results support the view that inhibition of protein synthesis by capdependent translation and S6K extends lifespan in C. elegans.
Fig. 2.
Lifespan extension by inhibiting ifg-1 and rsks-1 and their interaction with other longevity genes. (a) Mean lifespan was 16.4 days (n = 100) for N2 on control RNAi; 20 days (n = 116) for rsks-1(RNAi); and 22.3 days (n = 164) for ifg-1(RNAi) (P < 0.0001). (b) Mean lifespan was 19.2 days (n = 83) for N2 on control RNAi; 29.8 days (n = 83) for N2 on 1: 1 ifg-1(RNAi) and control RNAi; 21.2 days (n = 87) for 1: 1 let-363(RNAi) and control RNAi; and 32 days (n = 59) for N2 on 1: 1 ifg-1(RNAi) and let-363(RNAi) (P < 0.0001). (c) Mean lifespan was 19.2 days (n = 83) for N2 on control RNAi; 21 days (n = 78) for rsks-1(ok1255) on control RNAi; 30.6 days (n = 67) for rsks-1(ok1255) on 1: 1 ifg-1(RNAi) and control RNAi (P < 0.0001); and 16.1 days (n = 67) for 1: 1 let-363 (RNAi) and control RNAi. (d) daf-2(e1370) on control RNAi, rsks-1(RNAi), and ifg-1(RNAi). Mean lifespan was 30.8 days (n = 115) for daf-2(e1370) on control RNAi; 38.1 days (n = 119) for rsks-1(RNAi); and 52.3 days (n = 55) for ifg-1(RNAi) (P < 0.0001). (e) Mean lifespan was 12 days (n = 144) for daf-16(mu86) on control RNAi; 13.7 days (n = 136) for rsks-1(RNAi); and 15.2 days (n = 138) for ifg-1(RNAi) (P < 0.0001). (f) Mean lifespan was 19.6 days (n = 128) for clk-1(e2519) on control RNAi; 22.1 days (n = 95) for rsks-1(RNAi); and 24.1 days (n = 127) for ifg-1(RNAi) (P < 0.0001). (g) Mean lifespan was 19.5 days (n = 138) for eat-2(ad456) on control RNAi; 21.7 days (n = 133) for rsks-1(RNAi); and 30.1 days (n = 153) for ifg-1(RNAi) (P < 0.0001). (h) Mean lifespan was 15.2 days (n = 95) for sir-2.1(ok424) on control RNAi; 20.2 days (n = 105) for rsks-1(RNAi); and 29.3 days (n = 75) for ifg-1(RNAi) (P < 0.0001). Similar results were obtained in at least two separate experiments for each panel.
Inhibition of let-363 (worm homologue of TOR) has been shown to extend adult lifespan in C. elegans (Vellai et al., 2003). If effects of let-363 on lifespan are mediated by its influence on translation rate, one might expect non-additive effects of let-363 and ifg-1 on lifespan. We therefore examined whether the lifespan extension caused by inhibition of let-363 and ifg-1 is additive. Inhibition of let-363 increased lifespan in both the N2 background and when ifg-1 was inhibited (Fig. 2b). These results could imply that lifespan extension by let-363 and ifg-1 involves different mechanisms. We also find that lifespan extension by inhibition of ifg-1 and rsks-1 is additive (Fig. 2c). In the rsks-1(ok1255) mutant background, inhibition of ifg-1 led to a further extension of 46% in mean lifespan (Fig. 2c). However, inhibition of let-363 by RNAi in rsks-1 mutant background led to a slight shortening of lifespan (Fig. 2c). These results strongly imply that a common mechanism mediates lifespan extension due to inhibition of let-363 and rsks-1 but, surprisingly, suggests that this may not be the case for let-363 and ifg-1, or ifg-1 and rsks-1. Alternatively, additive effects could reflect a weak influence on a common mechanism in each case.
We tested the effects of inhibiting ifg-1 or rsks-1 on other lifespan extension pathways in worms. Down-regulation of daf-2 (insulin/IGF-1-like receptor) causes lifespan extension that is dependent on daf-16, a forkhead transcription factor (Kenyon et al., 1993; Lin et al., 1997; Ogg et al., 1997). In the daf-2(e1370) mutant background, we observed that inhibition of either ifg-1 or rsks-1 further extended lifespan by 70% and 24%, respectively (Fig. 2d). To further test interaction with the insulin-like signaling (ILS) pathway, we examined lifespan extensions by ifg-1or rsks-1 inhibition in daf-16(mu86) null mutant background. Inhibition of either ifg-1 or rsks-1 extended lifespan by 27% and 14%, respectively, in a daf-16(mu86) background (Fig. 2e). These findings show that knock-down of ifg-1 or rsks-1 does not increase lifespan by activating DAF-16, and also could imply that mutation of daf-2 also extends lifespan via a different mechanism.
Then we tested if lifespan extension by ifg-1 or rsks-1 inhibition is additive with that resulting from mutation of the Clock gene clk-1. Inactivation of clk-1, a gene required for ubiquinone biosynthesis (Ewbank et al., 1997), has been shown to increase lifespan independently of the ILS pathway and displays the associated phenotypes of delayed development, slow pharyngeal pumping and defecation rates (Lakowski & Hekimi, 1996). Inhibition of either ifg-1 or rsks-1 in the clk-1(e2519) mutant background led to a further lifespan extension by 19% and 13%, respectively (Fig. 2f). This could imply that clk-1(e2519) extends lifespan via mechanisms distinct to ifg-1 or rsks-1; however, it is worth noting that clk mutants show additive interactions with one another.
We also tested the role of ifg-1 and rsks-1 in lifespan extension by dietary restriction (DR) using the eat-2(ad465) mutant, which exhibits a mutationally induced form of DR (Lakowski & Hekimi, 1998). Inhibition of either ifg-1 or rsks-1 extended lifespan in the eat-2 background by 54% and 11%, respectively (Fig. 2g). This could imply that effects of ifg-1 and rsks-1 in lifespan are distinct from those of DR. But again, it remains possible that additive effects involve a shared mechanism.
SIR2 is a conserved regulator of lifespan in yeast (Kaeberlein et al., 1999), worms (Tissenbaum & Guarente, 2001), and flies (Rogina & Helfand, 2004). In a sir-2.1 mutant background, ifg-1 inhibition extended lifespan by 93% and rsks-1 inhibition by 33% (Fig. 2h). This clearly demonstrates that effects of ifg-1 and rsks-1 are not mediated by sir-2.1. Lifespan extension by inhibition of ifg-1 and rsks-1 is also independent of bec-1 which has been shown to regulate autophagy (Melendez et al., 2003) (data not shown). This implies that effects on lifespan here are not dependent in a change in levels of autophagy.
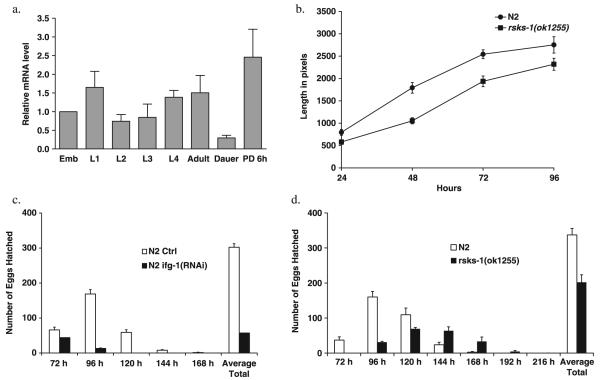
Quantification of ifg-1 mRNA levels at different developmental periods showed that it was significantly reduced in the dauer stage (Fig. 3a). These results suggest a down-regulation of cap-dependent translation due to inhibition of ifg-1 during the dauer stage. We observe that the rsks-1(ok1255) mutant developed more slowly than the controls (Fig. 3b). Mutant animals are smaller at the same chronological age compared to controls (Fig. 3b) and show a delay in reaching adulthood (data not shown). Inhibition of ifg-1 was reported earlier to lead to larval arrest in a manner similar to inhibition of let-363 (Long et al., 2002). Inhibition of either ifg-1 or rsks-1 during adulthood also led to a decrease in fecundity (Fig. 3c,d). Taken together, these results show that ifg-1 and rsks-1 are required for normal growth and development during the early part of life in C. elegans.
Fig. 3.
Pleiotropic effects on growth, development, reproduction and stress resistance upon inhibition of ifg-1 and rsks-1. (a) ifg-1 expression is reduced during dauer state. Quantification of relative levels of ifg-1 mRNA in N2 animals at different developmental stages at 20 °C using real-time reverse transcriptase-polymerase chain reaction (RT-PCR). pmp-2 expression was used for internal reference. Mean and standard error of the relative ifg-1/pmp-2 ratios based on three real-time RT-PCR trials are shown. Emb, embryos; PD, post dauer; 6 h, 6 h after dauer larvae were exposed to food. (b) rsks-1 regulates rate of development in C. elegans. Average length of 20 worms was measured every 24 h till 96 h after egg lay in the N2 vs. rsks-1(ok1255) strain. Similar results were obtained in five trials. (c,d) Reduction of fecundity by inhibition of ifg-1 and rsks-1. Progeny were counted every 24 h in (c) N2 on control RNAi and ifg-1(RNAi) (d) N2 and rsks-1(ok1255).
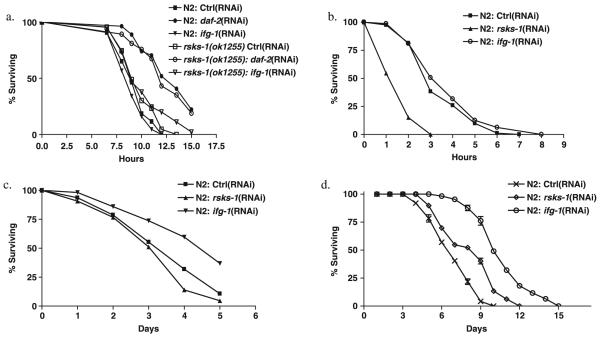
It has been proposed that mechanisms that increase resistance to various environmental stressors also extend lifespan (Johnson et al., 1996; Martin et al., 1996). We examined the effect of inhibition of ifg-1 and rsks-1 on several stressors. Inhibition of ifg-1 or rsks-1 had a negligible effect on thermotolerance while inhibition of daf-2 resulted in an increased thermotolerance (Fig. 4a) as previously reported (Lithgow et al., 1995). Similarly, no significant increase was observed in resistance to the oxidant juglone, which leads to increased levels of superoxide in vivo (Fig. 4b). In fact, inhibition of rsks-1 using RNAi (Fig. 4b) or the deletion mutant rsks-1(ok1255) caused increased sensitivity to juglone (data not shown). Increased resistance to UV stress was observed by inhibition of ifg-1 but not rsks-1 (Fig. 4c). In contrast to other stressors, we observed that inhibition of either ifg-1 or rsks-1 led to increased starvation resistance: a 56% and 28% increase, respectively, of median survival of starved animals compared to N2 (Fig. 4d). Selection for resistance to starvation leads to lifespan extension in Drosophila, implying a possible common mechanism for resistance to starvation and lifespan extension.
Fig. 4.
Effect of inhibition of ifg-1 and rsks-1 on resistance to various stresses. (a) One-day-old adult N2 and rsks-1(ok1255) worms were exposed to control RNAi, ifg-1(RNAi), and daf-2(RNAi) for 48 h in 20 °C before heat stressed at 35 °C. N2 on control RNAi (n = 43) N2 ifg-1(RNAi) (n = 44); N2 on daf-2(RNAi) (n = 21) rsks-1(ok1255) on control RNAi (n = 43), rsks-1(ok1255) ifg-1(RNAi) (n = 44), rsks-1(ok1255) daf-2(RNAi) (n = 30). Similar results were obtained in three experiments. (b) Adult worms were exposed to control RNAi, rsks-1(RNAi), and ifg-1(RNAi) for 48 h in 20 °C before being transferred to NGM agar plates containing 240 μm juglone. Mean survival was 3.5 h (n = 81) for N2 on control RNAi; 1.7 h (n = 66) for rsks-1(RNAi) (P < 0.0001); and 3.6 h (n = 74) for ifg-1(RNAi). (c) Worms treated with RNAi bacteria for 48 from the L4 stage were exposed to 2000 J m-2 of UV via a Stratalinker® 1800. Mean survival was 3.7 days (n = 47) for N2 on control RNAi; 3.4 days (n = 43) for rsks-1(RNAi); and 4.5 days (n = 57) for ifg-1(RNAi) (P < 0.0005). (d) N2 and rsks-1(ok1255) were both exposed to control RNAi, rsks-1(RNAi), and ifg-1(RNAi) for 48 h before being starved in S-basal. Standard deviations were calculated from five biological replicates. Between 50 and 100 worms per replicate were scored each day. Median survival was 6.4 days for N2 on control RNAi; 8.2 days for rsks-1(RNAi); and 10 days for ifg-1(RNAi). Similar results were obtained in at least three experiments.
Discussion
The nutrient-responsive TOR pathway is an evolutionarily conserved regulator of lifespan affecting yeast (Kaeberlein et al., 2005), worms (Vellai et al., 2003; Jia et al., 2004), and flies (Kapahi et al., 2004). Lifespan extension by inhibition of S6K was observed in C. elegans (Fig. 2a) and Drosophila melanogaster (Kapahi et al., 2004) suggesting that this enzyme also has a conserved role in determining lifespan. S6K1 null mice are smaller in size and show a reduction in ribosomal S6 phosphorylation (Pende et al., 2004). However, the mechanisms by which S6K regulates mRNA translation remain poorly understood (Pende et al., 2004). Mice lacking S6K1 are resistant to diet-induced obesity (Um et al., 2004). Moreover, both mice on high-fat diet and leptin-deficient mice show increased S6K activity, suggesting a role of S6K in fat metabolism.
To explore the role of rsks-1 and ifg-1 in known pathways affecting aging, we tested the effects of simultaneous knockdown of these and other genes. Such epistasis analysis is subject to certain limitations in terms of interpretation. While the absence of an additive effect between two treatments strongly implies that both act via a common mechanism, the occurrence of additive effects are consistent with both distinct and shared mechanisms (Gems et al., 2002). It is possible that the maximal lifespan extension possible due to perturbing a particular pathway may have not been achieved in a particular long-lived mutant. For example, daf-2 RNAi has been shown to further extend the lifespan of a daf-2 mutant (Arantes-Oliveira et al., 2003). We see additive effects of inhibition of rsks-1 and ifg-1 on longevity. Given than both regulate mRNA translation, this interaction may reflect additive effects on overall level of mRNA translation; alternatively, it could reflect distinct mechanisms. Similarly, our data also show that lifespan extension by inhibition of ifg-1 or rsks-1 is additive to the lifespan increases due to daf-2, clk-1 and eat-2; taken alone, these observations are difficult to interpret in terms of single or multiple underlying mechanisms. By contrast, the lifespan extension observed in daf-16 and sir-2.1 background clearly demonstrates that the effects on lifespan of ifg-1 and rsks-1 are not mediated by these genes. Recently, inhibition of translation factors ifg-1 and eIF2B were shown to extend lifespan in C. elegans (Henderson et al., 2006). The lifespan extension upon inhibition of ifg-1 was dependent on daf-16 when RNAi was applied during adulthood but not when applied throughout development. This contrasts with our results, where we see a significant increase in lifespan in a daf-16 background resulting from inhibition of ifg-1 during adulthood. Possibly, differences in RNAi constructs are responsible for this discrepancy.
It has been estimated that the fraction of genes devoted to translation may be as high as 35 to 45% (Sonenberg et al., 2000). We reason that inhibition of this costly process is likely to shift investment towards somatic maintenance and away from development, growth, and reproduction. The effects of inhibition of ifg-1 and rsks-1 on lifespan, growth, development and reproduction shown here are consistent with its role as an antagonistically pleiotropic mechanism that determines lifespan (Williams, 1957; Martin et al., 1996). Both S6K and eIF4G play a key role in growth in response to changes in nutrients in the environment in mammals (Sonenberg et al., 2000). Lifespan extension by DR has been proposed to be due to a shift in resources from growth and reproduction towards somatic maintenance allowing the animal to survive nutrient-poor conditions until they find conditions fit for reproduction. Hence, molecular mechanisms that lead to lifespan extension by DR may overlap with those that ensure increased survival during times of starvation. The data show that reduction in protein synthesis via inhibition of ifg-1 or rsks-1 leads to prolonged survival under starvation (Fig. 4d). Thus, reduction of protein synthesis may in part be responsible for the lifespan extension due to DR. The extension of lifespan that we observed in eat-2 mutants upon RNAi of ifg-1 or rsks-1 is not inconsistent with this.
Our data suggest a possible link between translation regulation and the longevity of dauer larvae. The dauer stage involves a nonfeeding, nonreproducing alternative developmental state under which animals have prolonged survival. Possibly, C. elegans has evolved a method to inhibit cap-dependent translation in the dauer state by reducing the expression of ifg-1, since we observe lowered expression of this gene (Fig. 3a), in agreement with a previous genome wide expression study of dauers (Wang & Kim, 2003). As the scaffold protein eIF4G forms a bridge between PABPs and eIF4Es it would be an effective target for inhibition of cap-dependent translation. Saccharomyces cerevisae have also evolved an eIF4G-dependent mechanism to reduce protein synthesis under conditions of starvation. In this case, eIF4G degradation is observed when yeast cells undergo a diauxic shift (Berset et al., 1998). Overexpression of eIF4E has recently been shown to increase cellular senescence in mice as measured by β-galactosidase staining, implicating its potential involvement in mammalian aging (Ruggero et al., 2004). The regulation of cap-dependent translation may therefore be a conserved response to nutrient limitation in different species.
Longevity is a trait which shows a very high degree of variation between animal species. Could protein synthesis play a role in determining species-specific lifespan? If it did one would expect to see an inverse correlation across species between rate of protein synthesis and longevity. In fact, such an inverse correlation has been observed (Munro, 1969; Finch, 1990). Taken together with our results, this supports the notion that differences in rate of protein synthesis may contribute to evolved differences in lifespan between species.
Experimental procedures
Strains
Caenorhabditis elegans strains N2 and rsks-1(ok1255) were cultured at 20 °C on NGM plates seeded with OP50 bacteria as described in Riddle et al. (1997). rsks-1(ok1255) was obtained from the Gene Knockout Project at OMRF, which is part of the International C. elegans Gene Knockout Consortium. rsks-1(ok1255) was backcrossed into control N2 strain five times for all the experiments. Other strains used, daf-2(e1370), daf-16(mu86), clk1(e2519), and eat-2(ad465) were obtained from the Caenorhabditis Genetics Center.
RNAi experiments
RNAi bacteria strains were cultured as previously described (Kamath et al., 2001b). 1 mm isopropyl-β-d-thiogalactopyranoside (IPTG) was used for induction in all cases except Fig. 2b,c where 2 mm was used. Inhibition of let-363 under 1 mm conditions did not extend lifespan (data not shown). Worms were staged at L4 and introduced to RNAi for 48 h using methods previously described (Kamath et al., 2001a). Clone identity of all RNAi bacteria was verified by sequencing. In addition, quantitative reverse transcriptase-PCR confirmed the inhibition of ifg-1 and rsks-1 (data not shown).
Polysomal profiling
Two-day-old adult worms were used to generate polysomal profiles. Previously used method (Dinkova et al., 2005) was optimized to resolve the polysomes and ribosomal subunits. One hundred microliter of gently pelleted worms were homogenized on ice in 300 μL of solublization buffer [300 mm NaCl, 50 mm Tris-HCl (pH 8.0), 10 mm MgCl2, 1 mm EGTA, 200 μg heparin per mL, 400 U RNAsin per mL, 1.0 mm phenylmethylsulfonyl fluoride, 0.2 mg cycloheximide per mL, 1% Triton X-100, 0.1% Sodium deoxycholate] by 60 strokes with a Teflon homogenizer. Seven hundred microliter additional solubilization buffer was added, vortexed briefly, and placed back on ice for 10 min before centrifuging the sample at 20 000 g for 15 min at 4 °C Approximately 0.9 mL of the supernatant was applied to the top of a 10-50% sucrose gradient in high salt resolving buffer [140 mm NaCl, 25 mm Tris-HCl (pH 8.0), 10 mm MgCl2] and centrifuged in a Beckman SW41Ti rotor (Beckman Coulter, Fullerton, CA, USA) at 180,000 g for 90 min at 4 °C. Gradients were fractionated with continuous monitoring of absorbance at 260 nm.
Lifespan analysis
Lifespan analysis was performed as previously described (Lithgow et al., 1995), except that 80 μL of 0.2 mm (+)-5-fluorodeoxyuridine (FUdR) was added onto plates daily during the reproductive phase to eliminate contamination from progeny. Worms were transferred onto fresh plates every 3 days. In all experiments, RNAi was introduced to 1-day-old adults except for the experiments in Fig. 2b, where RNAi was introduced to eggs. All trials were performed at 20 °C. Survival was scored by means of touch-provoked movement and pumping of the pharynx. All survival plots refer to lifespan beginning from adulthood including for rsks-1(ok1255) which develops slower than control. Animals that crawled off the plate, burrowed in the agar, or died from internally hatching progeny were censored.
Statistical analysis
Statistical analyses were performed using the Prism 4 software (Graphpad Software, Inc., San Diego, CA, USA). Kaplan-Meier survival curves were plotted for each lifespan and compared using the log-rank test.
35S-methionine incorporation
OP50 was cultured in LB containing 100 μCi mL-1 of 35S-methionine for 12 h and then spotted onto NGM plates, then spotted with approximately 200 worms in 0.2 mm FUdR and incubated at 20 °C for varying time points. We then measured radioactivity in protein extracts collected from these worms to determine translation rates. Worms were washed with S-basal and placed on normal OP50 for 30 min to purge undigested 35S-methionine-labelled OP50 out of their intestines. Worms were given another wash with S-basal and flash frozen in liquid nitrogen. Frozen samples were boiled in 1% SDS, centrifuged at 10,000 g, and the supernatant was precipitated in 5% TCA on ice for 1 h. Protein was collected by centrifugation at 13 000 r.p.m. and washed with icecold ethanol. The ethanol was then allowed to dry completely and the protein pellet was resuspended in 1% SDS. Protein concentration was then assayed by bicinchoninic acid (BCA) protein assay (Pierce Chemical Co., Rockford, IL, USA) in 96-well plates. 35S activity was measured by liquid scintillation with a Beckman LS6500.
Quantitative RT-PCR
Trizol reagent (Invitrogen Corp., Carlsbad, CA, USA) was used for total RNA preparations from C. elegans. Total RNA was treated with DNase I (New England Biolabs, Ipswich, MA, USA). The first strand cDNA was synthesized using the reverse transcription system (Promega, Madison, WI, USA). SYBR Green dye (Applied Biosystems, Foster City, CA, USA) was used for qRT-PCR. Reactions were run in triplicate on an ABI Prism 7000 real-time PCR machine (Applied Biosystems). Relative-fold changes were calculated using the 2-ΔΔCt method (Livak & Schmittgen, 2001). The sequences of primers used were as follows: for pmp-2 mRNA, forward, GAT TGT TTT ACC ACA AAC CAC; reverse, GTT CGA AAC GAT ATG ATC CAC; for ifg-1 mRNA, forward, AGC AAA GAG ATC GTA TGC AC; reverse, ACA AAC TTC AAT AAC CGC AG.
Nested PCR
The sequences of primers used were as follows: outer left GAG ATG CGG AAG CTA TGC TC, outer right GTT GAA TTC CTG CTC CTC CA, inner left ATT CAA CTG TGT GCC AGT GC, inner right TGG GGC TTC ACT ATT TGG TC. These primers were used to confirm the deletion in the rsks-1 mutant.
Fecundity analysis
Individual unmated hermaphrodites were transferred every 24 h and their daily progeny was scored.
Pump rate analysis
Pump rates were measured as previously described (Avery & Horvitz, 1989).
Body size measurement
N2 and rsks-1(ok1255) adults were allowed to lay eggs for 1 h on OP50. Eggs were then collected and grown at 20 °C. At 24 h intervals, twenty worms of each type were mounted onto slides in S-basal, heat killed at 65 °C for 2 min and then imaged on an Olympus 1 × 70 microscope. Worm lengths were measured from the center of the head to the tail, along the side of the worm, and converted into pixel numbers using Scion Image Beta 4.02 (Scion Corporation, Fredrick, MD, USA).
Stress resistance assays
Thermotolerance
Thermotolerance assays were performed as previously described (Lithgow et al., 1995). Synchronous 5-day-old adults were shifted from 20 °C to 35 °C and survival scored by means of touchprovoked movement. Young adult worms were exposed to RNAi treatment for 48 h before the assay. Worms not responding to touch were scored as dead.
Oxidative stress
Juglone resistance was performed as described previously (de Castro et al., 2004). Ethanol dissolved juglone solution was added to liquid NGM to a final concentration of 240 μm and poured immediately onto plates to dry. One hundred microliter of OP50 was spotted on the plates and dried. Three hours after the plates were poured, 30 5-day-old adults were transferred to each plate after RNAi treatment during adulthood. Worms were incubated at 20 °C and scored hourly until death.
UV resistance
Worms synchronized at L4 stage were put on RNAi bacteria for 48 h. Then worms are transferred onto a NGM agar plate without any bacteria and exposed to 2000 J m-2 UV using a UV Stratalinker® 1800 (Stratagene, La Jolla, CA, USA). Worms are then transferred back to plates containing their respective RNAi bacteria and scored for survival.
Starvation resistance
Synchronized L4 worms grown at 20 °C on OP50 were transferred onto large plates containing RNAi bacteria and 0.5 mL of 0.2 mm FUdR for 48 h. Then worms were washed from the plates and transferred to a 15-mL conical tube. Worms were washed five times in S-basal. Finally, worms were mixed in S-basal containing 0.2 mm FUdR and aliquots were placed into culture dishes. Worms were shaken continuously in this media. Scoring was done by placing 50 μL of the sample onto a NGM agar dish.
Supplementary Material
Fig. S1 Recruitment of the eIF4F complex to the 5′-cap structure. eIF4E binds to the 5′-cap of the mRNA. The binding of scaffold protein eIF4G to eIF4E allows the assembly of other factors such as eIF4A, eIF4B, and poly(A)-binding proteins (PABPs) to the mRNA. The PABPs are bound to both the poly(A) tail of the mRNA and eIF4G. The binding of eIF4G to both the PABPs and eIF4E circularizes the mRNA, which increases the rate of translation.
Fig. S2 Pumping rate upon inhibition of ifg-1 and rsks-1. Pump rates were measured in (a) rsks-1(ok1255) and N2 strains and (b) N2 on control RNAi and ifg-1(RNAi). At least 30 animals were used for each experiment. Animals were exposed to RNAi for 48 h from L4 stage and pump rate measured at day 2 of adulthood. Similar results were obtained in at least three separate experiments. Pumping rate was not significantly different to explain the large differences in methionine incorporation between strains, moreover, the measurement of the slopes of the curve, indicates the rate of methionine incorporation irrespective of the feeding rate.
Fig. S3 Inhibition of genes involved in cap-dependent translation extend lifespan in C. elegans. (a) Mean lifespan was 14.5 days (n = 103) for N2 on control RNAi; 16.8 days (n = 124) for pab-1(RNAi); 16.3 days (n = 112) for pab-2(RNAi); and 15.5 days (n = 112) for ife-1(RNAi) (P < 0.0002).
Acknowledgments
We thank members of the Kapahi and Lithgow labs, Brian Zid, David Walker, Judith Campisi and Dale Bredesen for useful discussions and comments on the manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH), National Center for Research Resources (NCRR). We also thank Xiaomeng Long and Joseph Avruch for providing RNAi clones. Aric N. Rogers is on a T32 NIH training grant fellowship (AG020495). Anders Olsen is supported by the Danish Cancer Society. Gordon J. Lithgow is supported by the Ellison Medical Foundation and NIH. Pankaj Kapahi is supported by a grant from the Ellison Medical Foundation and a gift from the Harold J. and Reta Haynes Family Foundation.
References
- Arantes-Oliveira N, Berman JR, Kenyon C. Healthy animals with extreme longevity. Science. 2003;302:611. doi: 10.1126/science.1089169. [DOI] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron. 1989;3:473–485. doi: 10.1016/0896-6273(89)90206-7. [DOI] [PubMed] [Google Scholar]
- Berset C, Trachsel H, Altmann M. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1998;95:4264–4269. doi: 10.1073/pnas.95.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E, Hegi de Castro S, Johnson TE. Isolation of long-lived mutants in Caenorhabditis elegans using selection for resistance to juglone. Free Radic. Biol. Med. 2004;37:139–145. doi: 10.1016/j.freeradbiomed.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Dinkova TD, Keiper BD, Korneeva NL, Aamodt EJ, Rhoads RE. Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol. Cell. Biol. 2005;25:100–113. doi: 10.1128/MCB.25.1.100-113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewbank JJ, Barnes TM, Lakowski B, Lussier M, Bussey H, Hekimi S. Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science. 1997;275:980–983. doi: 10.1126/science.275.5302.980. [DOI] [PubMed] [Google Scholar]
- Finch CE. Longevity, Senescence, and the Genome. University of Chicago Press; Chicago: 1990. [Google Scholar]
- Gems D, Pletcher S, Partridge L. Interpreting interactions between treatments affecting lifespan. Aging Cell. 2002;1:1–9. doi: 10.1046/j.1474-9728.2002.00003.x. [DOI] [PubMed] [Google Scholar]
- Harris TE, Lawrence JC., Jr TOR signaling. Sci. STKE 2003. 2003:re15. doi: 10.1126/stke.2122003re15. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Johnson TE, Lithgow GJ, Murakami S. Hypothesis: interventions that increase the response to stress offer the potential for effective life prolongation and increased health. J. Gerontol. A Biol. Sci. Med. Sci. 1996;51:B392–B395. doi: 10.1093/gerona/51a.6.b392. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001a:2. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid B. TOR pathway: linking nutrient sensing to life span. Sci. Aging Knowl. Environ. 2004. 2004:PE34. doi: 10.1126/sageke.2004.36.pe34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc. Natl Acad. Sci. USA. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long X, Spycher C, Han ZS, Rose AM, Muller F, Avruch J. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 2002;12:1448–1461. doi: 10.1016/s0960-9822(02)01091-6. [DOI] [PubMed] [Google Scholar]
- Martin GM, Austad SN, Johnson TE. Genetic analysis of ageing: role of oxidative damage and environmental stresses. Nat. Genet. 1996;13:25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Munro HN. Evolution of protein metabolism in mammals. In: Munro HN, Allison JB, editors. Mammalian Protein Metabolism. Vol. 3. Academic Press; New York: 1969. pp. 133–182. [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR. C. elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1997. [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl Acad. Sci. USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci. 2006;31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Shamji AF, Nghiem P, Schreiber SL. Integration of growth factor and nutrient signaling: implications for cancer biology. Mol. Cell. 2003;12:271–280. doi: 10.1016/j.molcel.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hershey JWB, Mathews BM, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2000. [Google Scholar]
- Thomas G. The S6 kinase signaling pathway in the control of development and growth. Biol. Res. 2002;35:305–313. doi: 10.4067/s0716-97602002000200022. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, Thomas G. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- Williams GC. Pleiotropy, natural selection, and evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Recruitment of the eIF4F complex to the 5′-cap structure. eIF4E binds to the 5′-cap of the mRNA. The binding of scaffold protein eIF4G to eIF4E allows the assembly of other factors such as eIF4A, eIF4B, and poly(A)-binding proteins (PABPs) to the mRNA. The PABPs are bound to both the poly(A) tail of the mRNA and eIF4G. The binding of eIF4G to both the PABPs and eIF4E circularizes the mRNA, which increases the rate of translation.
Fig. S2 Pumping rate upon inhibition of ifg-1 and rsks-1. Pump rates were measured in (a) rsks-1(ok1255) and N2 strains and (b) N2 on control RNAi and ifg-1(RNAi). At least 30 animals were used for each experiment. Animals were exposed to RNAi for 48 h from L4 stage and pump rate measured at day 2 of adulthood. Similar results were obtained in at least three separate experiments. Pumping rate was not significantly different to explain the large differences in methionine incorporation between strains, moreover, the measurement of the slopes of the curve, indicates the rate of methionine incorporation irrespective of the feeding rate.
Fig. S3 Inhibition of genes involved in cap-dependent translation extend lifespan in C. elegans. (a) Mean lifespan was 14.5 days (n = 103) for N2 on control RNAi; 16.8 days (n = 124) for pab-1(RNAi); 16.3 days (n = 112) for pab-2(RNAi); and 15.5 days (n = 112) for ife-1(RNAi) (P < 0.0002).