Abstract
The serpin endopin 2A inhibits the cysteine protease papain in cross-class inhibition. This study demonstrates the novel finding that both the non-RSL NH2-domain and the RSL domain with P1-P1′ residues participate in endopin 2A inhibition. Production of a chimeric mutant of endopin 2A with replacement of its NH2-domain with that of endopin 1 resulted in less effective inhibition of papain, indicated by its lower kass association rate constant compared to wild-type endopin 2A. This chimeric mutant formed complexes with papain, but at lower levels compared to that with wild-type endopin 2A. Papain degradation of a portion of the chimeric mutant suggested a role for the NH2-domain in regulating relative amounts of endopin 2A that enter the substrate pathway compared to the serpin inhibitory pathway. Furthermore, site-directed mutagenesis demonstrated that the RSL domain with intact P1-P1′ residues was necessary for inhibition. These findings indicate that the NH2-domain and the RSL region both participate in endopin 2A inhibition of papain.
Keywords: endopin, serpin, papain, cysteine protease, cross-class inhibition, non-RSL, active site residues, mutagenesis, cimera
Introduction
Endopin 2A is a member of the serpin protease inhibitor family that possesses the property of cross-class inhibition of cysteine and serine proteases [1, 2]. The secretory vesicle endopin 2A (bovine) has been identified as a serpin that effectively inhibits the cysteine protease papain and the serine protease elastase [1]. The related endopin 2C shows inhibition of the cysteine protease cathepsin L, as well as elastase [3]. The endopin 1 isoform, in contrast, inhibits only the serine protease trypsin and does not inhibit cysteine proteases [4-6]. These endopin isoforms are present in neurosecretory vesicles that contain cysteine and serine proteases. These proteases participate in processing proproteins into peptide neurotransmitters and hormones [1, 3].
Endopin 2A, like other serpins, contains a reactive site loop (RSL) domain that is recognized and cleaved by the target protease. During this process, complexes of endopin 2A with its target protease(s) are formed [1]. It is known that the P1-P1′ residues of serpins mimic the substrate cleavage site of the target protease so that the protease recognizes and cleaves the serpin's P1-P1′ site [2]. Indeed, peptide sequencing of endopin 2A fragments generated by papain indicated papain cleavage between the predicted Ser-Ser site as P1-P1′ residues [1]. These findings indicate the importance of the RSL domain for endopin 2A inhibition of papain.
In contrast to the RSL domain, little is known about the functional roles of non-RSL domains for endopin 2A and serpin cross-class inhibition. Therefore, this mutagenesis study investigated the role of the non-RSL NH2-domain combined with the RSL domain of endopin 2A for inhibition of papain. The predicted non-RSL and RSL domains were defined by primary sequence homology to the related serpins endopin 1 and α1-antichymotrypsin [1,4,7]. Mutagenesis generated chimeric forms of endopin 2A and endopin 1. Results demonstrated the novel finding that the non-RSL NH2-domain of endopin 2A participates in inhibition against papain. Furthermore, the RSL domain with specific P1-P1′ residues was important for endopin 2A inhibition of papain. These findings demonstrate the dual roles of the NH2-domain, a non-RSL region, and the RSL region with P1-P1′ residues for mediating endopin 2A cross-class inhibition of papain.
Materials and Methods
Construction and expression of endopin 2A chimeras
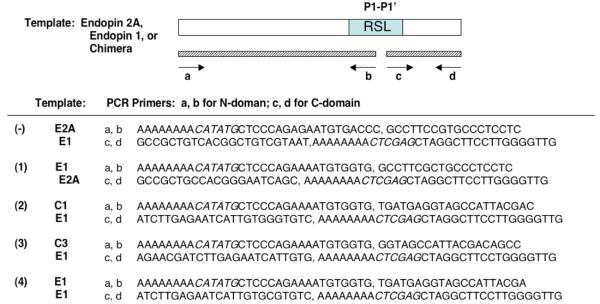
Endopin 2A chimeras with selected regions of endopin 1 were constructed by PCR under cycle conditions of 94° C, 44° C, and 72° C for 1 minute at each temperature, for a total of 25 cycles. Oligonucleotide primers and templates (endopin 2A, endopin 1, or chimera) used for PCR mutagenesis are summarized in Figure 1a. PCR amplified cDNA segments corresponding to 5′- and 3′-domains of endopin 2A were blunt-end ligated and ligated to the pET19b expression vector (NdeI and XhoI restriction sites). All constructs were subjected to DNA sequencing (UC San Diego DNA Sequencing Facility) to confirm mutations.
Figure 1. Endopin 2A chimeras and mutant P1-P1′ variants.
(a) Chimeras of endopin 2A with endopin 1. Chimeras of endopin 2A with endopin 1 that exchanged non-RSL domains, as well as RSL domains, were generated by PCR with the indicated primers and templates consisting of endopin 2A (E2A), endopin 1 (E1), chimera #1 (C1), or chimera #3 (C3). PCR reactions generated 5′- and 3′-domains of cDNAs with primer pairs a/b and c/d, respectively, which were blunt-ligated to generate the mutant endopin 2A cDNAs, subcloned into the expression vector (NdeI and Xho I sites, shown in italics for primers a and d). Expression of recombinant endopins (containing N-His tags) was conducted in E. coli and purification was achieved by nickel affinity chromatography.
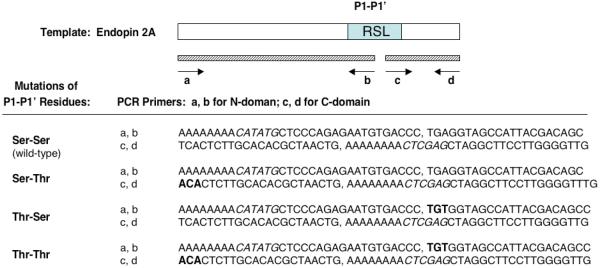
(b) Mutant P1-P1′ variants of endopin 2A. Site-directed mutagenesis of P1-P1′ residues of endopin 2A was performed by PCR of 5′- and 3′-regions with endopin 2A with the illustrated PCR primers and template. PCR reactions generated 5′- and 3′-domains of cDNAs with primer pairs a/b and c/d, respectively, which were blunt-ligated to generate the mutant endopin 2A cDNA that was subcloned into the expression vector (NdeI and Xho I sites). Alterations in P1 and/or P1′ residues resulting from site-directed mutagenesis by PCR are indicated.
These constructs yielded recombinant endopin 2A mutant proteins with His-tag at NH2-termini. Expression was conducted in BL21 (DE3) codon Plus RP E. coli cells (Stratagene, La Jolla, CA). Recombinant mutant endopin proteins were purified by nickel affinity column chromatography (nickel resin from Novagen, Madison, WI), as we have described previously [1,3,4]. Purified endopin 2A variants were demonstrated by single bands on SDS-PAGE gels (12% acrylamide gels).
Construction and expression of endopin 2A mutants with modified P1-P1′ residues
Endopin 2A mutants with modified residues at P1-P1′ were generated by PCR site-directed mutagenesis, as described previously [1,3,4]. Oligonucleotide primers used for PCR mutagenesis, with endopin 2A as template, are illustrated in Figure 1b. Constructs were generated by PCR of 5′- and 3′-domains of endopin 2A with blunt-end ligation, followed by ligation to the pET19b plasmid vector (NdeI and XhoI digested). All constructs were subjected to DNA sequencing to confirm mutations. Expression in E. coli and purification on nickel affinity columns was performed as described in the previous paragraph.
Protease assay and inhibition by endopin 2A
Papain was evaluated for inhibition by endopin 2A chimeras and mutants. Papain (Worthington Biochemical Corp, Freehold, NJ) was assayed with the fluorogenic substrate Z-Phe-Arg-MCA (Bachem, Torrance, CA), in duplicate for each experimental condition. Papain activity was titrated with E64 (Roche, Indianapolis, IN). To assess inhibition of papain, recombinant mutant endopins were preincubated with papain at room temperature for 15 minutes at a molar ratio of serpin/protease of 50/1, and substrate was added for a final concentration of 200 μM with final buffer conditions of 50 mM Na-phosphate, pH 6.4, 200 mM NaCl, 1 mM EDTA, and 2 mM DTT. The remaining protease activity was assayed by incubation at room temperature for 30 minutes and measurement of fluorescent AMC product at excitation/emission wavelengths of 365/450 nm. Protease activity detected in the presence of inhibitor was expressed as percent of control activity (100%, with no inhibitor).
Association rate constant, kass, for inhibition of papain
Inhibition of papain by mutant endopin 2A was assessed by measuring kass, the association rate constant. The constant kass was determined based on the equations ln[E] = −kobs × t and kass = kobs/[I] [8] under pseudo-first-order conditions. The kass rate constant was calculated based on plots of ln[E] vs. incubation time.
Serpin/protease complexes detected by SDS-PAGE gels
Formation of serpin/protease complexes was detected by western blots [1,3,4]. Specifically, mutant endopin 2A was combined with papain at a ratio of endopin/papain of 5/1 or 10/1 in 20 μl assay buffer at room temperature for 10 minutes. The reaction was then stopped by addition of E64 (to 140 μM E64) and 6 μl 5X SDS-PAGE loading buffer (0.062 M Tris-HCl, pH 6.8, 2% SDS, 10% glycerol), and an aliquot was subjected to SDS-PAGE (Tris-glycine gels, 12% acrylamide) for western blots with anti-endopin 2A, as described previously [1,3,4]. Protease complex formation with wild-type endopin 2A was included as positive controls.
Results and Discussion
Role of the non-RSL NH2-domain of endopin 2A for inhibition of papain
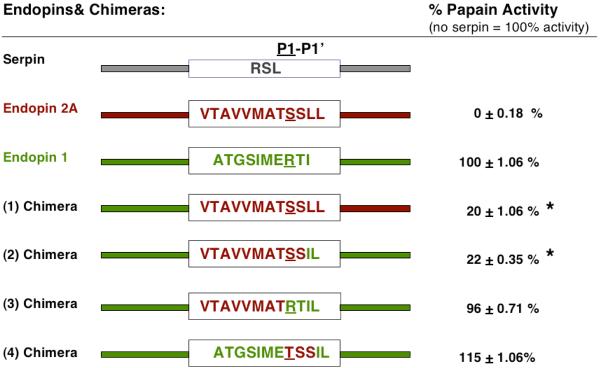
Endopin 2A mutant variants were generated to evaluate the roles of the non-RSL NH2-domain, as well as the RSL region, for inhibition of papain. Evaluation of the NH2-domain of endopin 2A for inhibition of papain utilized mutant chimera #1, which consisted of replacement of the NH2-domain of endopin 2A with the NH2-domain of endopin 1 (Figure 2). The NH2-domain of endopin 1 was defined based on its primary sequence homology with endopin 2A; notably, endopin 1 does not inhibit papain [4]. Chimera #1 showed diminished inhibition of papain (Figure 2). Chimera #1 inhibited papain by 80%, compared to inhibition of papain by 100% by wild-type endopin 2A (with inhibitor/protease at molar ratios of 50/1).
Figure 2. Inhibitory activities of endopin chimeras against papain.
Wild-type endopin 2A, endopin 1, and chimeras #1-4 of endopin 2A were assessed for inhibition of papain. Endopin 2A domains are shown in red, and endopin 1 domains are shown in green. Papain activity was expressed as percent control activity in the absence of inhibitor (100% activity). Mutant endopins were assessed at a molar ratio of endopin/protease of 50/1. The amount of papain activities in the presence chimeric mutants of endopin 2A are expressed as the mean ± sem with the * symbol indicating statistical significance with p < 0.005 (by student's t-test).
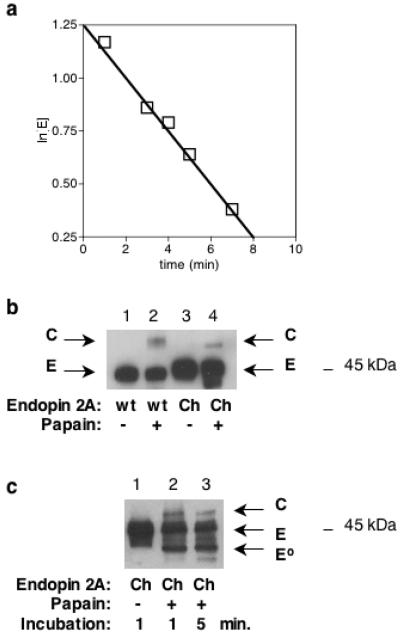
To compare the relative inhibitory activity of chimera #1 compared to wild-type endopin 2A, the association rate constant, kass, was measured (Table 1). The kass kinetic constant was determined by the equations ln[E] = −kobs × t and kass = kobs/[I] [8]. The plot of ln[E] vs incubation time (Figure 3a) provided determination of kass for chimera #1 as 3.3 × 103 M−1s−1. The kass for chimera #1 was found to be substantially lower than the kass for wild-type endopin 2, determined as 1.4 × 106 M−1s−1 [1] (Table 1). These results indicated that chimera #1 was substantially less effective than wild-type endopin 2A for inhibition of papain, indicating a functional role for the non-RSL NH2-domain of endopin 2A for inhibition.
Table 1.
Association rate constants, kass, for chimeric and wild-type forms of endopin 2A.
| Endopin Form |
kass, M−1s−1 |
|---|---|
| Wild-type Endopin 2A | 1.4 × 106 M−1s−1 |
| Chimera #1 | 3.3 × 103 M−1s−1 |
Figure 3. Chimeric endopin 2A with NH2-domain of endopin 1: association rate constant, kass, and complex formation.
(a) Determination of kass constant for inhibition of papain. The association rate constant, kass, for inhibition of papain by chimera #1 was determined by plotting ln[E] vs. time of incubation. The constant kass was calculated based on the equations ln[E] = −kobs × t and kass = kobs/[I] [8]. Chimera #1 inhibited papain with kass of 3.3 × 103 M−1s−1. Values for ln[E] at at all time points showed means with standard error of the mean (sem) of less than 1.5% of mean values. These standard error values were smaller than the symbols shown in figure 3a.
(b) Chimeric endopin 2A complexes with papain. The mutant chimera #1 was evaluated for complex (C) formation with papain enzyme (E) after 10 minutes incubation (as described in materials and methods), with analyses of complexes by western blots with anti-endopin 2A. The molar ratio of endopin/protease was 10/1.
(c) Time-course for incubation of mutant chimera #1 with endopin 2A. The mutant chimera #1 form of endopin 2A (E) was incubated with papain for 1, 5, and 10 minutes under the identical conditions described for figure 3b. Analyses of endopin and papain complexes (C), as well as degraded endopin 2A (E°), were conducted by western blots with anti-endopin 2A.
The ability of chimera #1 to form complexes with papain was assessed, since serpins typically form SDS-stable complexes with target protease(s) [2]. These experiments showed that SDS-stable complexes of chimera #1 with papain were generated (after 10 min incubation, with complexes detected by western blots, shown in Figure 3b). It was noted that complexes of mutant chimera #1 with papain were present at lower levels compared to complexes of wild-type endopin 2A and papain. Analyses at different times points (Figure 3b and 3c) showed that degradation of chimera #1 occurred immediately with one minute of incubation with papain, but wild-type endopin 2A was not degraded by papain (Figure 3b). These results indicated that the mutant NH2-terminal domain of chimera #1 promoted the substrate pathway for papain degradation of the mutant serpin, while the wild-type NH2-domain of endopin 2A maintained the serpin's inhibitory function known to occur through the substrate suicide pathway [2].
Evaluation of the RSL domain for endopin 2A inhibitory activity
Experiments assessed the role of the RSL domain with chimera #2, a mutant consisting of the RSL domain of endopin 2A with non-RSL NH2- and COOH-domains of endopin 1 (illustrated in Figure 2). Chimera #2 showed effective inhibition of papain by 78%, indicating a functional role for the RSL domain. Further assessment of the RSL region at P1-P1′residues was assessed with chimera #3 that was generated by mutagenesis of chimera #2 to contain mutant P1-P1′ residues of Arg-Thr. Chimera #3 showed no inhibition of papain, which contrasts with much inhibition of papain by chimera #2. Chimera #4, containing mutant P2-P1′ residues of endopin 2A within the RSL of endopin 1 also showed no inhibition of papain. These results indicated that the RSL domain of endopin 2A with intact P1-P1′ residues was necessary for inhibition of papain.
Further evaluation of P1-P1′ residues of endopin 2A was achieved by site-directed mutagenesis of endopin 2A. Comparison of the P1-P1′ region of endopin 2A to other serpins that inhibit cysteine proteases -- headpin, SCCA1, SQN-5, and MENT [9-13] showed Ser or Thr residues at or near these positions (Table 2). Mutagenesis of endopin 2A with Ser-Thr as P1-P1′ residues (instead of wild-type Ser-Ser) retained its ability to inhibit papain by 99% (Table 3). In contrast, the endopin 2A mutant with Thr-Ser as P1-P1′ residues resulted in diminished inhibition of papain by 49%, and the mutant with Thr-Thr as P1-P1′ residues showed absence of inhibitory activity. These results demonstrated the importance of Ser as P1 and P1′ residues for inhibition.
Table 2.
RSL Domains of Endopins and Serpins that Inhibit Papain-Like Cysteine Proteases.
| Serpins |
P10 to P4' residues of RSL Domains |
Target Protease# |
|---|---|---|
| Secreted Serpins: | P1–P1′ | |
| Endopin 2A, 2B* | A V T A V V M A T S S L | papain |
| Endopin 2C | A V T A V I M F T S L P | cathepsin L/papain, elastase |
| Cytosolic Serpins: | ||
| Headpin | A A T G I G F T V T S A | cathepsin L |
| SCCA1 | A A T A E V G F G S S P | cathepsin L, papain, cathepsin G. |
| SQN–5 | A A T G V E V S L T S A | cathepsins L and G |
| MENT | A A T A V I I S F T T S | cathepsin L |
Table 3.
Mutagenesis of Endopin 2A at P1-P1′ Residues and Inhibition of Papain
| P1 - P1′ |
Percent Control Papain Activity. |
|---|---|
| No inhibitor | 100 ± 1.06 % |
| Ser-Ser (wild-type) | 0 ± 0.18 % * |
| Ser-Thr | 1 ± 0.35 % * |
| Thr-Ser | 51 ± 2.5 % * |
| Thr-Thr | 118 ± 0.35 % |
Mutant forms of endopin 2A at P1-P1′ residues were generated by site-directed mutagenesis, as described in the methods. Inhibitory activities of purified recombinant endopin 2A mutants was tested at a constant molar serpin/protease ratio of 50/1. Protease activities for papain were expressed as percent of control in the absence of inhibitor. Results are shown as the mean ± sem, and the
symbol indicates statistically significant compared to no inhibitor control (100%) with p < 0.005 (by student's t-test).
Conclusions
The combined results of these mutagenesis studies demonstrated the functional roles of both the non-RSL NH2-domain and the RSL region. Studies with mutant chimeric forms of endopin 2A demonstrated the novel finding that the non-RSL NH2-domain participates in endopin 2A inhibition of papain (chimera #1). The mutant chimera #1 with the replacement of the NH2-domain of endopin 2A with that of endopin 1 showed reduced inhibitory activity indicated by its lower kass association rate constant compared to wild-type endopin 2A for inhibition of papain.
Analyses of complex formation demonstrated that the wild-type NH2-domain of endopin 2A was important for maintaining the inhibitory pathway for endopin 2A complex formation and inhibition of papain. In contrast, the mutant NH2-domain of chimera #1 facilitated the substrate pathway for its degradation by papain, with a lower level of complex formation compared to wild-type endopin 2A. This novel finding suggests that the NH2-domain of endopin 2A may be important for regulating the distribution of serpin-protease interactions to enter the inhibitory pathway with complex formation, compared to entering the substrate pathway for protease degradation of the serpin.
Identification of endopin 2A by molecular cloning in an earlier study provided primary sequence information of the predicted RSL domain ([1]. Therefore, this study assessed features of the RSL domain for inhibitory activity of endopin 2A through mutagenesis experiments. Results from this study showed that the RSL domain of endopin 2A, with intact P1-P1′ residues, was necessary for inhibition of papain. Chimeric mutant #2, consisting of the RSL domain of endopin 2A with NH2- and COOH-domains of endopin 1, showed inhibition of papain. But P2-P1′ residues of endopin 2A alone within endopin 1, represented by chimera #4, had no inhibitory activity; such results demonstrated that P2-P1′ residues alone were not sufficient for inhibition. Additional mutagenesis of endopin 2A P1-P1′ residues indicated the requirement for Ser at the P1 position. At the P1′ position, Thr or Ser allowed inhibition of papain. These results illustrate the functional roles of the intact RSL domain with Ser-Ser as P1-P1′ residues for endopin 2A inhibition of papain.
The secretory vesicle localization of endopin 2A predicts that it functions as an endogenous inhibitor of target proteases within this subcellular organelle, or in RER and Golgi apparatus that are involved in the biosynthesis and cellular trafficking of secretory proteins [18] such as endopin 2A. Upon translation of endopin 2A from its mRNA at the rough endoplasmic reticulum (RER), the signal sequence would be cleaved at the RER. The resultant mature endopin 2A, like other proteins targeted to secretory vesicles, would be predicted to undergo trafficking from RER to the Golgi apparatus prior to packaging into secretory vesiscles. Neuroendocrine secretory vesicles possess important protease functions for the biosynthesis of active neuropeptide transmitters and hormones that are produced by proteolytic processing of precursor proteins. Based on the subcellular localization of endopin 2A, endopin 2A may play a role in the regulation of proteases that support secretory vesicle functions. It is likely that papain-like cysteine proteases, or elastase-like serine proteases, may be regulated by endopin 2A in vivo which has been shown to possess cross-class inhibition [1]. Ongoing studies are in progress to identify endogenous mammalian protease targets of endopin 2A to gain knowledge of the biological role of endopin 2A in secretory vesicle functions.
In summary, this study is among the first to demonstrate a functional role for the non-RSL NH2-domain, combined with the RSL domain with P1-P1′ residues, for endopin 2A serpin cross-class inhibition of the cysteine protease papain. Notably, results with chimeric mutant forms of endopin 2A suggest that the NH2-domain of endopin 2A may be involved in designation of the endopin 2A serpin to the inhibitory substrate suicide pathway for papain inhibition, or to the non-inhibitory pathway whereby endopin 2A serves as substrate and undergoes degradation by papain. Overall, the NH2-domain and RSL participate in the inhibitory mechanism of the serpin endopin 2A for cross-class inhibition of papain.
Acknowledgments
The authors appreciate technical assistance by Jennifer Rattan. This work was supported by grants from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hwang S-R, Steineckert B, Toneff T, Bundey R, Logvinova AV, Goldsmith P, Hook VYH. Biochemistry. 2002;41:10397–10405. doi: 10.1021/bi020088o. [DOI] [PubMed] [Google Scholar]
- 2.Gettins PGW. Serpin structure, mechanism, and function. Chem. Rev. 2002;102:4751–4803. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 3.Hwang S-R, Stoka V, Turk V, Hook VYH. Biochemistry. 2005;44:7757–7767. doi: 10.1021/bi050053z. [DOI] [PubMed] [Google Scholar]
- 4.Hwang S-R, Steineckert B, Yasothornsrikul S, Sei CA, Toneff T, Rattan J, Hook VYH. J. Biol. Chem. 1999;274:34164–34173. doi: 10.1074/jbc.274.48.34164. [DOI] [PubMed] [Google Scholar]
- 5.Tassy C, Herrera-Mendez CH, Sentandreu MA, Aubry L, Brémaud L, Pélissier P, Delourme D, Brillard M, Gauthier F, Levéziel H, Ouali A. Biochem. J. 2005;388:273–280. doi: 10.1042/BJ20041921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrera-Mendez CH, Brémaud L, Coulis G, Pélissier P, Sentandreu MA, Aubry L, Delourme D, Chambon C, Maftah A, Levéziel H, Ouali A. FEBS Lett. 2006;580:3477–3484. doi: 10.1016/j.febslet.2006.04.099. [DOI] [PubMed] [Google Scholar]
- 7.Rubin H, Wang ZM, Nickbarg EB, McLarney S, Naidoo N, Schoenberger OL, Johnson JL, Cooperman BS. J. Biol. Chem. 1990;265:1199–1207. [PubMed] [Google Scholar]
- 8.Salvesen G, Hagase H. In: Proteolytic Enzymes, a Practical Approach. Beynon RJ, Bond JS, editors. IRL Press; Oxford: 1989. pp. 83–104. 1989. [Google Scholar]
- 9.Jayakumar A, Kang Y, Frederick MJ, Pak SC, Henderson Y, Holton PR, Mitsudo K, Silverman GA, EL-Naggar AK, Brömme D, Clayman GL. Arch. Biochem. Biophys. 2003;409:367–374. doi: 10.1016/s0003-9861(02)00635-5. 2003. [DOI] [PubMed] [Google Scholar]
- 10.Welss T, Sun J, Irving JA, Blum R, Smith AI, Whisstock JC, Pike RN, von Mikecz A, Ruzicka T, Bird PI, Abts HF. Biochemistry. 2003;42:7381–7389. doi: 10.1021/bi027307q. [DOI] [PubMed] [Google Scholar]
- 11.Schick C, Pemberton PA, Shi G-P, Kamachi Y, Çataltepe S, Bartuski AJ, Gornstein ER, Brömme D, Chapman HA, Silverman GA. Biochemistry. 1998;37:5258–5266. doi: 10.1021/bi972521d. [DOI] [PubMed] [Google Scholar]
- 12.Al-Khunaizi M, Luke CJ, Askew YS, Pak SC, Askew DJ, Çataltepe S, Miller D, Mills DR, Tsu C, Brömme D, Irving JA, Whisstock JC, Silverman GA. Biochemistry. 2002;41:3189–3199. doi: 10.1021/bi015999x. [DOI] [PubMed] [Google Scholar]
- 13.Irving JA, Shushanov SS, Pike RN, Popova EY, Brömme D, Coetzer TH, Bottomley SP, Boulynko IA, Grigoryev SA, Whisstock JC. J. Biol. Chem. 2002;277:13192–13201. doi: 10.1074/jbc.M108460200. J.C. [DOI] [PubMed] [Google Scholar]
- 14.Schick C, Brömme D, Bartuski AJ, Uemura Y, Schechter NM, Silverman GA. Proc. Natl. Acad. Sci. USA. 1998;95:13465–13470. doi: 10.1073/pnas.95.23.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luke C, Schick C, Tsu C, Whisstock JC, Irving JA, Brömme D, Juliano L, Shi G-P, Chapman HA, Silverman GA. Biochemistry. 2000;39:7081–7091. doi: 10.1021/bi000050g. [DOI] [PubMed] [Google Scholar]
- 16.Irving JA, Pike RN, Dai W, Brömme D, Worrall DM, Silverman GA, Coetzer TH, Dennison C, Bottomley SP, Whisstock JC. Biochemistry. 2002;41:4998–5004. doi: 10.1021/bi0159985. [DOI] [PubMed] [Google Scholar]
- 17.Hwang S-R, Garza CZ, Wegrzyn JL, Hook VY. Biochem. Biophys. Res. Commun. 2005;327:837–844. doi: 10.1016/j.bbrc.2004.12.053. V.Y. [DOI] [PubMed] [Google Scholar]
- 18.Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. Molecular Cell Biology. W. H. Freeman and Company; New York: 2000. [Google Scholar]