Abstract
Human travel to malaria endemic lowlands from epidemic highlands has been shown to increase the risk of malaria infections in the highlands. In order to gain insight on the impact of human travel, we examined prevalence, genetic variability and population genetic structure of Plasmodium falciparum in asymptomatic children from one highland site and three surrounding malaria endemic lowland sites in western Kenya, using multilocus microsatellite genotyping. We further analyzed the frequencies of mutations at the genes conferring resistance to chloroquine and sulphadoxine-pyrimethamine. We found a significant decrease in malaria prevalence in the highland site from 2006 to 2007, one year after the introduction of the artemisinin-based combination therapy as first-line treatment for uncomplicated malaria and the scale-up of insecticide treated bed nets. Population genetic diversity, measured by the number of observed and effective microsatellite alleles and Nei’s unbiased genetic diversity, was high and comparable for both highland and lowland populations. Analysis of molecular variance did not detect a significant genetic structure across highland and lowland regions. Similarly, mutations at key antimalarial -resistance codons of the pfcrt, pfmdr1, pfdhfr and pfdhps genes were found at comparable high frequencies in all four sites. High level of gene flow and lack of significant genetic structure in malaria parasites between highland and lowland areas suggest the importance of human travel in shaping parasite population structure.
Index Key Words: Plasmodium falciparum, Kenya, highlands, lowlands, human travel, antimalarial-drug resistance
1. Introduction
Malaria remains the leading cause of morbidity and mortality in Africa, with an estimated death toll exceeding one million people each year (WHO, 2000). Stable endemic malaria thrives throughout the African continent, but epidemic malaria occurs at the fringe of the endemic areas, particularly in areas of higher altitudes, in the arid regions of North Africa and in communities living at the most southern latitudes (Hay et al., 2002). The densely populated and agriculturally significant highlands of western Kenya are among these epidemic prone-areas. In the past century, the highlands of Western Kenya suffered from two periods of epidemic malaria. The first occurred in the early 1930s and was contained by the 1960s through the use of indoor residual spray with DDT and mass-drug administration (Shanks et al., 2005b). However, epidemics have resurged since the late 1980s (Shanks et al., 2005a). A number of hypotheses have been proposed to explain the mechanisms for the re-emergence of malaria in African highlands: among these are antimalarial drug resistance (Zhong et al., 2008) and parasite introduction from surrounding endemic lowlands through human travel (Shanks et al., 2005a).
Currently, the central tools of malaria prevention and treatment are insecticide-treated bed nets (ITNs) and artemisinin-based combination therapy (ACT). ACT, in the form of artemether-lumefantrine combination, has replaced sulfadoxine-pyrimethamine (SP) and since 2006 has become the first-line antimalarial drug in rural Kenya communities (Amin et al., 2007). Nonetheless, SP is still used for intermittent preventive treatment in pregnancy while quinine is the drug of choice for treating children <5 kg and pregnant women (Zurovac et al., 2008). Despite the fact that human factors, such as the immune response, influence treatment outcome (White, 2002), indications of the level of resistance to antimalarial drugs can be estimated from the frequency of resistance allele types in known drug target genes. Specifically, codon 76 of the chloroquine resistance transporter gene (pfcrt) and codons 86, 1042, and 1246 of the Plasmodium falciparum multidrug resistance gene 1 (pfmdr1) are markers for resistance to chloroquine (CQ) (Babiker et al., 2001; Dorsey et al., 2001). The pfcrt (K76T) mutation is essential to CQ resistance (Babiker et al., 2001; Dorsey et al., 2001), but mutations in the pfmdr1 gene modulate the degree of CQ resistance (Sanchez and Lanzer, 2000) and are also thought to play a role in lumefantrine resistance (Sisowath et al., 2005). Mutations at codons 437 and 540 of the dihydropteroate synthetase (dhps) gene and at codons 51, 59, and 108 of the dihydrofolate reductase (dhfr) gene are markers for resistance to SP (Ouellette, 2001; Peterson et al., 1990). Clinical studies show that SP treatment failure is associated with the quintuple mutant haplotype dhfr-108N/51I/59R dhps-437G/540E (Kublin et al., 2002) and begins to appear with the triple mutant haplotype dhfr-108N /51I/59R (Talisuna et al., 2003). Mutations for resistance to the artemisinin derivates have not been identified yet (Nosten and White, 2007).
In this paper, we examined the level of genetic polymorphism of asymptomatic P. falciparum infections from three lowland localities and one highland site in western Kenya, and the frequencies of gene mutations for SP-and CQ-resistance. Our goal is to determine the genetic structure and gene flow of malaria parasite populations, and to provide baseline mutation frequencies at the onset of large-scale ACT application for malaria control. The baseline mutation frequency information is useful in tracking the evolution of genes associated with SP-and CQ-resistance when SP and CQ selection pressure on malaria parasites is relaxed, and in the deployment of appropriate antimalarial drugs (Laufer et al., 2007).
2. Materials and Methods
2.1. Study sites and sample collection
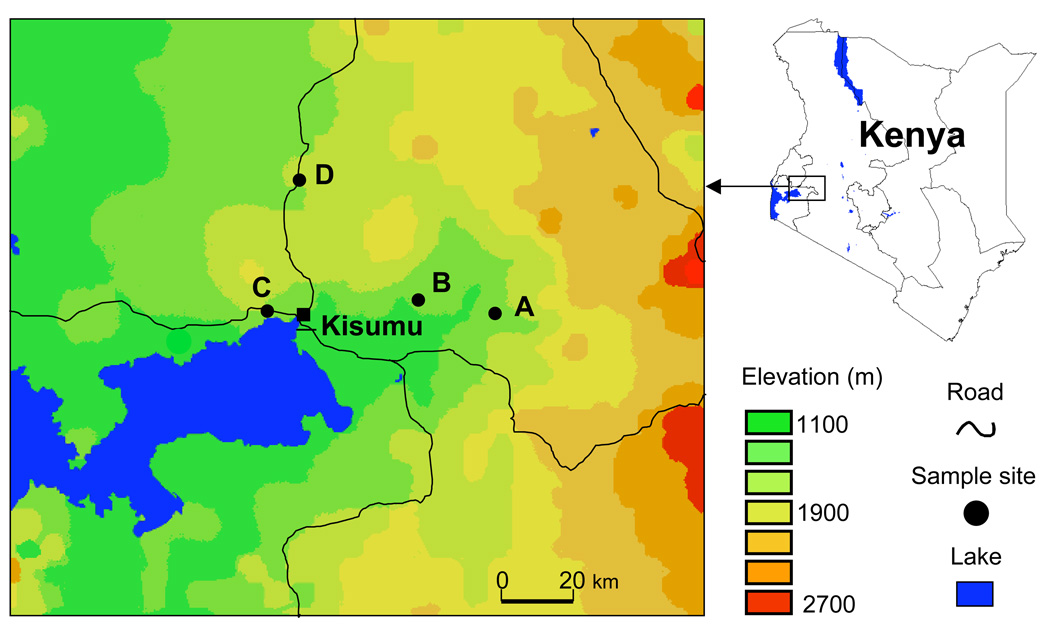
As a part of malaria surveillance activities, blood samples were taken from a total of 599 primary school children (age 6–14) in three lowland sites: Chemelil (35°08”E, 0°05”S, altitude 1248m), Miwani (34°58”E, 0°03”S, altitude 1214m) and Kisian (34°40”E, 0°04”S, altitude 1164m); and in the village of Iguhu, in the highland district of Kakamega (34°–35°E, 0°010”S, altitude 1480–1580m) (Fig. 1). Chemelil and Miwani were sampled in November 2006; Kisian was sampled in October 2006. Iguhu was sampled in October 2006 and June 2007; hereafter the 2006 sample is referred as Iguhu06, the 2007 as Iguhu07. We chose these three representative lowland sites because there is extensive human travel to and from the highland site. If the lowland sites are the source of infection to the highland site, these lowland sites could best represent the main source of infection to the highland site. However, other lowland sites that were not sampled in the present study could also be a source of infection to the highland site.
Figure 1.
A map showing the distribution of sampling sites in Nyanza and Western provinces of Kenya. The three lowland sites are: Chemelil (A), Miwani (B), and Kisian (C). The highland site is Iguhu (D) in the Kakamega district. A total of 599 asymptomatic children (age 6–14 years), including 299 from the lowlands and 300 from the highlands, were enrolled in this study.
Malaria transmission in the lowland sites is perennial, with the main vectors being A. gambiae s. s., A. arabiensis and A. funestus (Githeko et al., 1996; Mutuku et al., 2006). Chemilil and Miwani are within Nyando district, where the temperature ranges from 16.2 to 29.3 C. Average annual rainfall in these two sites is about 1600 mm. Kisian is a village on the shores of Lake Victoria in Kisumu district with temperature ranging from 15.0 to 28.4 C and an average annual rainfall of 1400 mm during the period 1970–2000. In this site, the entomological inoculation rate (EIR) was estimated at 31.1 infectious bites per person per year in 2003–2004 (Ndenga et al., 2006) and the average malaria prevalence among primary school children during the sampling period was 55%. The village of Iguhu experiences two rainy seasons and averages about 1800 mm rainfall per year. The long rainy season usually occurs between mid-March and May while the short rainy season occurs between September and October. The mean annual temperature is 20.8°C. The predominant malaria vector species in the area is A. gambiae s.s. EIR has been estimated at 16.6 and malaria prevalence at 34% in 2003–2004 (Ndenga et al., 2006).
From each child, approximately 200 µl of finger-pricked blood was spotted onto a filter paper, air-dried and stored at −20°C until parasite DNA extraction. P. falciparum asymptomatic infections were diagnosed by microscopy as previously reported (Munyekenye et al., 2005). The human subject protocol involved in this study was approved by the University of California at Irvine, USA and the Kenya Medical Research Institute, Kenya.
2.2 Parasite DNA extraction and species identification
DNA was extracted from the blood filters using the Saponin/Chelex method (Wooden et al., 1993). Parasite DNA was extracted from one quarter of a blood spot of about 1 cm in diameter and dissolved in ~200 µl of distilled water. P. falciparum infections were identified by a species-specific nested PCR method as previously described (Singh et al., 1999). Only samples positive for P. falciparum DNA were used for genotyping analysis.
2.3 Molecular analysis of P. falciparum genetic variability
Ten microsatellite markers (TA1, TA42, TA81, TA87, TA109, ARA2, 2490, Polyα, PfPK2, Pfg377) previously described (Anderson et al., 1999) were used in this study, with the aid of the 4300 automated DNA analyzer (Li-Cor, Lincoln, NE) (Zhong et al., 2007). An allele was declared null after at least three amplification failures. The infection was considered to contain multiple clones if one or more loci showed more than one allele, with minor peaks showing more than one third the height of the predominant peak. In the presence of mixed-clone infections, only the predominant allele (i.e., the alleles with the highest amplification intensity) was used to define haplotypes and calculate microsatellite allele frequencies (Anderson et al., 2000; Ferreira et al., 2002, 2007; Zhong et al., 2007; Joy et al., 2008).
2.4 Molecular analysis of mutations in genesconferring resistance to CQ and SP
The restriction fragment length polymorphism-polymerase chain reaction (RFLP) technique was applied to detect mutations in the pfcrt (K76T), pfmrd1 (N86Y, N1042D and D1246Y), pfdhfr (N51I; C59R and S108N) and pfdhps (A437G and K540E) genes, as described previously (Duraisingh et al., 1998; Flueck et al., 2000; Lopes et al., 2002; Plowe et al., 1995; Schneider et al., 2002). For each codon, with the exception of pfdhps 437, nested PCR was conducted. The analysis of pfmdr1 codon 184 was based on a specific PCR as previously described (Adagu and Warhurst, 1999). Purified genomic DNA from P. falciparum clones Hb3 and Dd2 (MR4, Manassas, VA, USA) were used as positive controls.
2.5 Data analysis
The sensitivity and the specificity of the microscopic detection versus the PCR based identification of P. falciparum were calculated (Banoo et al., 2006). Because of the small number of P. falciparum-infected blood-filters (14) from Iguhu07, the samples collected in 2007 in Iguhu was excluded from the subsequent analysis of the population genetic diversity, structure, and frequencies of mutations at genes associated with antimalarial drug resistance.
To determine the genetic diversity of P. falciparum infections, the number of observed alleles per locus (Na), the number of effective alleles per locus (Ne) and the unbiased genetic diversity (uD) were calculated using GenAlEX6.1 (Peakall R, 2006). Ne is calculated as 1/Σpi , and uD as (n/(n-1))*(1-Σpi ), where pi is the frequency of the ith allele in the population and n is the number of samples. The software ARLEQUIN (Schneider S, 2000) was used to estimate the gene flow (Nm) between samples using the Slatkin's private allele method and to compute the hierarchical analysis of molecular variance (AMOVA), genetic variation portioned at three levels: among individuals within populations, among populations within groups (highland vs. lowland) and among groups. Nei’s genetic distance among population pairs (Nei et al., 1983) was calculated using TFPGA (http://www.marksgeneticsoftware.net/). Bayesian population assignment of haploid organisms was used to assess the potential source population of each isolate using multilocus genotypes through the program BAYEASS-analyses (Fisher et al., 2002). A sample was assigned to a population if the posterior probability that it belonged to that population was greater than 0.95, when all source populations were considered (Fisher et al., 2002).
For among-population variations in the frequencies of mutations at genes conferring resistance to CQ and SP, Fisher’s exact two-tailed test was used to determine the statistical significance. The double/triple/ quadruple/quintuple mutation haplotype frequency at the pfdhfr and pfdhps genes were calculated. For mixed-clone infection, the infection was classified as carrying resistant mutation for the codon as long as resistant alleles are present regardless of the presence of susceptible alleles (e.g., Ferreira et al., 2007; Zhong et al., 2007; Schonfeld et al., 2007; Bacon et al., 2009; Enosse et al., 2008).
3. Results
3.1 P. falciparum infection prevalence and density in asymptomatics from lowland and highland sites
In all the 599 samples examined, the presence of P. falciparum infection was investigated both microscopically and by a species-specific PCR reaction. PCR analyses identified the presence of P. falciparum infections in 44.1% (132/299) of the samples from lowland sites, and in 41.7% (35/84) of the samples from the highland site of Iguhu in 2006 (Iguhu06). Only 6.5% (14/216) of the Iguhu samples collected in 2007 (Iguhu07) were infected with P. falciparum; this is a highly significant reduction in prevalence in comparison to 2006 (P < 0.0001) (Table 1). Microscopic tests found a much lower prevalence rate, which is likely caused by parasite density beyond the threshold of microscopic detection. Microscopic tests showed a high specificity (88.99%), but a low sensitivity (<40%), particularly in those infections with low density. The geometric mean density of P. falciparum ranged from 134.5 to 471.88 infected blood cells per µl blood, the lowest value being found in Iguhu07 and the highest in Kisian.
Table 1.
Comparison between the microscopic and PCR based detection of Plasmodium falciparum from asymptomatic infections from highland and lowland sites in Western Kenya.
| Microscopic method | PCR method | ||||||
|---|---|---|---|---|---|---|---|
| No. positive | Mean density | Sensitivity | Specificity | No. positive | |||
| Region | Site | Samples tested | (%) | (range)1 | (95% CI)2 | (95% CI)2 | (%) |
| Lowland | Chemelil | 102 | 18 (17.7) | 233.8 (80–640) | 0.32 (0.18–0.51) | 0.88 (0.79–0.94) | 28 (27.5) |
| Miwani | 92 | 12 (13.0) | 388.7 (40–10280) | 0.25 (0.14–0.41) | 0.95 (0.85–0.98) | 36 (39.1) | |
| Kisian | 105 | 30 (28.6) | 471.8 (80–7200) | 0.40 (0.29–0.52) | 0.92 (0.79–0.97) | 68 (64.8) | |
| Highland | Iguhu06 | 84 | 8 (9.5) | 329.1 (120–1000) | 0.17 (0.08–0.33) | 0.96 (0.86–0.99) | 35 (41.7) |
| Iguhu07 | 216 | 3 (1.4) | 134.5 (40–760) | 0 (0–0.22) | 0.99 (0.896–1.00) | 14 (6.5) | |
Geometric mean intensity of P. falciparum is calculated only for microscopic-positive infections, and is expressed as the number infected red blood cells per µl blood.
95% CI refers to 95% confidence interval.
3.2 P. falciparum genetic variability between lowland and highland sites
Two isolates in Iguhu06 failed to be amplified and were thus excluded from the analysis, giving a total of 179 isolates examined at 10 microsatellite loci. Genetic variability results are summarized in Table 2. The number of observed alleles (Na) ranged between 1 and 17, and the number of effective alleles (Ne) was between 1.00 and 13.44. Markers Polyα and TA42 were the most and least polymorphic loci over all the samples. In general, Ne was much lower than Na, suggesting that most of the alleles were present at low frequency. The unbiased genetic diversity ranged between 0.62 and 0.80, with the lowest value being found in Chemelil, and the highest in Iguhu06. The proportion of infections with more than one clone among the four populations analyzed ranged from 82.1 to 95.8%. There was no significant difference in the percentage of infections with multiple clones between highland and lowland sites.
Table 2.
Microsatellite variability of Plasmodium falciparum from lowland and highland sites in Western Kenya.
| Region | Site | n | % multiple infections |
Loci | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARA2 | 2490 | Pfg377 | PfPK2 | Polyα | TA42 | TA87 | TA109 | TA81 | TA1 | Mean ± SE | |||||
| Lowland | Chemelil | 28 | 82.1% | Na1 | 8 | 2 | 4 | 6 | 15 | 1 | 11 | 6 | 6 | 9 | 6.80±1.32 |
| Ne2 | 2.53 | 1.07 | 1.62 | 3.53 | 13.44 | 1.00 | 6.88 | 3.98 | 4.46 | 6.76 | 4.53±1.19 | ||||
| uD3 | 0.63 | 0.07 | 0.40 | 0.74 | 0.97 | 0 | 0.89 | 0.78 | 0.80 | 0.88 | 0.62±0.11 | ||||
| Miwani | 36 | 94.4% | Na | 6 | 6 | 4 | 10 | 12 | 2 | 10 | 9 | 11 | 9 | 7.90±1.03 | |
| Ne | 4.77 | 1.73 | 2.55 | 5.31 | 7.28 | 1.06 | 6.82 | 3.82 | 5.54 | 5.89 | 4.48±0.67 | ||||
| uD | 0.81 | 0.44 | 0.63 | 0.84 | 0.89 | 0.06 | 0.88 | 0.76 | 0.84 | 0.85 | 0.70±0.08 | ||||
| Kisian | 68 | 96.8% | Na | 8 | 7 | 6 | 16 | 14 | 2 | 17 | 16 | 12 | 7 | 10.50±1.64 | |
| Ne | 4.06 | 1.90 | 1.87 | 6.66 | 5.75 | 1.03 | 10.61 | 6.78 | 5.56 | 5.36 | 4.96±0.91 | ||||
| uD | 0.77 | 0.48 | 0.47 | 0.86 | 0.84 | 0.03 | 0.92 | 0.87 | 0.83 | 0.83 | 0.69±0.09 | ||||
| Highland | Iguhu06 | 33 | 91.4% | Na | 10 | 5 | 5 | 9 | 13 | 6 | 10 | 14 | 9 | 11 | 9.20±0.99 |
| Ne | 7.34 | 2.60 | 3.47 | 5.57 | 8.44 | 1.73 | 8.08 | 8.31 | 5.89 | 7.08 | 5.85±0.78 | ||||
| uD | 0.89 | 0.63 | 0.74 | 0.85 | 0.91 | 0.44 | 0.91 | 0.91 | 0.86 | 0.89 | 0.80±0.05 | ||||
Na is the number of observed alleles
Ne is the number of effective alleles
uD is the unbiased estimation of genetic diversity.
3.3 Gene flow and population structure
High levels of gene flow (Nm>3) were detected among all the four populations analyzed (Table 3). The highest gene flow was detected between Iguhu06 and Miwani (Nm = 15.34). Nei’s genetic distance estimates were consistent with this notion. AMOVA found insignificant between-group (highland vs. lowland) variance (P = 1.00), suggesting lack of genetic structure between highland and lowland populations. Results from the Bayesian assignment test found that 30.6 – 57.1% isolates were assigned to the sampling localities among the three lowland sites (Table 4). However, the highland site had a very low assignment rate (18.2%), and the majority of the isolates (81.8%) could not be assigned. This suggests that the majority of the parasites in the highland site originated from other localities not analyzed in the current study.
Table 3.
Genetic distance (upper matrix) and gene flow (lower matrix) among the Plasmodium falciparum populations in western Kenya highland and lowland sites.
| Chemelil | Miwani | Kisian | Iguhu06 | |
|---|---|---|---|---|
| Chemelil | 0.429 | 0.185 | 0.394 | |
| Miwani | 3.09 | 0.340 | 0.155 | |
| Kisian | 7.82 | 4.42 | 0.266 | |
| Iguhu06 | 4.37 | 15.34 | 7.72 |
Table 4.
Percentage of Plasmodium falciparum samples assigned to particular geographic populations based on multi-locus BAYESASS- analysis.
| Percentage of samples | |||||
|---|---|---|---|---|---|
| assigned to population | not assigned | ||||
| Samples from | Chemelil | Miwani | Kisian | Iguhu06 | |
| Chemelil | 57.1 | 0 | 0 | 0 | 42.9 |
| Miwani | 0 | 30.6 | 0 | 0 | 69.4 |
| Kisian | 0 | 0 | 55.9 | 1.5 | 42.6 |
| Iguhu06 | 0 | 0 | 0 | 18.2 | 81.8 |
3.4 Molecular analysis of drug resistance
A total of 181 P. falciparum samples were analyzed for polymorphism at 10 key codons involved in antimalarial drug resistance in pfcrt, pfmdr1, pfdhfr and pfdhps genes. Mixed (wild/mutant) genotypes were detected in 8 of the 10 codons analyzed. Mixed genotypes were included in the resistance mutation frequency calculation below.
Mutations in CQ resistance genes
The frequency of the pfcrt 76T mutant allele was found in >80% of samples analyzed for both lowland and highland populations (Table 5). For pfmdr1, no mutations were detected in codon 1042, which has been associated with CQ resistance in South America; whereas single mutations in codons 86 and 184 were present at a frequency >70%. The triple mutation haplotype 86Y/184F/1246Y at pfmdr1 was observed in >40% of the samples (Table 6). Since pfmdr1 alleles were also found associated with CQ resistance, high frequencies of pfcrt 76T mutation and triple mutations (86Y/184F/1246Y) at pfmdr1 suggest that CQ resistance still remains high 11 years after the Ministry of Health of Kenya changed the first line of malaria treatment from CQ to SP (Anabwani et al., 1996). There was no statistical difference in the mutation frequencies at codon 76 of pfcrt and at codon 1246 of pfmdr1 between highland and lowland populations. However, significant differences were detected at codon86 of the pfmdr1 gene between Miwani and Chemelil, Iguhu06 and Kisian, p<0.05 for the first two comparisons and p<0.01 for the third. Significant differences (p<0.05) were also detected at codon 184 of the pfmdr1 gene in comparisons between Kisian and the other three localities tested.
Table 5.
Frequency of mutations in genes associated with resistance to chloroquine and sulfadoxine-pyrimethamine in Plasmodium falciparum from asymptomatic infections from lowland and highland sites of Western Kenya.
| Lowland | Highland | |||||
|---|---|---|---|---|---|---|
| Gene | Mutation | Polymorphism | Chemelil | Miwani | Kisian | Iguhu06 |
| pfcrt | K76T | Mutant | 0.76 | 0.65 | 0.50 | 0.78 |
| Mixed | 0.12 | 0.20 | 0.39 | 0.06 | ||
| Wild-type | 0.12 | 0.15 | 0.11 | 0.16 | ||
| pfmdr1 | N86Y | Mutant | 0.37 | 0.47 | 0.26 | 0.24 |
| Mixed | 0.37 | 0.45 | 0.55 | 0.46 | ||
| Wild-type | 0.26 | 0.08 | 0.19 | 0.30 | ||
| Y184F | Mutant | 0 | 0 | 0 | 0.00 | |
| Mixed | 0.71 | 0.72 | 0.55 | 0.70 | ||
| Wild-type | 0.29 | 0.28 | 0.45 | 0.30 | ||
| N1042D | Mutant | 0 | 0 | 0 | 0 | |
| Mixed | 0 | 0 | 0 | 0 | ||
| Wild-type | 100 | 1.00 | 100 | 100 | ||
| D1246Y | Mutant | 0.58 | 0.42 | 0.12 | 0.54 | |
| Mixed | 0.38 | 0.48 | 0.82 | 0.37 | ||
| Wild-type | 0.04 | 0.10 | 0.06 | 0.09 | ||
| pfdhfr | N51I | Mutant | 0.86 | 0.75 | 0.90 | 1.00 |
| Mixed | 0.07 | 0.22 | 0.07 | 0 | ||
| Wild-type | 0.07 | 0.03 | 0.03 | 0 | ||
| C59R | Mutant | 0.86 | 0.67 | 0.69 | 0 | |
| Mixed | 0.07 | 0.33 | 0.16 | 1.00 | ||
| Wild-type | 0.07 | 0 | 0.15 | 0 | ||
| S108N | Mutant | 1.00 | 1.00 | 1.00 | 1.00 | |
| Mixed | 0 | 0 | 0 | 0 | ||
| Wild-type | 0 | 0 | 0 | 0 | ||
| pfdhps | A437G | Mutant | 0.04 | 0.09 | 0.84 | 0.73 |
| Mixed | 0.96 | 0.85 | 0.03 | 0.10 | ||
| Wild-type | 0 | 0.06 | 0.13 | 0.17 | ||
| K540E | Mutant | 0.43 | 0.75 | 0.07 | 0.36 | |
| Mixed | 0.36 | 0.17 | 0.72 | 0.36 | ||
| Wild-type | 0.21 | 0.08 | 0.21 | 0.28 |
Table 6.
Percentage of haplotypes of Plasmodium falciparum parasites collected from asymptomatic infections in the lowland and highland sites of western Kenya.
| Lowland | Highland | |||
|---|---|---|---|---|
| Haplotypes* | Chemelil | Miwani | Kisian | Iguhu06 |
| pfcrt | ||||
| 86Y/76K | 8.7 | 10.0 | 5.4 | 7.4 |
| 86Y/76T | 60.9 | 83.3 | 76.8 | 63.0 |
| 86N/76K | 4.3 | 3.3 | 3.6 | 3.70 |
| 86N/76T | 26.1 | 3.3 | 14.3 | 25.9 |
| pfmdr1 | ||||
| 86Y/184Y/1246Y | 17.4 | 23.3 | 32.1 | 14.8 |
| 86Y/184F/1246Y | 47.8 | 56. 7 | 48.2 | 48.1 |
| 86N/184F/1246D | 0 | 0 | 1.8 | 0 |
| 86Y/184F/1246D | 0 | 6. 7 | 0 | 3.7 |
| 86Y/184Y/1246D | 4.3 | 3.3 | 1.8 | 3.7 |
| 86N/184Y/1246D | 0 | 0 | 3.6 | 0 |
| 86N/184F/1246Y | 30.4 | 6. 7 | 7.1 | 22.2 |
| 86N/184Y/1246Y | 0 | 3.3 | 5.4 | 7.4 |
| pfdhfr/pfdhps | ||||
| quintuple | ||||
| 51I/59R/108N/437G/540E | 82.6 | 26.7 | 66.1 | 63.0 |
| quadruple | ||||
| 51I/59C/108N/437G/540E | 0 | 0 | 5.4 | 0 |
| 51N/59R/108N/437G/540E | 0 | 0 | 0 | 0 |
| 51I/59R/108N/437G/540K | 8.7 | 66.7 | 14.3 | 18.5 |
| 51I/59R/108N/437A/540E | 0 | 0 | 5.4 | 18.5 |
| triple | ||||
| 51I/59C/108N/437G/540K | 0 | 0 | 3.6 | 0 |
| 51I/59R/108N/437A/540K | 0 | 3.3 | 1.8 | 0 |
| 51N/59C/108N/437G/540E | 4.3 | 0 | 1.8 | 0 |
| 51N/59R/108N/437G/540K | 0 | 3.3 | 0 | 0 |
| 51I/59R/108S/437G/540K | 0 | 0 | 0 | 0 |
| double | ||||
| 51I/59C/108N/437A/540K | 0 | 0 | 1.8 | 0 |
| 51N/59C/108N/437G/540K | 4.3 | 0 | 0 | 0 |
The mutation amino acids are underlined.
Mutations in antifolate drug resistance genes
After a decade using SP as the first-line antimalarial drug in Kenya, mutations in the drug target genes, pfdhfr and pfdhps, were highly prevalent. For example, the mutation at codon 108 (allele 108N) of the pfdhfr gene, a major determinant of pyrimethamine resistance (Ouellette, 2001; Peterson et al., 1990), was fixed in all four populations collected in 2006 (Table 5). Frequency of mutation at the other two codons (51 and 59) of this gene exceeded 85%. Similarly, frequency of mutations at the two key sulfadoxine-resistance codons (437 and 540) of the pfdhps gene was over 72% in the 2006 samples analyzed (Table 5). Because increased numbers of mutations at the two antifolate target genes are associated with increased resistance to SP, we examined the frequency of all mutations in the pfdhfr and pfdhps genes. Overall, double or triple mutations were rarely encountered (<5%), whereas the quintuple mutation was most frequent (>63%), except in one lowland population (Miwani) where the quadruple mutation (pfdhfr 51I/59R/108N-pfdhps437G) was prevalent (Table 6).
4. Discussion
In this study, we found that the prevalence of P. falciparum asymptomatic infections among school children detected by PCR was 2–4 folds higher than that detected by the microscopic test. The low sensitivity but high specificity of microscopic tests for malaria infection has been documented in other studies (Alves et al., 2005; Baliraine et al., 2009). Based on the PCR detection method, a high prevalence of P. falciparum infections among school children was found in the lowlands and highlands (28–65%) in western Kenya in 2006, and there was a dramatic reduction of malaria prevalence between 2006 and 2007 in the highlands. In a survey conducted at the Iguhu study site between 2003 and 2004, malaria prevalence among asymptomatic school children detected by microscopic tests was 68% (Githeko et al., 2006). Compared to that period, malaria infection prevalence was decreased by about 10 folds in 2007. Such a reduction in malaria prevalence corresponded to the period of rapidly increased coverage of ITNs and the introduction of the artemisinin-based combination therapy (ACT) in the study area. Artemisinin derivates have been showed to have a strong activity against gametocytes (Okell et al., 2008), the infectious stage to the mosquito vectors.
Because asymptomatic infections are major malaria reservoirs (Bousema et al., 2004), population genetic variability and structure based on asymptomatic infections can provide information on the potential for parasite spread. We found comparable genetic diversity, as measured by the number of microsatellite alleles, Nei’s unbiased genetic diversity and the percentage of multiple clone infections, between parasites from lowlands and highlands. Thus, despite low transmission intensity in the Iguhu highland, parasite genetic variability has not been significantly reduced. Two possibilities may account for this phenomenon. First, transmission intensity is not sufficiently low to cause a reduction in parasite genetic variability in the highlands. Second, the high levels of gene flow between lowland and highland populations facilitate the introduction of new alleles from endemic lowland sites. Lack of genetic structure between highland and lowland populations, as determined by the analysis of molecular variance, may result from human travel between highland and lowland sites. Travel of highland residents to malaria-endemic areas was linked to increased malaria risk in highland residents (Shanks et al., 2005a). The Bayesian assignment test supports the notion that malaria parasites in the highlands represent a mixture of parasite genotypes from other localities. Because only a small proportion of infections (18.2%) from the highland site were assigned, and the majority of infections could not be assigned to any particular localities, the origins of most parasite genotypes identified in the highland site have not been identified. This result indicates the importance of gene flow in shaping parasite genetic structure in the highland site with relatively low indigenous transmission intensity.
We observed a high frequency of mutations in genes conferring resistance to CQ and SP in both highland and lowland parasite populations. More than 80% of infections carried the pfcrt 76T mutation eleven years after CQ was phased out as the first-line antimalarial in Kenya. Such a high frequency of the pfcrt 76T mutation suggests that the fitness costs associated with this mutation is not high enough to cause dramatic reduction in its frequency. Alternatively, infrequent but continuous use of amodiaquine, a 4-aminoquinoline compound related to chloroquine, alone or in combination with SP or artesunate to treat uncomplicated malaria infections in children and pregnant women (Nosten et al., 2006; Orton and Omari, 2008) may have maintained the pfcrt 76T frequency in the populations. Lack of significant difference in pfcrt 76T and pfmdr1 mutation frequencies between highland and lowland sites is consistent with the notion of large gene flow between the two regions.
Despite the fact that ACT has been designated as the first-line antimalarial drug in Kenya since 2006 (Amin et al., 2007) and residents have been offered free or subsidized treatment, ACT has not often been available in the hospitals in our study sites. As a result, SP along with AQ, and also antibiotics, are still being used as antimalarials in rural Kenya (Zurovac et al., 2008). As such, selection pressure for a high frequency of pfdhfr/pfdhps mutations remains in place. At the same time, concerns have been raised that SP resistance could be generated and maintained through cross-resistance with co-trimoxazole, which is used in the treatment of opportunistic infections in immune compromised patients in most parts of Kenya and in all of our study sites (Hamel et al., 2008). Co-trimoxazole is a sulfonamide antibacterial combination of trimethoprim and sulfamethoxazole, which inhibits successive steps in the folate synthesis pathway (Malamba et al., 2006). Cross-resistance between trimethoprim and pyrimethamine has been shown in vitro in laboratory isolates containing the pfdhfr triple mutation 108N/51I/59R (Jelinek et al., 1999). However, the issue as to whether cross-resistance occurs in vivo is controversial (Hamel et al., 2008; Malamba et al., 2006). Data reported in the present study suggest the need to continuously monitor P. falciparum resistance to SP and the use of cotrimoxazole.
In conclusion, the dramatic fall in the prevalence of asymptomatic infections in 2007 from the previous period in the highland site suggests the effectiveness of the new ACT antimalarial treatment regime and of the increased coverage of insecticide-treated bed nets. Continuous monitoring of infection prevalence is needed to verify if this decreasing trend is sustained. High level of gene flow and lack of significant genetic structure in malaria parasites between highlands and lowlands suggest the importance of human travel in shaping parasite population structure. Cohort studies with detailed measurement of travel history would help untangle the effect of human travel to malaria endemic areas from indigenous transmission on the genetic makeup of malaria parasites in the highlands.
Acknowledgements
This work was supported by NIH grants R01 AI050243 and D43 TW001505. We are grateful to the field staff at Center for Vector Biology and Control Research, Kenya Medical Research Institute for their assistance. We thank Goufa Zhou for help with data analysis and two anonymous reviewers for valuable comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Mariangela Bonizzoni, Email: mbonizzo@uci.edu.
Yaw Afrane, Email: yaw_afrane@yahoo.com.
Frederick N. Baliraine, Email: fbalirai@uci.edu.
Dolphine A. Amenya, Email: damenya@uci.edu.
Andrew K. Githeko, Email: AGitheko1@kisian.mimcom.net.
Guiyun Yan, Email: guiyuny@uci.edu.
References
- Adagu IS, Warhurst DC. Allele-specific, nested, one tube PCR: application to Pfmdr1 polymorphisms in Plasmodium falciparum. Parasitology. 1999;119(Pt 1):1–6. doi: 10.1017/s0031182099004382. [DOI] [PubMed] [Google Scholar]
- Alves FP, Gil LH, Marrelli MT, Ribolla PE, Camargo EP, Da Silva LH. Asymptomatic carriers of Plasmodium spp. as infection source for malaria vector mosquitoes in the Brazilian Amazon. J. Med. Entomol. 2005;42:777–779. doi: 10.1093/jmedent/42.5.777. [DOI] [PubMed] [Google Scholar]
- Amin AA, Zurovac D, Kangwana BB, Greenfield J, Otieno DN, Akhwale WS, Snow RW. The challenges of changing national malaria drug policy to artemisinin based combinations in Kenya. Malar. J. 2007;6:72–82. doi: 10.1186/1475-2875-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anabwani GM, E.F, Menya DA. A randomised controlled trial to assess the relative efficacy of chloroquine, amodiaquine, halofantrine and Fansidar in the treatment of uncomplicated malaria in children. East Afr. Med. J. 1996;73:155–158. [PubMed] [Google Scholar]
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119(Pt 2):113–125. doi: 10.1017/s0031182099004552. [DOI] [PubMed] [Google Scholar]
- Babiker HA, Pringle SJ, Abdel-Muhsin A, Mackinnon M, Hunt P, Walliker D. High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance Gene pfmdr1. J. Infect. Dis. 2001;183:1535–1538. doi: 10.1086/320195. [DOI] [PubMed] [Google Scholar]
- Bacon DJ, McCollum AM, Griffing SM, Salas C, Soberon V, Santolalla M, Haley R, Tsukayama P, Lucas C, Escalante AA, Udhayakumar V. Dynamics of malaria drug resistance patterns in the Amazon basin region following changes in Peruvian national treatment policy for uncomplicated malaria. Antimicrob. Agents Chemother. 2009 doi: 10.1128/AAC.01677-08. 0: AAC.01677-08v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliraine FN, A.Y, Amenya DA, Bonizzoni M, Menge DM, Goufa Z, Zhong D, Vardo-Zalik AM, Githeko AK, Yan G. High prevalence of asymptomatic Plasmodium falciparum infections in a highland area of western Kenya: a cohort study. J. Infect. Dis. 2009 doi: 10.1086/599317. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banoo S, Bell D, Bossuyt P, Herring A, Mabey D, Poole F, Smith PG, Sriram N, Wongsrichanalai C, Linke R, O'Brien R, Perkins M, Cunningham J, Matsoso P, Nathanson CM, Olliaro P, Peeling RW, Ramsay A. Evaluation of diagnostic tests for infectious diseases: general principles. Nat. Rev. Microbiol. 2006;4:S21–S31. doi: 10.1038/nrmicro1523. [DOI] [PubMed] [Google Scholar]
- Bousema JT, Gouagna LC, Drakeley CJ, Meutstege AM, Okech BA, Akim IN, Beier JC, Githure JI, Sauerwein RW. Plasmodium falciparum gametocyte carriage in asymptomatic children in western Kenya. Malar. J. 2004;3:18–23. doi: 10.1186/1475-2875-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey G, Kamya MR, Singh A, Rosenthal PJ. Polymorphisms in the Plasmodium falciparum pfcrt and pfmdr-1 genes and clinical response to chloroquine in Kampala, Uganda. J. Infect. Dis. 2001;183:1417–1420. doi: 10.1086/319865. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, Curtis J, Warhurst DC. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp. Parasitol. 1998;89:1–8. doi: 10.1006/expr.1998.4274. [DOI] [PubMed] [Google Scholar]
- Enosse S, Magnussen P, Abacassamo F, Gómez-Olivé X, Rønn AM, Thompson R, Alifrangis M. Rapid increase of Plasmodium falciparum dhfr/dhps resistant haplotypes, after the adoption of sulphadoxine-pyrimethamine as first line treatment in 2002, in southern Mozambique. Malar. J. 2008;7:115. doi: 10.1186/1475-2875-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MU, Nair S, Hyunh TV, Kawamoto F, Anderson TJC. Microsatellite characterization of Plasmodium falciparum from cerebral and uncomplicated malaria patients in southern Vietnam. J. Clin. Microbiol. 2002;40:1854–1857. doi: 10.1128/JCM.40.5.1854-1857.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MU, Karunaweera ND, da Silva-Nunes M, da Silva NS, Wirth DF, Hartl DL. Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. J. Infect. Dis. 2007;195:1218–1226. doi: 10.1086/512685. [DOI] [PubMed] [Google Scholar]
- Fisher MC, Rannala B, Chaturvedi V, Taylor JW. Disease surveillance in recombining pathogens: multilocus genotypes identify sources of human Coccidioides infections. Proc. Natl. Acad. Sci. USA. 2002;99:9067–9071. doi: 10.1073/pnas.132178099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flueck TP, Jelinek T, Kilian AH, Adagu IS, Kabagambe G, Sonnenburg F, Warhurst DC. Correlation of in vivo-resistance to chloroquine and allelic polymorphisms in Plasmodium falciparum isolates from Uganda. Trop. Med. Int. Health. 2000;5:174–178. doi: 10.1046/j.1365-3156.2000.00543.x. [DOI] [PubMed] [Google Scholar]
- Githeko AK, Service MW, Mbogo CM, Atieli FK. Resting behaviour, ecology and genetics of malaria vectors in large scale agricultural areas of Western Kenya. Parassitologia. 1996;38:481–489. [PubMed] [Google Scholar]
- Githeko AK, Ayisi JM, Odada PK, Atieli FK, Ndenga BA, Githure JI, Yan G. Topography and malaria transmission heterogeneity in western Kenya highlands: prospects for focal vector control. Malar. J. 2006;5:107–115. doi: 10.1186/1475-2875-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel MJ, Greene C, Chiller T, Ouma P, Polyak C, Otieno K, Williamson J, Shi YP, Feikin DR, Marston B, Brooks JT, Poe A, Zhou Z, Ochieng B, Mintz E, Slutsker L. Does cotrimoxazole prophylaxis for the prevention of HIV-associated opportunistic infections select for resistant pathogens in Kenyan adults? Am. J. Trop. Med. Hyg. 2008;79:320–330. [PubMed] [Google Scholar]
- Hay SI, Noor AM, Simba M, Busolo M, Guyatt HL, Ochola SA, Snow RW. Clinical epidemiology of malaria in the highlands of western Kenya. Emerg. Infect. Dis. 2002;8:543–548. doi: 10.3201/eid0806.010309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek T, Kilian AH, Curtis J, Duraisingh MT, Kabagambe G, von Sonnenburg F, Warhurst DC. Plasmodium falciparum: selection of serine 108 of dihydrofolate reductase during treatment of uncomplicated malaria with co-trimoxazole in Ugandan children. Am. J. Trop. Med. Hyg. 1999;61:125–130. doi: 10.4269/ajtmh.1999.61.125. [DOI] [PubMed] [Google Scholar]
- Joy DA, Gonzalez-Ceron L, Carlton JM, Gueye A, Fay M, McCutchan TF, Su XZ. Local adaptation and vector-mediated population structure in Plasmodium vivax malaria. Mol. Biol. Evol. 2008;25:1245–1252. doi: 10.1093/molbev/msn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, Mukadam RA, Rogerson SJ, Lescano AG, Molyneux ME, Winstanley PA, Chimpeni P, Taylor TE, Plowe CV. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J. Infect. Dis. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- Laufer MK, Djimde AA, Plowe CV. Monitoring and deterring drug-resistant malaria in the era of combination therapy. Am. J. Trop. Med. Hyg. 2007;77:160–169. [PubMed] [Google Scholar]
- Lopes D, Rungsihirunrat K, Nogueira F, Seugorn A, Gil JP, do Rosario VE, Cravo P. Molecular characterization of drug-resistant Plasmodium falciparum from Thailand. Malar. J. 2002;14:1–12. doi: 10.1186/1475-2875-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamba SS, Mermin J, Reingold A, Lule JR, Downing R, Ransom R, Kigozi A, Hunt BM, Hubbard A, Rosenthal PJ, Dorsey G. Effect of cotrimoxazole prophylaxis taken by human immunodeficiency virus (HIV)-infected persons on the selection of sulfadoxine-pyrimethamine-resistant malaria parasites among HIV-uninfected household members. Am. J. Trop. Med. Hyg. 2006;75:375–380. [PubMed] [Google Scholar]
- Munyekenye OG, Githeko AK, Zhou G, Mushinzimana E, Minakawa N, Yan G. Plasmodium falciparum spatial analysis, western Kenya highlands. Emerg. Infect. Dis. 2005;11:1571–1577. doi: 10.3201/eid1110.050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutuku FM, Alaii JA, Bayoh MN, Gimnig JE, Vulule JM, Walker ED, Kabiru E, Hawley WA. Distribution, description, and local knowledge of larval habitats of Anopheles gambiae s.l. in a village in western Kenya. Am. J. Trop. Med. Hyg. 2006;74:44–53. [PubMed] [Google Scholar]
- Ndenga B, Githeko A, Omukunda E, Munyekenye G, Atieli H, Wamai P, Mbogo C, Minakawa N, Zhou G, Yan G. Population dynamics of malaria vectors in western Kenya highlands. J. Med. Entomol. 2006;43:200–206. doi: 10.1603/0022-2585(2006)043[0200:pdomvi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Nei M, Tajima F, Tateno Y. Accuracy of estimated phylogenetic tree from molecular data. II. Gene frequency data. J. Mol. Evol. 1983;19:153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- Nosten F, McGready R, d'Alessandro U, Bonell A, Verhoeff F, Menendez C, Mutabingwa T, Brabin B. Antimalarial drugs in pregnancy: a review. Curr. Drug Saf. 2006;1:1–15. doi: 10.2174/157488606775252584. [DOI] [PubMed] [Google Scholar]
- Nosten F, White NJ. Artemisinin-based combination treatment of falciparum malaria. Am. J. Trop. Med. Hyg. 2007;77:181–192. [PubMed] [Google Scholar]
- Okell LC, Drakeley CJ, Ghani AC, Bousema T, Sutherland CJ. Reduction of transmission from malaria patients by artemisinin combination therapies: a pooled analysis of six randomized trials. Malar. J. 2008;7:125–137. doi: 10.1186/1475-2875-7-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orton LC, Omari AA. Drugs for treating uncomplicated malaria in pregnant women. Cochrane Database Syst. Rev. 2008 doi: 10.1002/14651858.CD004912.pub3. CD004912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M. Biochemical and molecular mechanisms of drug resistance in parasites. Trop. Med. Int. Health. 2001;6:874–882. doi: 10.1046/j.1365-3156.2001.00777.x. [DOI] [PubMed] [Google Scholar]
- Peakall R, S.P GENALEX 6: genetic analyses in Excel. Population generic software for teaching and research. Mol. Ecol. Notes. 2006;6:288–295. [Google Scholar]
- Peterson DS, Milhous WK, Wellems TE. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc. Natl. Acad. Sci. U S A. 1990;87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am. J. Trop. Med. Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- Sanchez CP, Lanzer M. Changing ideas on chloroquine in Plasmodium falciparum. Curr. Opin. Infect. Dis. 2000;13:653–658. doi: 10.1097/00001432-200012000-00013. [DOI] [PubMed] [Google Scholar]
- Schneider AG, Premji Z, Felger I, Smith T, Abdulla S, Beck HP, Mshinda H. A point mutation in codon 76 of pfcrt of P. falciparum is positively selected for by Chloroquine treatment in Tanzania. Infect. Genet. Evol. 2002;1:183–189. doi: 10.1016/s1567-1348(01)00021-1. [DOI] [PubMed] [Google Scholar]
- Schneider S RD, Excofier L. User Manual. Geneva: Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva; 2000. ARLEQUIN: A software for population genetics data analyses. [Google Scholar]
- Schonfeld M, Barreto Miranda I, Schunk M, Maduhu I, Maboko L, Hoelscher M, Berens-Riha N, Kitua A, Löscher T. Molecular surveillance of drug-resistance associated mutations of Plasmodium falciparum in south-west Tanzania. Malar. J. 2007;6:2. doi: 10.1186/1475-2875-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks GD, Biomndo K, Guyatt HL, Snow RW. Travel as a risk factor for uncomplicated Plasmodium falciparum malaria in the highlands of western Kenya. Trans. R. Soc.Trop. Med. Hyg. 2005a;99:71–74. doi: 10.1016/j.trstmh.2004.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks GD, Hay SI, Omumbo JA, Snow RW. Malaria in Kenya's western highlands. Emerg. Infect. Dis. 2005b;11:1425–1432. doi: 10.3201/eid1109.041131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus-and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am. J. Trop. Med. Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, Gil JP. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J. Infect. Dis. 2005;191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- Talisuna AO, Langi P, Mutabingwa TK, Watkins W, Van Marck E, Egwang TG, D'Alessandro U. Population-based validation of dihydrofolate reductase gene mutations for the prediction of sulfadoxine-pyrimethamine resistance in Uganda. Trans. R. Soc. Trop. Med. Hyg. 2003;97:338–342. doi: 10.1016/s0035-9203(03)90163-5. [DOI] [PubMed] [Google Scholar]
- White NJ. The assessment of antimalarial drug efficacy. Trends Parasitol. 2002;18:458–464. doi: 10.1016/s1471-4922(02)02373-5. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Geneva, Switzerland: World Health Organization, Technical Report Series; Malaria. 2000:892.
- Wooden J, Kyes S, Sibley CH. PCR and strain identification in Plasmodium falciparum. Parasitol. Today. 1993;9:303–305. doi: 10.1016/0169-4758(93)90131-x. [DOI] [PubMed] [Google Scholar]
- Zhong D, Afrane Y, Githeko A, Yang Z, Cui L, Menge DM, Temu EA, Yan G. Plasmodium falciparum genetic diversity in western Kenya highlands. Am. J. Trop. Med. Hyg. 2007;77:1043–1050. [PubMed] [Google Scholar]
- Zhong D, Afrane Y, Githeko A, Cui L, Menge DM, Yan G. Molecular epidemiology of drug-resistant malaria in western Kenya highlands. BMC Infect. Dis. 2008;8:105–129. doi: 10.1186/1471-2334-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurovac D, Njogu J, Akhwale W, Hamer DH, Larson BA, Snow RW. Effects of revised diagnostic recommendations on malaria treatment practices across age groups in Kenya. Trop. Med. Int. Health. 2008;13:784–787. doi: 10.1111/j.1365-3156.2008.02072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]