Abstract
Formaldehyde is considered carcinogenic to humans by the International Agency for Research on Cancer, but there are no previous reports of formaldehyde-DNA adducts in humans. In this study, we used liquid chromatography-electrospray ionization-tandem mass spectrometry to quantify the formaldehyde DNA adduct N6-hydroxymethyldeoxyadenosine (N6-HOMe-dAdo) in leukocyte DNA samples from 32 smokers of 10 or more cigarettes per day and 30 non-smokers. Clear peaks co-eluting with the internal standard in two different systems were seen in samples from smokers, but rarely in non-smokers. N6-HOMe-dAdo was detected in 29 of 32 smoker samples, mean ± SD, 179 ± 205 fmol/µmol dAdo. In contrast, it was detected in only 7 of 30 non-smoker samples, 15.5 ± 33.8 fmol/µmol dAdo (P<0.001). The results of this study demonstrate remarkable differences between smokers and non-smokers in levels of a leukocyte formaldehyde-DNA adduct, suggesting a potentially important and previously unrecognized role for formaldehyde as a cause of cancer induced by cigarette smoking.
Introduction
Formaldehyde is considered “carcinogenic to humans” by the International Agency for Research on Cancer (IARC) (1). It is genotoxic and cytotoxic, and both properties are believed to play important roles in its carcinogenicity (1). Several formaldehyde-DNA adducts and cross-links have been identified in vitro (2–5). The most abundant of these is N6-hydroxymethyldeoxyadenosine (N6-HOMe-dAdo). However, there are no data in the literature on specific formaldehyde DNA adducts in humans.
In the study reported here, we have refined and extended our liquid chromatography-electrospray ionization-tandem mass spectrometry-selected reaction monitoring (LC-ESI-MS/MS-SRM) methodology (6) to allow quantitation of N6-HOMe-dAdo in human leukocyte DNA. The method was applied for the analysis of leukocyte DNA from smokers and non-smokers. The data provide the first demonstration of a specific formaldehyde-DNA adduct in humans and illustrate a remarkable difference in its levels between smokers and non-smokers.
Materials and Methods
Subjects
Subjects were smokers of more than 10 cigarettes per day, or non-smokers This study was approved by the University of Minnesota Research Subjects’ Protection Programs Institutional Review Board Human Subjects Committee.
LC-ESI-MS/MS-SRM Analysis
The analysis was carried out with either a Quantum Ultra AM (with Ion Max ESI source) or Discovery Max triple quadrupole mass spectrometer (Thermo Scientific, San Jose, CA) interfaced with an Agilent 1100 capillary flow HPLC and a 150 mm × 0.5 mm Zorbax SB C18 column (Agilent Technologies, Palo Alto, CA). The column was operated at 50 °C. For analysis of N6-Me-dAdo, we used isocratic elution by 15 mM ammonium acetate buffer (pH 6.6) for 10 min, then a gradient to 25% CH3CN over the course of 29 min, then a gradient from 25–100% CH3CN in 2 min, then held at 100% for 10 min, and finally returning to buffer in 3 min, at a flow rate of 15 µl/min. The first 15 min of eluant was directed to waste, and the 15–35 min fractions were diverted to the ESI source.
A second HPLC system was used for confirmation of identity. A 150 mm × 0.5 mm, 4 µm, Synergi Polar-RP column (Phenomenex, Torrance, CA) was eluted with a gratdient from 5 to 35% CH3OH in 15 mM ammonium acetate buffer in 35 min at a flow rate of 10 µl/min at 50 °C. The first 15 min of eluant was directed to waste, and the 15 to 35 min fraction was diverted to the ESI source.
The MS parameters were set as follows: spray voltage, 4kV; sheath gas pressure, 30 psi; capillary temperature, 250 °C; collision energy, 18 V; scan width, 0.1 amu; scan time, 0.1 s; Q1 peak width, 0.7; Q3 peak width, 0.7; Q2 pressure, 1.0 mTorr; source CID, 10V; and tube lens offset, 70V. Transitions monitored were as follows: N6-Me-dAdo and [15N5]N6-Me-dAdo m/z 266 [M+H]+→m/z 150 [BH]+ and m/z 271→ m/z 155, respectively.
Chemicals and Enzymes
[15N5]dAdo was purchased from Spectra Stable Isotopes (Columbia, MD). [15N5]N6-Me-dAdo was prepared as described (6). Puregene DNA purification solution was obtained from Qiagen (Morris Plains, NJ). Alkaline phosphatase was purchased from Roche Diagnostic Corp (Indianapolis, IN). N6-Me-dAdo and all other chemicals and enzymes were obtained from Sigma-Aldrich (St. Louis, MO).
Isolation of Buffy coat and Separation of Lymphocytes and Neutrophils
Buffy coat isolation was performed following the protocol described in the Genomic DNA Handbook (Qiagen). Briefly, 10 – 30 ml of whole blood was collected in tubes containing EDTA and centrifuged at 2500 rpm for 15 min. A thin layer of white blood cells, buffy coat, was obtained between the upper plasma layer and the lower red blood cell layer. The upper layer was removed and the buffy coat was carefully collected with a pipet, and kept on ice until use. Separation of lymphocytes and neutrophils, where necessary, was performed essentially as described (7).
DNA Isolation from Leukocytes
DNA isolation was performed using the DNA purification from buffy coat protocol described in the Gentra Puregene Handbook (Qiagen) with several modifications. Briefly, 9 ml of RBC cell lysis solution was added to 3 ml of buffy coat prepared from 30 ml of whole blood. The white blood cell pellet was collected by centrifugation and treated with 15 ml of cell lysis solution and 150 µl of RNase A (4 mg/ml). To the cell lysate was added 5 ml of protein precipitation solution, and the mixture was centrifuged to remove protein. DNA was precipitated from the supernatant by addition of 15 ml of isopropanol. The DNA was washed with 2 ml of 70% ethanol and twice with 2 ml of 100 % ethanol, and dried with a stream of nitrogen.
Analysis of DNA for N6-HOMe-dAdo, as N6-Me-dAdo
For enzyme hydrolysis, DNA (224 ± 169 µg, range 16 – 777 µg) was dissolved in 0.5 ml of 10 mM PIPES /5 mM MgCl2 buffer (pH 7.0) containing [15N5]N6-Me-dAdo (200 fmol) and NaBH3CN (30 mg). The pH was adjusted to 7 with 8 µl of 1N HCl. The DNA was initially digested overnight at room temperature with 500 units of DNase I (type II, from bovine pancreas). Then to the resulting mixture were added 500 additional units of DNase I, 0.02 units of phosphodiesterase I (type II, from Crotalus adamanteus venom) and 150 units of alkaline phosphatase (from calf intestine). The mixture was incubated at 37 °C for 60 min. The hydrolysate, after removal of a 10 µl aliquot for dAdo quantitation (by calculation upon HPLC measurement of dG), was desalted and purified using a solid-phase extraction cartridge [Strata-X, 30 mg/1ml (Phenomenex)]. The cartridge was conditioned with 1 ml of CH3OH and 1 ml of H2O. The hydrolysate was loaded and the column was washed with 1 ml of H2O and 1ml of 10% CH3OH in H2O, which were discarded. It was then washed with 1 ml of 50% CH3OH. The 50% CH3OH fraction was collected and evaporated to dryness on a SpeedVac. The residue was dissolved in 200 µl of CH3OH and transferred to a 300 µl Snap seal vial (Chrom Tech), then dried using a Speed Vac. This residue was dissolved in 100 µl of H2O and 8 µl aliquots were analyzed by LC-ESI-MS/MS-SRM.
Samples were analyzed without knowledge of their origin from smokers or non-smokers. Negative controls, consisting of PIPES buffer without DNA, were run with each set of samples using the same method employed for the samples. None showed an analyte peak.
Statistical Analysis
Adduct levels in smokers versus non-smokers were compared using the non-parametric Wilcoxon rank sum test and the chi-square test. Geometric means were calculated using a value of 0.1 for samples in which N6-Me-dAdo was not detected.
Results
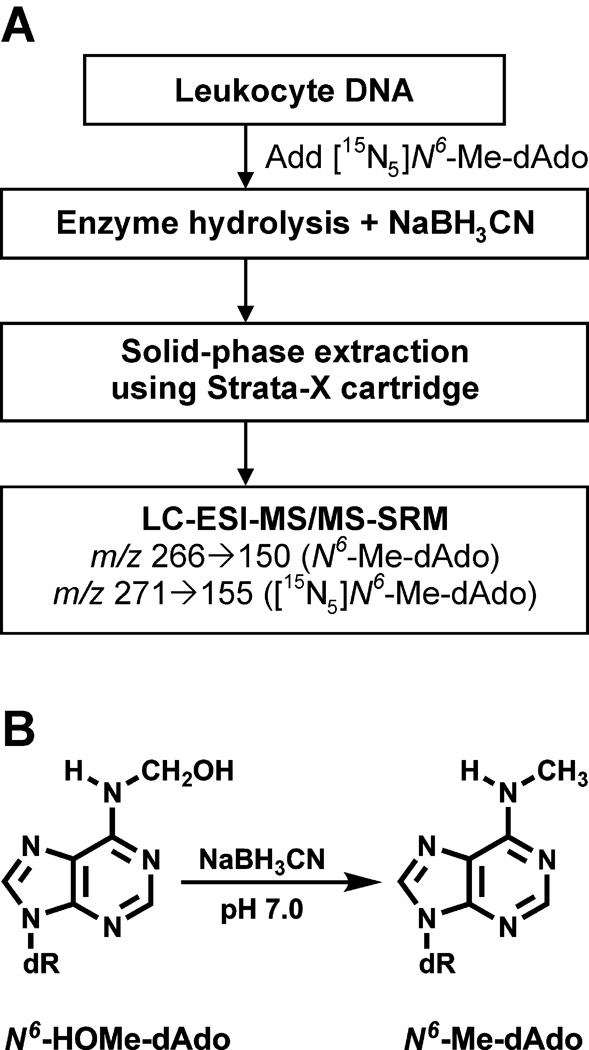
The analytical method is summarized in Figure 1A. [15N5]N6-Me-dAdo is added to leukocyte DNA as the internal standard. Treatment with NaBH3CN during enzyme hydrolysis of the DNA converts N6-HOMe-dAdo to N6-Me-dAdo (Figure 1B). This is necessary because N6-Me-dAdo is stable while N6-HOMe-dAdo is somewhat unstable at the nucleoside level (3,6). Control of pH is critical during this step, with the highest yields being obtained at pH 7. Tris-buffer, sometimes used for washing the buffy coat and as a solvent for DNA hydrolysis, contributes to chemical background and to potentially artificially high results. This is logical because the structure of Tris suggests facile release of, or contamination with, formaldehyde. This was not a problem when we used PIPES buffer, which, based on its structure, would be unlikely to release formaldehyde. Solid-phase extraction enriches the adducts, and the appropriate fraction is analyzed by LC-ESI-MS/MS-SRM.
Figure 1.
Schematic representation of analytical method for N6-HOMe-dAdo (as N6-Me-dAdo) in human leukocyte DNA (A), Structures of N6-HOMe-dAdo and N6-Me-dAdo (B).
Calibration curves for N6-Me-dAdo were linear in the range measured (R2 = 1.0). Accuracy and precision were excellent as determined by analysis of human leukocyte DNA to which N6-Me-dAdo was added (Table 1). Recoveries were approximately 60%. Limits of detection were 0.3 fmol N6-Me-dAdo injected on column and 0.8 fmol (signal/noise = 5) when analyzing 10 µg DNA.
Table 1.
Analysis of human leukocyte DNA to which N6-Me-dAdo was added.
|
N6-Me-dAdo (fmol) |
|||
|---|---|---|---|
| Addeda | Detectedb | Accuracy (%) | CV (%) |
| 10 | 10.6 ± 0.4 | 106 | 4 |
| 20 | 20.3 ± 0.5 | 102 | 3 |
| 50 | 53.3 ± 0.3 | 107 | 0.6 |
| 100 | 105.0 ± 0.2 | 105 | 0.2 |
| 150 | 150.4 ± 1.5 | 100 | 1.0 |
Added to 75 µg of leukocyte DNA. N6-Me-dAdo was not detected in 6 µg of this DNA treated with NaBH3CN.
Mean ± S.D. (N=3)
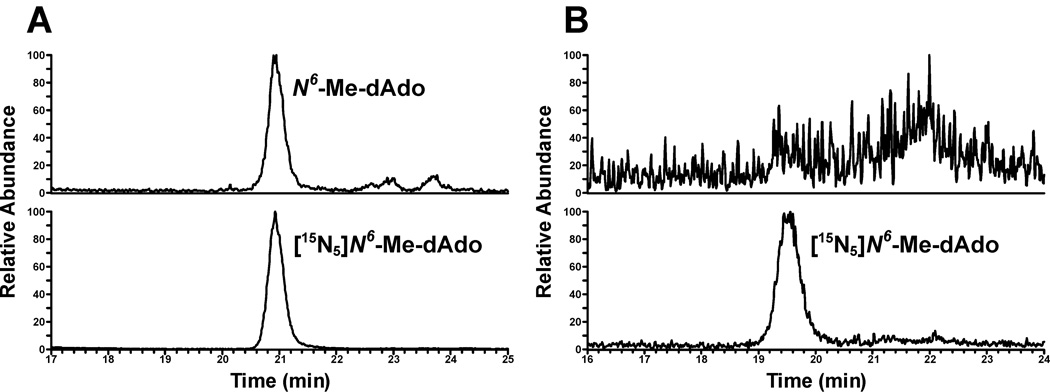
Thirty-two smokers (16 male, age range 26–66 years, mean ± S.D. 42 ± 11 years) and 30 non-smokers (15 male, age range 21–78 years, mean ± S.D. 48 ± 15 years) were recruited. LC-ESI-MS/MS-SRM traces obtained upon analysis of leukocyte DNA from a smoker and a non-smoker are illustrated in Figure 2. Co-elution of the internal standard and analyte was also observed using a different HPLC column and conditions. Levels of N6-HOMe-dAdo (as N6-Me-dAdo) in leukocyte DNA of 32 smokers and 30 non-smokers are summarized in Table 2. Twenty-nine of 32 (91%) smoker samples were positive for N6-Me-dAdo while only seven of 30 (23%) of non-smoker samples were positive (P<0.001; detection limit, approximately 10 fmol/µmol dAdo). The mean level ± S.D. of N6-Me-dAdo in DNA samples from smokers was 179 ± 205 fmol/µmol dAdo (5.4 adducts/108 nucleotides) while that in non-smokers was 15.5 ± 33.8 fmol/µmol dAdo (0.47 adducts/108 nucleotides; P<0.001). The corresponding geometric means were 66.7 fmol/µmol and 0.44 fmol/µmol dAdo. There was no significant difference in adduct levels between male and female smokers, and there was no effect of age on adduct levels.
Figure 2.
Typical LC-ESI-MS/MS-SRM chromatograms of N6-Me-dAdo from leukocyte DNA of A) a smoker and B) a non-smoker.
Table 2.
Levels of N6-HOMe-dAdo (as N6-Me-dAdo) in leukocyte DNA of smokers and non-smokers
| fmol N6-Me-dAdo/µmol dAdo | |||||
|---|---|---|---|---|---|
| Smokers |
Non-smokers |
||||
| 1 | F | 76 | 1 | F | NDa |
| 2 | M | 118 | 2 | F | 36 |
| 3 | M | 72 | 3 | F | ND |
| 4 | F | 151 | 4 | F | ND |
| 5 | M | 116 | 5 | F | ND |
| 6 | M | ND | 6 | F | ND |
| 7 | M | 116 | 7 | F | ND |
| 8 | M | 144 | 8 | M | ND |
| 9 | F | 168 | 9 | F | ND |
| 10 | M | ND | 10 | M | ND |
| 11 | M | 30 | 11 | M | 74 |
| 12 | M | 41 | 12 | M | ND |
| 13 | F | 66 | 13 | M | ND |
| 14 | F | 128 | 14 | M | 23 |
| 15 | M | 25 | 15 | M | ND |
| 16 | M | 303 | 16 | M | ND |
| 17 | F | 94 | 17 | M | ND |
| 18 | M | 180 | 18 | M | ND |
| 19 | F | 195 | 19 | M | 122 |
| 20 | F | 452 | 20 | M | ND |
| 21 | F | 284 | 21 | M | ND |
| 22 | F | 631 | 22 | M | 65 |
| 23 | M | 929 | 23 | F | ND |
| 24 | F | 35 | 24 | F | ND |
| 25 | F | 75 | 25 | F | ND |
| 26 | F | 141 | 26 | F | 32 |
| 27 | F | 33 | 27 | F | ND |
| 28 | F | ND | 28 | F | ND |
| 29 | M | 181 | 29 | M | ND |
| 30 | M | 557 | 30 | F | 114 |
| 31 | M | 302 | Mean ± S.D. | 15.5 ± 33.8 | |
| 32 | F | 95 | (7/30 positive) | ||
| Mean ± S.D. | 179 ± 205 | ||||
| (29/32 positive) | |||||
Levels in smokers were significantly higher than in non-smokers, P<0.001
ND = not detected. A value of zero was used to calculate the arithmetic mean shown. Detection limit ≅ 10 fmol/µmol dAdo
We considered the possibility that the adduct in smokers’ leukocyte DNA was in fact N6-Me-dAdo, and that NaBH3CN reduction was not necessary. To address this possibility, buffy coat samples from two smokers were split, and the DNA was isolated. One half of the DNA samples were treated with NaBH3CN while the others were not. In the samples treated with NaBH3CN, levels of N6-Me-dAdo were 460 and 143 fmol/µmol dAdo while the corresponding levels in the untreated samples were 60 and 30 fmol/µmol dAdo, respectively, demonstrating that 83–88% of the adduct was present as N6-HOMe-dAdo in each case.
It was also possible that formaldehyde present in smokers’ buffy coat could react with DNA during the DNA isolation procedure. To address this possibility, buffy coat samples from two smokers were split, and one half was treated with NaBH3CN immediately, which would reduce formaldehyde to methanol, while the other half was untreated. Each half was then subjected to the procedure outlined in Figure 1. The results demonstrated no differences in DNA adduct levels.
We also used a different isolation procedure to further validate our results, and exclude the possibility that unstable formaldehyde protein adducts that might be present in red cells potentially contaminating the buffy coat were transferring formaldehyde to DNA during isolation. Buffy coat samples from three smokers were split, and half were isolated by the usual procedure while the other half were processed by a Ficoll-Hypaque double gradient to separate lymphocytes and neutrophils. The results demonstrated no differences in DNA adduct levels in buffy coat versus the lymphocyte plus neutrophil fraction.
Discussion
The results of this study clearly demonstrate that the formaldehyde-DNA adduct N6-HOMe-dAdo is present in human leukocyte DNA. Highly significant differences between smokers and non-smokers were observed based on both adduct detectability and adduct levels. Such clear differences in leukocyte DNA adduct levels between smokers and non-smokers have rarely been reported. These results indicate a previously unrecognized and potentially important role for formaldehyde-DNA damage in smoking induced cancer.
Smoking-related DNA adducts in blood cells were comprehensively reviewed by Phillips (8). Most studies were carried out by the non-specific and semi-quantitative 32P-postlabelling and immunoassay methods. The results of these studies were inconsistent when comparing adduct levels in DNA samples from smokers and non-smokers. In studies using chemically specific methods, some showed increases in levels of the oxidative damage adduct 8-hydroxy-dG in blood DNA while others found decreases. We found that the mean level of the acetaldehyde adduct N2-ethylidene-dG in smokers was about 1200 fmol/µmol dG, and this decreased by only 28% to about 700 fmol/µmol dG after four weeks of abstinence from smoking and alcohol consumption (9). A somewhat larger difference between smokers and non-smokers in specific DNA adduct levels in blood cells was reported in a recent study of anti-benzo[a]pyrene diol epoxide-DNA adducts in lymphomonocytes of 128 smokers and 457 non-smokers (10). The reported levels were 2.03 ± 3.68 adducts/108 nucleotides (66% detectability) in smokers and 1.07 ± 2.47 adducts/108 nucleotides (36% detectability) in non-smokers (P<0.001). In the study reported here, we observed a 10-fold difference in levels of N6-HOMe-dAdo between smokers and non-smokers.
What is the source of an elevated formaldehyde-DNA adduct in smokers? The simplest explanation is inhalation of formaldehyde in cigarette smoke. Mainstream cigarette smoke contains 14 –28 µg/cigarette of formaldehyde, based on analysis of 48 brands under ISO conditions (11), which means a smoker of 20 cigarettes per day might be exposed to about 400 µg of formaldehyde by inhalation daily. The concentration of formaldehyde in the blood of volunteers exposed to 2.3 mg/m3 for 40 min, which would entail inhalation of about 600 µg (based on fifteen 500 ml breaths per min), was however unchanged from pre-exposure levels (1). Formaldehyde reacts quickly at the site of contact and is rapidly metabolized by human erythrocytes (1). Formaldehyde is also an endogenous compound, with a concentration (of free plus bound) in human blood of 2–3 µg/g blood (or about 100 µM) (1). Another source of N6-HOMe-dAdo could be formaldehyde released upon metabolism of a tobacco-specific compound such as NNK or nicotine. We have shown that rats treated with NNK have N6-HOMe-dAdo in their liver and lung DNA, but the doses used in that study were far higher than human exposure levels (6). Unstable formaldehyde-histone adducts, which may be elevated in smokers, might also transfer formaldehyde to DNA (12). Another possibility is that formaldehyde is a secondary metabolite generated during lipid peroxidation or inflammation associated with smoking.
The results of this study provide some potentially important new insights on mechanisms of cancer induced by smoking. It is possible that the role of formaldehyde has been overlooked previously. Formaldehyde causes squamous cell carcinoma of the nasal cavities upon inhalation exposure of rats, and is considered a cause of nasopharyngeal cancer in humans, with weaker evidence for leukemia (1). These two cancers are among the many caused by smoking and arguably could be due to formaldehyde in cigarette smoke (13).
There are some limitations to this study. First, it is relatively small, with samples from only 62 subjects having been analyzed. Larger studies comparing smokers and non-smokers, and studies of smokers who stopped smoking are required. Second, the internal standard is [15N5]N6-Me-dAdo, not [15N5]N6-HOMe-dAdo. The latter cannot be used as internal standard because of its moderate stability at the deoxyribonucleoside level. We estimate that the conversion of N6-HOMe-dAdo to N6-Me-dAdo is 50–80% (6). Thus, the actual levels of N6-HOMe-dAdo in DNA may be underestimated. Third, we do not know the relationship of leukocyte DNA adduct levels to those in potential target tissues such as lung.
In summary, our results show for the first time that a formaldehyde-DNA adduct is present in human leukocyte DNA and demonstrate remarkable differences in its levels in smokers and non-smokers. These results provide potentially important new insights on mechanisms of human carcinogenesis by formaldehyde and cigarette smoke.
Acknowledgements
This study was supported by grant no.CA-81301 and contract NO1-CP-64402 from the U.S. National Cancer Institute. Mass spectrometry was carried out in the Analytical Biochemistry Core Facility of the Masonic Cancer Center, supported in part by Cancer Center Support Grant CA-77598. SSH is an American Cancer Society Research Professor, supported by grant RP-00-138.
References
- 1.International Agency for Research on Cancer. Formaldehyde, 2-butoxyethanol and 1-tert-butoxypropan-2-ol. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. vol. 88. Lyon, FR: IARC; 2006. p. 39-325. [PMC free article] [PubMed] [Google Scholar]
- 2.Chaw YF, Crane LE, Lange P, Shapiro R. Isolation and identification of cross-links from formaldehyde-treated nucleic acids. Biochemistry. 1980;19:5525–5531. doi: 10.1021/bi00565a010. [DOI] [PubMed] [Google Scholar]
- 3.Beland FA, Fullerton NF, Heflich RH. Rapid isolation, hydrolysis and chromatography of formaldehyde-modified DNA. J Chromatogr. 1984;308:121–131. doi: 10.1016/s0021-9673(01)87539-7. [DOI] [PubMed] [Google Scholar]
- 4.Huang H, Hopkins PB. DNA interstrand cross-linking by formaldehyde: nucleotide sequence preference and covalent structure of the predominant cross-link formed in synthetic oligonucleotides. J Am Chem Soc. 1993;115:9402–9408. [Google Scholar]
- 5.Cheng G, Shi Y, Sturla S, et al. Reactions of formaldehyde plus acetaldehyde with deoxyguanosine and DNA: formation of cyclic deoxyguanosine adducts and formaldehyde cross-links. Chem Res Toxicol. 2003;16:145–152. doi: 10.1021/tx025614r. [DOI] [PubMed] [Google Scholar]
- 6.Wang M, Cheng G, Villalta PW, Hecht SS. Development of liquid chromatography electrospray ionization tandem mass spectrometry methods for analysis of DNA adducts of formaldehyde and their application to rats treated with N-nitrosodimethylamine or 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem Res Toxicol. 2007;20:1141–1148. doi: 10.1021/tx700189c. [DOI] [PubMed] [Google Scholar]
- 7.Whitehouse RC, Prasad AS, Rabbani PI, Cossack ZT. Zinc in plasma, neutrophils, lymphocytes, and erythrocytes as determined by flameless atomic absorption spectrophotometry. Clin Chem. 1982;28:475–480. [PubMed] [Google Scholar]
- 8.Phillips DH. Smoking-related DNA and protein adducts in human tissues. Carcinogenesis. 2002;23:1979–2004. doi: 10.1093/carcin/23.12.1979. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Wang M, Villalta PW, et al. Quantitation of an acetaldehyde adduct in human leukocyte DNA and the effect of smoking cessation. Chem Res Toxicol. 2007;20:108–113. doi: 10.1021/tx060232x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavanello S, Pulliero A, Saia BO, Clonfero E. Determinants of anti-benzo[a]pyrene diol epoxide-DNA adduct formation in lymphomonocytes of the general population. Mutat Res. 2006;611:54–63. doi: 10.1016/j.mrgentox.2006.06.034. [DOI] [PubMed] [Google Scholar]
- 11.Counts ME, Hsu FS, Laffoon SW, Dwyer RW, Cox RH. Mainstream smoke constituent yields and predicting relationships from a worldwide market sample of cigarette brands: ISO smoking conditions. Regul Toxicol Pharmacol. 2004;39:111–134. doi: 10.1016/j.yrtph.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Lu K, Boysen G, Gao L, Collins LB, Swenberg JA. Formaldehyde-induced histone modifications in vitro. Chem Res Toxicol. 2008;21:1586–1593. doi: 10.1021/tx8000576. [DOI] [PubMed] [Google Scholar]
- 13.International Agency for Research on Cancer. Tobacco Smoke and Involuntary Smoking. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. vol. 83. Lyon, FR: IARC; 2004. pp. 1179–1187. [PMC free article] [PubMed] [Google Scholar]