Abstract
Aggressive tumor cells express a plastic, multipotent phenotype similar to embryonic stem cells. However, the absence of major regulatory checkpoints in these tumor cells allows aberrant activation of embryonic signaling pathways, which appears to contribute to their plastic phenotype. Emerging evidence showing the molecular cross-talk between two major stem cell signaling pathways, Nodal and Notch, suggest a promising therapeutic strategy that could target aggressive tumor cells based on their unique plasticity, and provide new insights into the mechanisms underlying the re-emergence of developmental signaling pathways during tumor progression.
Introduction
It is becoming increasingly clear that regulators of cell fate during embryonic development may also play a role during tumorigenesis. Notch and Nodal are two examples of such regulators. Nodal, a member of the TGF-beta family of proteins, is involved in stem cell maintenance and differentiation, and shown to be associated with cancer progression (1, 2). Notch is an evolutionary highly conserved transmembrane receptor involved in cellular proliferation and differentiation that is often exploited by cancer cells to promote tumorigenesis and metastasis (3). This review will summarize the role of Notch and Nodal during normal development and cancer, and speculate about the molecular cross talk between these two signaling pathways in cancer resulting in possible targets for novel therapy.
Nodal and Notch signaling
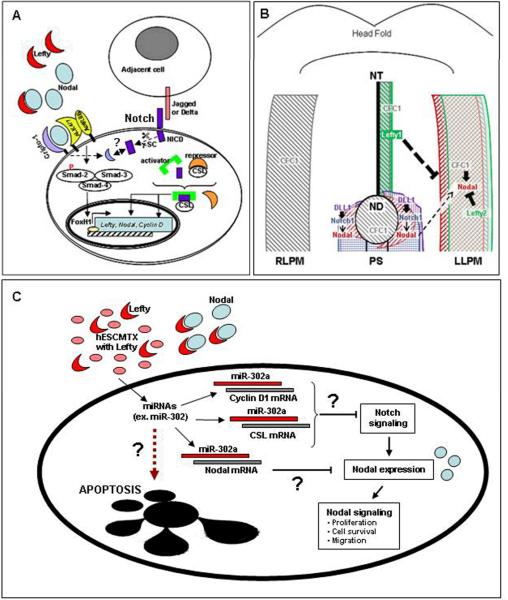
Typically, Nodal signals by binding to its coreceptor Cripto-1 and with type I (ALK4/7) and type II (ActRIIB) activin-like kinase receptors (1) (Figure 1A). This results in activation of the smad2/3/4 complex which translocates to the nucleus where it regulates gene expression by associating with transcription factors including FoxH1 and Mixer (Figure 1A). Nodal can induce its own transcription as well as the transcription of its inhibitor Lefty (4). The Lefty isoforms, A and B, like Nodal, are TGF-beta family members, and specifically antagonize Nodal signaling by directly binding to Nodal and/or to Cripto-1, thus preventing activation of the ALK receptor complex (Figure 1A).
Figure 1. Convergence of Nodal and Notch signaling.
(A) Nodal can bind to ALK4/7 and ActRIIB receptors and induce smad-2/3/4-dependent signaling in a Cripto-1 dependent or independent manner, leading to activation of target genes including FoxH1, Nodal and Lefty. Binding of Notch receptors to ligands expressed on adjacent cells leads to enzymatic cleavage by gamma-secretase (γ-SC) and intracytoplasmic release of the active Notch intracellular domain (NICD). The CSL DNA binding protein can function as a transcriptional repressor. However, when NCID binds to CSL it forms an activator complex leading to target gene transcription. Cripto-1 may also play a role in regulating Notch expression in melanoma cells (broken arrows). (B) During the establishment of left-right asymmetry in mouse, Nodal expression around the node (ND) is directly regulated by Notch/Delta-like-1 (DLL1) signaling. This domain of Nodal expression is required for the subsequent expression of Nodal in the left lateral plate mesoderm (LLPM) which autoregulates its own expression, as well as the expression of Lefty2 in the LLPM and Lefty1 in the left presumptive floor plate. Together these proteins function to restrict Nodal signaling to the left side of the embryo where it induces left-sided structures. The Nodal coreceptor, CFC1 (Cryptic), an ortholog of human Cripto-1, is symmetrically expressed in both the left and right lateral plate mesoderm (RLPM), as well as in the node and along the left midline. Thin arrows indicate regulation at the transcriptional level. Block symbols indicate regulation at the protein level. Abbreviations: ND, node; NT, notochord, PS, primitive streak; LLPM, left lateral plate mesoderm; RLPM, right lateral plate mesoderm. (C) Human embryonic stem cell conditioned matrix (hESCMTX) contains factors, such as Lefty, capable of repressing Nodal expression in aggressive human melanoma cells (C8161). These cells also show increased miR-302a levels that could inhibit Notch signaling and Nodal expression. Inhibition of Nodal signaling may lead to redifferentiation or apoptosis in C8161 cells.
Canonical Notch signaling is activated upon binding of Notch receptors (Notch 1-4) with their ligands (Delta-like1, 3 and 4 and Jagged1 and 2) expressed on adjacent cells (Figure 1A) (3). The binding of Notch with its ligands triggers a series of proteolytic cleavages resulting in the release of the Notch intracellular domain (NICD) capable of binding to the CSL (CBF1/RBPjk in vertebrates, Suppressor of Hairless in Drosophila, Lag-1 in C. elegans) DNA binding protein (3). In the absence of NICD, CSL inhibits transcription by associating with co-repressor proteins. NICD activates transcription by competing with these repressor proteins and facilitating the binding of activator complexes to CSL (Figure 1A). This activating complex is then capable of transcribing target genes including Hes and Hey, and genes implicated in cancer development, such as c-myc, cyclin D1 and p21/Waf1 (3).
Evidence from developmental studies has shown that Notch signaling regulates Nodal expression during the establishment of vertebrate left-right asymmetry (5). In fact, Nodal has been identified as a direct target of Notch activity, and, through promoter analysis, common putative CSL binding sites required for specific Nodal expression around the node have been described (and subsequently in the left lateral plate mesoderm) (Figure 1B).
Nodal and Notch in cancer
Nodal was shown to be expressed in human breast carcinoma cell lines, but poorly expressed in normal mammary epithelial and myoepithelial cells, and intensity of Nodal staining was found to correlate with breast carcinoma progression (6). While Nodal was not detected in normal melanocytes and poorly expressed in non-invasive melanoma, Nodal was robustly expressed in invasive and metastatic melanoma (2). Notch signaling has also been associated with a number of human cancers including leukemia and cancers of the colon and breast (7). However, the oncogenic role for Notch appears to be more complex. For instance, in breast cancer, Notch-2 functions as a tumor suppressor while other Notch receptors are oncogenic (8), yet, in brain tumorigenesis, Notch-2 acts as an oncogene while Notch-1 has the opposite effect (9). In the skin, Notch can induce cell cycle arrest and differentiation of keratinocytes (10), but promote melanocytic stem cell survival and melanoma tumorigenesis (11). Given the role for Nodal in melanoma and the observation that perturbation of Notch signaling in the skin can result in malignancies including melanoma (11), it is possible that molecular cross-talk exists between Nodal and Notch signaling in human melanoma. Evidence from developmental studies describe a role for Notch in regulating expression of Nodal that is supported by the presence of two CSL binding sites in the Node Specific Enhancer, a transcriptional regulatory region for Nodal shown to respond to Notch signaling (3).
Since Notch and Nodal signaling can be inhibited in cancer cells, pre-clinical and clinical studies have examined the potential for targeting Notch signaling at different points of its pathway in various Notch-expressing cancers including breast cancer, myeloma and leukemia (12). A commonly studied target in these studies is gamma-secretase, whose inhibitors (GSIs) can affect Notch signaling by preventing cleavage of the active NICD. For example, GSI was capable of inducing apotosis in notch expressing human aggressive melanoma cells but not in melanocytes (13). However, the potential for undesirable effects, given the physiologic role of gamma-secretase (or Notch signaling) in normal tissues or organs, remains a concern.
Other studies have targeted Nodal signaling pathway using small molecule ALK receptor inhibitors, or by directly inhibiting Nodal with human embryonic stem cell (hESC)-derived Lefty or anti-Nodal morpholino and showed redifferentiation and suppression of tumorigenesis in Nodal-expressing human metastatic melanoma cells (5, 6). More recently, an anti-Nodal function blocking antibody was shown to inhibit vasculogenic mimicry of aggressive melanoma cells in vitro. Long term exposure to the anti-Nodal antibody induced apoptosis in human melanoma cells in an in vivo mouse lung colonization assay (14). Since Nodal expression appears to be limited predominantly to cancer cells in adult tissues (5) and there is evidence for cross-talk between the Nodal and Notch signaling pathways, it may be possible to specifically target both these pathways in cancer cells in a “two-for-one” hit strategy whereby inhibition of the Notch pathway may affect Nodal-dependent signaling and vice versa. In fact, knockdown experiments using specific Notch siRNAs showed that Notch4 knockdown was associated with reduced Nodal levels in human aggressive melanoma cells (15). Furthermore, ongoing studies from our laboratory have shown a connection between expression of Cripto-1, the Nodal co-receptor, and increased expression of Notch-4 in human melanoma cells (personal communication).
miRNA targets and potential implication with Notch and Nodal signaling/function
MicroRNAs (miRNAs) are endogenous non-coding RNA sequences of ~22 nucleotides found in vertebrates and non-vertebrates capable of post-transcriptional control of gene expression by binding to specific protein coding mRNAs (16). MicroRNA-430 (miR-430) has been shown to inhibit the translation of the zebrafish Nodal homolog, squint (17). Target sites of miR-430 are detected in the mammalian Nodal gene suggesting that miRNA-dependent regulation of Nodal could occur in humans. In fact, web based analyses of potential miRNA targets predict that members of the miR-302 cluster, the human homolog of miR-430, may regulate Nodal expression. Notch signaling is also regulated by miRNAs in mammalian cells. Specifically, miR-1 negatively affects Notch signaling by repressing the expression of the Notch ligand, Delta-like1 (18). Thus, it seems plausible that if Nodal and Notch signaling are interrelated, miRNAs capable of regulating one pathway may affect the other.
Particular miRNAs involved in tumorigenesis and progression of certain human cancers are continuously being identified. Recently, numerous miRNAs that are upregulated or downregulated in melanoma cells and specific miRNAs associated with malignant progression of melanoma have been described (19). When human melanoma cells were cultured on hESC conditioned matrix (CMTX) containing hESC-derived Lefty (Figure 1C), the expression of Nodal in the melanoma cells was dramatically downregulated as these cells acquired a less aggressive and more differentiated phenotype (5). This significant reduction of Nodal was also accompanied by reduced expressions of Notch4 and miR-145, and increased levels of miR-302a (personal communication). Interestingly, miR-145 is one of several miRNAs associated with early progression of melanoma (19). In addition to potential regulation of Nodal signaling, as mentioned above, members of the miR-302 cluster have been shown to reprogram cancer cells into slowly proliferating, ES-like cells, by inhibiting cell cycle regulators, such as cyclin D1, a target of Notch signaling (Figure 1A) (20). Further studies will identify other candidate miRNAs induced by the stem cell microenvironment that may be involved in regulating plasticity and aggressiveness of melanoma cells by disrupting Nodal- or Notch-dependent oncogenic effects.
Significance and implications
The importance of embryonic signaling pathways in cancer biology has gained considerable attention. Studies show that cancer cells can exploit developmental signaling pathways to facilitate proliferation and metastatic spread. Nodal and Notch are involved in normal development and have emerged as markers and mediators of tumor progression as well. Importantly, studies suggest that these critical pathways converge at the transcriptional level and that epigenetic factors, including miRNAs, may regulate the aberrant expression of these proteins. Preliminary evidence suggests that it is possible to target Nodal-expressing cancer cells with anti-Nodal function blocking antibodies. The specific expression of Nodal in cancer cells could be exploited to target delivery of anti-Notch or other therapeutics directly to cancer cells without affecting normal surrounding cells. Newly emerging technologies such as nanoparticles hold promise as delivery tools for simultaneously targeting Nodal and Notch. The identification of epigenetic and genomic alterations that facilitate the reemergence of developmental signaling pathways and levels of potential cross-talk adds a new dimension to the targeting strategy for therapeutic interventions of cancer.
Acknowledgements
This work is supported by the U.S. National Institutes of Health (CA59702 and CA121205), the Eisenberg Scholar Research Award and by the Maeve McNicholas Memorial Foundation. We apologize to authors whose work was not directly mentioned. Due to space limitations primary reviews were cited.
References
- 1.Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- 2.Topczewska JM, Postovit LM, Margaryan NV, et al. Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nat Med. 2006;12(8):925–32. doi: 10.1038/nm1448. [DOI] [PubMed] [Google Scholar]
- 3.Borggrefe T, Oswald F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009 doi: 10.1007/s00018-009-8668-7. DOI 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabibzadeh S, Hemmati-Brivanlou A. Lefty at the crossroads of “stemness” and differentiative events. Stem Cells. 2006;24(9):1998–2006. doi: 10.1634/stemcells.2006-0075. [DOI] [PubMed] [Google Scholar]
- 5.Krebs LT, Iwai N, Nonaka S, et al. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 2003;17(10):1207–12. doi: 10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Postovit LM, Margaryan NV, Seftor EA, et al. Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc Natl Acad Sci U S A. 2008;105(11):4329–34. doi: 10.1073/pnas.0800467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64(21):2746–62. doi: 10.1007/s00018-007-7164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Neill CF, Urs S, Cinelli C, et al. Notch2 signaling induces apoptosis and inhibits human MDA-MB-2 3 1 xenograft growth. Am J Pathol. 2007;171(3):1023–36. doi: 10.2353/ajpath.2007.061029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan X, Mikolaenko I, Elhassan I, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64(21):7787–93. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 10.Rangarajan A, Talora C, Okuyama R, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20(13):3427–36. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinnix CC, Herlyn M. The many faces of Notch signaling in skin-derived cells. Pigment Cell Res. 2007;20(6):458–65. doi: 10.1111/j.1600-0749.2007.00410.x. [DOI] [PubMed] [Google Scholar]
- 12.Rizzo P, Osipo C, Foreman K, et al. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27(38):5124–31. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- 13.Nickoloff BJ, Hendrix MJC, Pollock PM, et al. Notch and NOXA-related pathways in melanoma cells. J Investig Dermatol Symp Proc. 2005;10(2):95–104. doi: 10.1111/j.1087-0024.2005.200404.x. [DOI] [PubMed] [Google Scholar]
- 14.Strizzi L, Postovit LM, Margaryan NV, et al. Nodal as a biomarker for melanoma progression and a new therapeutic target for clinical intervention. Exp 14 Rev Dermatol. 2009;4(1):67–78. doi: 10.1586/17469872.4.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Postovit LM, Seftor EA, Seftor REB, Hendrix MJC. Targeting Nodal in malignant melanoma cells. Expert Opin Ther Targets. 2007;11(4):497–505. doi: 10.1517/14728222.11.4.497. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318(5848):271–4. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 18.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A. 2005;102(52):18986–91. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mueller DW, Rehli M, Bosserhoff AK. miRNA Expression Profiling in Melanocytes and Melanoma Cell Lines Reveals miRNAs Associated with Formation and Progression of Malignant Melanoma. J Invest Dermatol. 2009 doi: 10.1038/jid.2008.452. DOI:10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- 20.Lin SL, Chang DC, Chang-Lin S, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14(10):2115–24. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]