Abstract
DNA methylation patterns are established and maintained by three DNA methyltransferases (DNMTs): DNMT1, DNMT3A, and DNMT3B. While essential for development, methylation patterns are frequently disrupted in cancer and contribute directly to carcinogenesis. Recent studies linking polycomb group (PcG) repression complexes (PRC1 and PRC2) to the DNMTs have begun to shed light on how methylation is targeted. We previously identified a panel of genes regulated by DNMT3B. Here we compare these with known PcG targets to show that nearly 47% of DNMT3B regulated genes are also bound by PRC1 or PRC2. We chose 44 genes co-regulated by DNMT3B and PRC1/PRC2 to test whether these criteria would accurately identify novel targets of epigenetic silencing in colon cancer. Using RT-PCR, bisulfite genomic sequencing, and pyrosequencing, we show that the majority of these genes are frequently silenced in colorectal cancer cell lines and primary tumors. Some of these, including HAND1, HMX2, and SIX3 repressed cell growth. Finally, we analyzed the histone code, DNMT1, DNMT3B, and PRC2 binding by ChIP at epigenetically silenced genes to reveal a novel link between DNMT3B and the mark mediated by PRC1. Taken together, these studies suggest that patterns of epigenetic modifiers and the histone code influence a gene’s propensity to become hypermethylated in cancer and that DNMT3B plays an important role in regulating PRC1 function.
INTRODUCTION
DNA methylation is an essential epigenetic mark for mammalian embryonic development and transcriptional regulation. Global patterns of DNA methylation are established and regulated via the action of four DNA methyltransferases (DNMTs): DNMT1, DNMT3A, DNMT3B, and DNMT3L (1). DNMT1 associates with S-phase replication foci and acts primarily as a maintenance methyltransferase (2). DNMT3A and DNMT3B are essential for de novo methylation during embryonic development (3). Hypomorphic mutations in the DNMT3B gene result in Immunodeficiency, Centromere Instability, and Facial Anomalies (ICF) syndrome (3–5). Dnmt3L forms a complex with Dnmt3a and Dnmt3b in embryonic stem (ES) cells and stimulates their activity (6, 7).
Aberrant DNA methylation patterns, including CpG island hypermethylation and repetitive region hypomethylation, are hallmarks of transformed cells (8). Misregulation of DNMT expression clearly contributes to tumorigenesis as demonstrated recently by the work of Linhart et al., which showed that elevated Dnmt3b1 expression in the ApcMin/+ model enhanced colorectal carcinogenesis and caused tumor suppressor gene methylation (9). DNA methylation, however, does not act alone in repressing gene activity. A complex and intertwined set of post-translational modifications of the core histone tails dynamically impart both transcriptionally repressive and activating signals. These marks, and the cellular machinery regulating them, are also disrupted in cancer (10). DNA methyltransferases, such as DNMT3B, interface directly with the histone code by interacting with the histone methylases SUV39H1 (11) and EZH2 (12), which impart trancriptionally repressive H3K9 and H3K27 trimethylation marks, respectively.
In mammals, there are two principal polycomb group (PcG) complexes, PRC1 and PRC2. PRC2 is composed of EED, EZH2, and SUZ12, while PRC1 is composed of HPH1–3, RING1/2, BMI1/MEL-18, and HPC1–4. A widely accepted model posits that PRC2 initiates gene silencing by trimethylating H3K27, followed by recruitment of PRC1 and its associated H2AK119 monoubiquitination activity, resulting in stable maintenance of gene silencing (13). PRC1 and PRC2 are essential for mammalian development and maintenance of ES cell pluripotency (14). PcG complexes, like DNMTs, have strong links to cancer. For example, EZH2 is overexpressed in tumors and is predictive of poor prognosis (15). BMI1 cooperates with MYC to promote lymphomas by repressing the INK4a/ARF tumor suppressor locus (16).
Interesting connections between DNA methylation and PcG have emerged recently. DNMT1, DNMT3A, and DNMT3B interact with EZH2 and EZH2 targets DNA methylation to certain promoters (12). Schlesinger et al. showed that genes subject to tumor-specific hypermethylation in colon cancer were significantly more likely to be marked by H3K27 methylation in normal tissues than genes lacking H3K27 methylation (17). Laird’s group showed that PcG targets in ES cells were 12-fold more likely to sustain cancer-specific hypermethylation (18). Nearly 49% of genes methylated in colon cancer are PcG targets in ES cells in another recent study (19). In contrast, Gal-Yam et al. reported that many genes hypermethylated in a prostate cancer cell line were bound by PcG in normal cells but lost PcG binding upon acquisition of DNA methylation in the cancer (20). These studies demonstrate compelling connections between DNA methylation and PcG, yet there is clearly much that we do not understand regarding the molecular mechanisms linking these two systems.
Using cell lines derived from ICF syndrome patients we recently identified a large number of DNMT3B-regulated genes (21). While the DNA methylation changes in ICF cells were small in many cases, changes in the histone code were dramatic and this study provided additional support for a link between PcG complexes and DNMT3B in particular (21). In the present manuscript, we have expanded upon these studies in the context of colon cancer. We show that there is significant overlap between DNMT3B and PRC1/PRC2 targets. We then asked the question, can binding of DNMT3B and PRC1 and/or PRC2 predict genes that sustain aberrant DNA hypermethylation in colon cancer? Nearly 80% of 44 randomly chosen genes co-regulated by DNMT3B and PRC1 and/or PRC2 were indeed expressed in normal colon but silenced in at least one cell line. Closer examination of 24 of these genes revealed that they were subject to high frequency epigenetic silencing in colorectal cancer cell lines and primary tumors. Several of these genes, including HAND1, SIX3, HMX2, and TBX4 modulated cell growth in a colony formation assay. Finally, we examined the relationship between DNMT1, DNMT3B, and a large panel of histone marks by chromatin immunoprecipitation. Our analysis revealed an intriguing and hitherto unknown relationship between DNMT3B and the PRC1 mark H2AK119 monoubiquitination, suggesting a functional link between these two important epigenetic modifiers.
MATERIALS AND METHODS
Cell lines, drug treatments, and human tissues
Human colon tumor cell lines HCT116, HCT15, HT29, SW48, SW480, T84, RKO, and LoVo (American Type Culture Collection) were grown in McCoy’s 5-a media (Mediatech). 5-aza-2'-deoxycytidine (5-azadC) was purchased from Sigma. For drug treatments, 5-azadC was added to cultures at a final concentration of 5 µM for 3 days. Fresh-frozen tumor and normal colon samples were obtained from the University of Florida Shands Cancer Center Molecular Tissue Bank. Specimens were analyzed histologically by a surgical pathologist. DNA and RNA were prepared from these samples as previously described (22). The normal colonic mucosa used in ChIP analysis was obtained from a 52-year old female patient undergoing surgery for neoplastic disease of the right colon.
Reverse transcriptase (RT)-PCR, bisulfite genomic sequencing (BGS), and pyrosequencing
Semi-quantitative RT-PCR and BGS were carried out as reported previously (21). For BGS, multiple independent clones from at least two independent PCR reactions were sequenced. For pyrosequencing, bisulfite-treated DNA was amplified by PCR with one biotinylated primer. Pyrosequencing reactions were performed according to the manufacturer’s specifications using the PyroMark MD System (Biotage AB), and the degree of methylation was calculated using the PSQ HS 96A 1.2 software (Biotage AB) as we have reported previously (23). Primer sequences are listed in Table S1.
Colony formation analysis
HCT15 or HCT116 cells were seeded at 1.8×105/well in six-well plates. Twenty four hours later they were transfected using TransIT-LT1 according to the manufacturer's instructions (Mirus). At 24 hours post-transfection, cells were trypsinized, counted, and re-seeded at 1.8×105/well in six-well plates with fresh medium containing 800U/ml G418 (Mediatech) to kill untransfected cells. Cells were grown for 14 days with fresh media and G418 added every three days. Colonies were stained with 0.2% methylene blue and photographed. Details on the construction of expression vectors used for transfection are provided in the supplemental material.
Chromatin immunoprecipitation (ChIP) and western blotting
ChIP was performed essentially as described (21). Briefly, cells were seeded at 1.5×106/150-mm dish and grown for 72 hrs. Normal colorectal tissue was cut into small pieces with a scalpel then transferred to a tube containing 10 ml of tissue culture medium. Formaldehyde was added to a final concentration of 1% and cells/tissues were fixed for 10 minutes. Crosslinked chromatin was sheared by sonication using a Branson Sonifier 450 (tip model 102, output 5, 50% duty, 12 × 1 min. bursts). Fifty micrograms of chromatin was used in each ChIP reaction. Supernatant from the irrelevant antibody served as a positive control (‘Input’, 1% of the ChIP material). Purified, immunoprecipitated DNA was analyzed by semi-quantitative PCR (see Table S1 for primer sequences). For western blotting, cells were fractionated as described in the supplemental methods and the soluble chromatin fraction (P3) was used. Antibodies used in ChIP and western blotting are described in the supplemental material.
RESULTS
Overlap between DNMT3B and PcG target genes
We previously used microarray expression profiling in normal and ICF-derived lymphoblastoid cell lines (LCLs) to identify genes regulated by DNMT3B (21). We further analyzed and refined this data (Supplemental material, Fig. 1), to yield a list of 506 DNMT3B targets. Using data derived from published ChIP-chip and ChIP-seq studies that mapped genome-wide binding of PRC1 and PRC2 subunits and the marks they mediate, we compiled a list of all known PRC1 and/or PRC2 target genes (Supplemental material). Comparison of these lists revealed that nearly 47% of DNMT3B target genes were also targets of PRC1 or PRC2, a significant non-random enrichment (p<10−8). Approximately 36% and 35% of DNMT3B target genes were also PRC1 and PRC2 targets, respectively, in ES cells or embryonic fibroblasts (Fig. 1A).
Fig. 1.
Relationship between genes regulated by DNMT3B and genes bound by PRC1 and PRC2. (A) Left. Venn diagram comparing the overlap between DNMT3B and PRC1/PRC2 target genes. Right. Table summarizing the percent overlap between DNMT3B, PRC1, and PRC2 along with results of our frequency of gene methylation in cancer predictive analysis. (B) Expression of forty-four DNMT3B+PRC1 and/or PRC2 target genes in normal human colon (N. colon) and HCT116 cell lines proficient or deficient for DNMT1 and DNMT3B by semi-quantitative RT-PCR. We defined expression at 35 cycles as moderate and at 50 (but not 35) cycles as low for normal colon. Reactions were repeated three times. Amplification of GAPDH serves as a loading control. ‘*’ homeobox gene.
DNMT3B plus PcG binding in normal cells as a predictor of genes epigenetically silenced in cancer
Recent reports have noted compelling links between genes bound by PcG in normal cells and genes targeted for aberrant DNA methylation in cancer cells (17, 18, 24). For example, Schlesinger et al. showed that nearly 50% of cancer-methylated genes are pre-marked by PRC2 and H3K27 methylation in colon cancer and these marks were highly specific because the same genes were not selectively associated with other repressive histone modifications (17). We compiled our own list of genes subject to DNA hypermethylation in human colorectal tumors using recent publications (1531 genes, Supplemental material). Comparing our list of genes methylated in colon cancer with the list of PRC1/PRC2 targets we generated revealed that 42% and 44% of cancer methylated genes are targets of PRC1 and PRC2, respectively, results which are comparable to those of others (17), thus validating our methodology. Use of PRC1 and/or PRC2 binding specifically, or regulation of expression by a DNA methyltransferase as a predictor of genes undergoing promoter hypermethylation in human tumor cells, however, has not been examined to our knowledge. We therefore performed such an analysis using our data sets, revealing that DNMT3B regulation of gene activity alone (17.2%) is a slightly better predictor of genes methylated in colon cancer than PRC1 only (12.5%, p<0.01) or PRC2 only (13.5%, p<0.05). Furthermore, the propensity of a gene for sustaining cancer-hypermethylation was very significantly different between the targets of PRC2 combined with DNMT3B (25.6%) compared to PRC2 only (13.5%, p<0.01). Interestingly, genes regulated by DNMT3B and marked by PRC1 (28.9%, p<10−5), or by both PRC1 and PRC2 (30.8%, p<10−3) were the most prone to cancer-specific DNA hypermethylation (Fig. 1A).
Our in silico analyses revealed that DNMT3B regulation and PRC1/PRC2 binding were good predictors of genes susceptible to acquiring aberrant DNA hypermethylation in cancer. To validate these predictions experimentally and determine if these criteria could identify novel cancer-methylation targets, we randomly chose 44 genes from the region of overlap between DNMT3B and PRC1 or PRC2 (Fig. 1A, 236 genes) and examined their expression in normal colon and the HCT116 colon cancer cell line. Among the 44 genes selected from the region of overlap with DNMT3B, 37 were PRC1 targets, 35 were PRC2 targets, and 28 were marked by both PRC1 and PRC2 (Fig. 1A). Using semi-quantitative RT-PCR, we determined that expression of approximately 13%, 48%, and 39% of the genes was absent, low, and moderate, respectively, in normal colon (Fig. 1B). These results were reproducible in two other normal colon samples (data not shown). To determine whether these genes are also be subject to epigenetic silencing in colon cancer, and examine the role of DNMT1 and DNMT3B in this process, we monitored their expression in parental HCT116 (WT), HCT116 DNMT1 (1KO), HCT116 DNMT3B (3BKO), and HCT116 DNMT1+DNMT3B (DKO) knockout cells (25). Interestingly, parental HCT116 cells lacked expression of nearly 80% of the 44 (PRC1 or PRC2) + DNMT3B targets (Fig. 1B). Of the 34 genes expressed at some level in normal colon but silent in WT HCT116 cells, six were reactivated in 1KO and DKO, seven were reactivated in 3BKO and DKO, 11 were reactivated in 1KO, 3BKO, and DKO, and ten were reactivated only in DKO cells. Taken together, these results support our in silico analyses and suggest that use of DNMT3B and polycomb binding accurately identifies novel targets of epigenetic silencing in a colon cancer cell line. These results also show that DNMT1 and DNMT3B play both separate and overlapping roles in mediating the epigenetic silencing of these genes.
Frequent silencing and DNA hypermethylation of DNMT3B plus PcG target genes in colon cancer cell lines and primary tissues
To obtain a better estimate of the frequency of gene silencing, we examined expression of 24 of the 44 genes from Fig. 1 in a panel of eight colon tumor cell lines untreated or treated with the DNA methylation inhibitor 5-azadC (5 µM) for 72 hours. Of these 24, twenty were silenced in untreated cells and transcriptionally reactivated by 5-azadC-treatment in 50% or more of the lines tested (Fig. S1, Table S2). The majority of the 24 genes were silenced at even higher frequency (13 were silenced in 7 or 8 out 8 lines, Fig. S1). The ability to reactivate expression by 5-azadC strongly suggests the involvement of DNA hypermethylation in their repression.
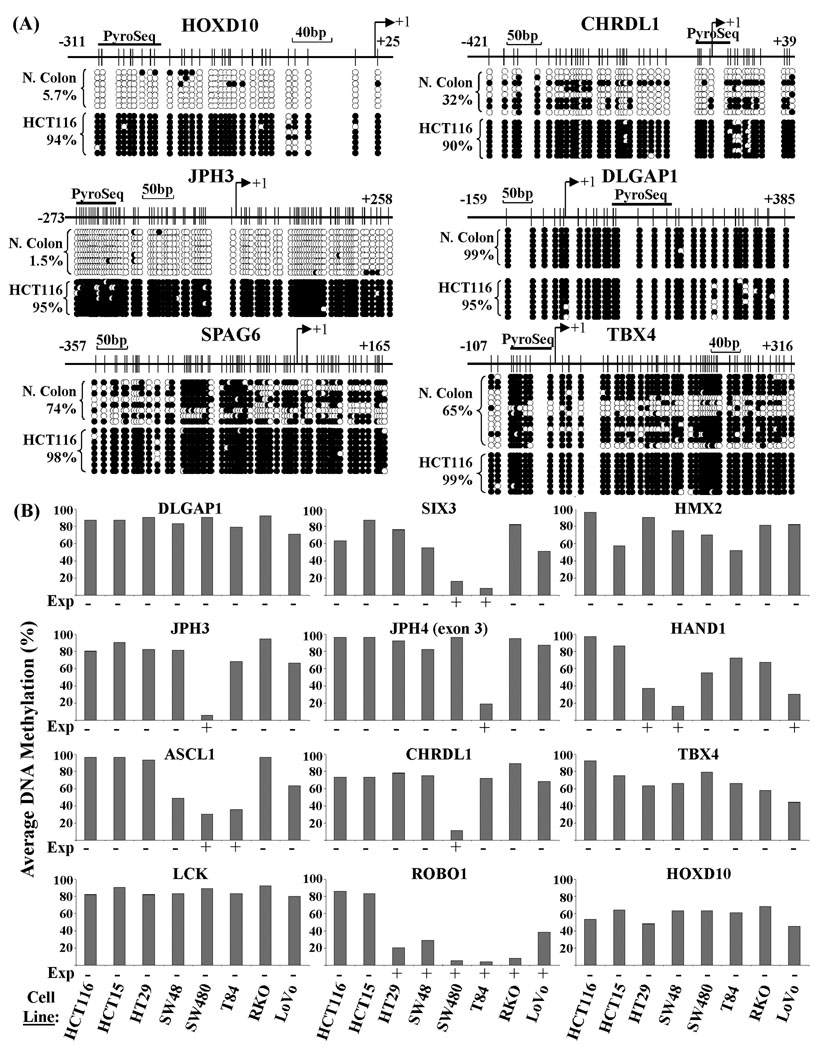
To directly examine the role of DNA methylation is repressing gene activity, we performed bisulfite genomic sequencing (BGS) on 13 of them from normal colon and HCT116 cells (Fig. 2A and Fig. S2). For eight genes (HOXD10, JPH3, JPH4 (2 regions), CHRDL1, ASCL1, FGF12, HMX2, and SIX3), DNA methylation levels were generally low in normal colon (<30%) but they were densely methylated in HCT116 cells, consistent with their patterns of expression. Although expressed in normal colon, the LCK promoter was nearly 60% methylated (Fig. S2). Four other genes, SPAG6, DLGAP1, TBX4, and TDRD12, were densely hypermethylated in both normal colon and HCT116 cells, suggesting that hypermethylation is part of their normal biology (Fig. 2A and Fig. S2). We extended the DNA methylation analysis to our full panel of seven additional colon cell lines using bisulfite pyrosequencing, revealing complete correlation between promoter DNA hypermethylation and lack of expression (Fig. 2B, Table S2). Generally, methylation levels in excess of 40% were incompatible with transcription.
Fig. 2.
DNA methylation analysis of DNMT3B+PRC1/PRC2 target genes in colon tumor cell lines. (A) DNA methylation patterns were determined for a subset of the genes examined in Fig. 1 using bisulfite genomic sequencing (BGS). The gene promoter region is shown, with the bent arrow denoting the transcription start site. CpG sites are indicated with tick marks and the region analyzed by pyrosequencing is indicated with a thick horizontal bar (‘Pyroseq’). Results are summarized below with open (unmethylated) or filled (methylated) circles for each CpG site. Each row is an individually cloned and sequenced molecule. Numbering is relative to the transcription start site. The total percent methylation over all clones and CpG sites is given at the left. (B) Quantitative bisulfite pyrosequencing DNA methylation analysis for twelve genes in eight colon tumor cell lines. Results are presented as the percent methylation over all CpG sites within the pyrosequenced region (part A and Fig. S2). Below each graph the expression status (‘Exp’) of the gene, derived from Fig. 1, is indicated with a ‘+’ for expressed and ‘−‘ for no expression.
We extended our DNA methylation and expression analyses of six genes (HAND1, HMX2, JPH3, JPH4, ASCL1, and TBX4) out of the total 44 to 12 fresh-frozen primary colorectal tumor and adjacent normal tissue pairs. With the exception of TBX4, there was an excellent correlation between reduced/absent expression and DNA hypermethylation in the tumor relative to the adjacent normal (Fig. 3, Table S2, representative programs are provided in Fig. S3). TBX4 was an interesting exception because it was not expressed in normal colon, but was re-expressed in 7/12 tumor tissues with a concomitant small, but significant reduction in methylation. We also utilized an extensive microarray expression database to further analyze expression of select genes (HAND1, JPH3, DLGAP1, CHRDL1, ROBO1, IL1R2, HOXD10, and LCK) in silico (26). Decreased expression of all eight was observed in colorectal tumors (or metastasis) relative to normal tissue (or the primary site) in at least one microarray study (Fig. S4). Furthermore, significant downregulation occurred in other tumor types, suggesting that epigenetic silencing of these genes is not restricted to colon cancer. Taken together, reduced expression of genes in tumor relative to normal tissue is suggestive of a growth suppressive role (i.e. tumor suppressors), while the findings for TBX4 indicate possible oncogenic function.
Fig. 3.
Epigenetic silencing of DNMT3B+PcG target genes in primary colorectal tissues. For six of the genes analyzed in previous figures, we determined expression by semi-quantitative RT-PCR (representative gel photos at the bottom of each graph, GAPDH is shown in the JPH3 panel) and DNA methylation levels by bisulfite pyrosequencing in twelve fresh frozen human colorectal cancer (‘T’) and adjacent normal (‘N’) tissue pairs. Regions analyzed by pyrosequencing are shown in Fig. 2 and Fig. S2. Results are presented as the percent methylation for all CpG sites within the analyzed region. The error bar is the standard error and significance (*) is determined using the T-test (P < 0.05).
DNMT3B+PRC1/PRC2 target genes regulate cell growth
Most of the genes we identified as targets of epigenetic silencing have not been previously shown to influence colon tumor cell growth. To investigate this more directly, we cloned the full-length open reading frames of ASCL1, CHRDL1, HAND1, HMX2, JPH3, SIX3, and TBX4 into an expression vector and transfected them into HCT15 cells (where the endogenous copy of all of these genes is epigenetically silenced, Fig. S1). Ectopic expression of HAND1, HMX2, and SIX3 caused significant growth suppression (Fig. 4). In contrast, expression of TBX4 resulted in a small but significant increase in cell growth, consistent with its possible role as a proto-oncogene. Similar results were obtained with HCT116 cells (data not shown). Taken together, these experiments show that a subset of the genes we identified possess properties consistent with them acting as tumor suppressors, while the others may regulate aspects of differentiation or cell motility that are not detectable in a colony formation assay.
Fig. 4.
Genes co-regulated by DNMT3B, PRC1, and/or PRC2, and subject to epigenetic silencing in cancer, regulate cell growth. Colony formation assays were used to assess growth regulatory potential of seven genes. The full-length open reading frame was cloned into an expression vector and transfected into HCT15 cells. Colonies surviving G418 selection were counted and set relative to transfection with empty parental expression vector (at 1.0). (A) Summary of average growth suppression or enhancement relative to empty vector. The bar is the standard error and the asterisk indicates significance at the P-values indicated. (B) Representative wells from transfections following selection. A p16INK4a expression plasmid was used as a positive control for growth suppression.
Role of specific DNA methyltransferases in modulating aberrant methylation and the histone code at PRC1/PRC2+DNMT3B target genes
Based on our expression studies (Fig. 1B), we identified genes regulated by the activity of DNMT1, DNMT3B, or both. We are particularly interested in DNMT3B’s role in gene regulation because of our prior work with ICF syndrome, therefore we examined the HAND1, IL1R2, and ROBO1 gene promoters because they were clear DNMT3B-regulated genes in HCT116 cells (Fig. 1B) and are established targets of PRC1 and PRC2 (27–32). BGS revealed that the HAND1 and IL1R2 CpG islands (Fig. 5A–5B) were hypomethylated in normal colon but hypermethylated in both HCT116 and 1KO cells. DNA methylation was reduced overall in 3BKO cells and nearly absent in DKO cells at both promoters (Fig. 5). Interestingly, there was a clearly defined region upstream of the HAND1 transcription start site that lost DNA methylation in 3BKO cells (boxed region, Fig. 5A), consistent with HAND1 becoming re-expressed in 3BKO cells. Two CpG sites and a small cluster of CpGs just downstream of the IL1R2 and ROBO1-B start sites, respectively, behaved similarly (boxed regions, Fig. 5B, Fig. S5A) suggesting that DNMT3B plays a critical role in methylating and repressing these genes.
Fig. 5.
Analysis of DNA methylation and the histone code at genes regulated by DNMT3B+PRC1/PRC2 reveals novel connections between H2A ubiquitination and DNMT3B. (A) BGS analysis of the HAND1 promoter defines a region of DNA methylation mediated by DNMT3B (boxed). Labeling is as in Fig. 2A. Methylation of the boxed region only is underlined. (B) BGS analysis of the IL1R2 promoter. (C) ChIP followed by semi-quantitative PCR analysis for the histone marks, PcG proteins, and DNMTs listed at the left of the gel photos for HAND1. (D) ChIP analysis of the IL1R2 promoter. Expression status of each gene is derived from Fig. 1B. ‘Me’ - methyl, ‘2x’ – dimethylated, ‘3x’ – trimethylated, ‘Ac’ – acetylated, ‘Ub’ – monoubiquitin, ‘NC’ – normal colon.
We next examined aspects of the histone code by ChIP. For all three genes, a mixture of activating and repressive marks was present at the promoter in normal colon, similar to the bivalent pattern at PcG targets in ES cells and some genes in differentiated cells like T cells (14, 33). This result could also be because we are examining a mixture of cell types from the colon. DNMT1 and DNMT3B were also bound, despite the low level of DNA methylation present, suggesting that their catalytic activities may in some way be restrained (Figs. 5A–5B). Two regions of ROBO1 were examined, an upstream promoter (ROBO1-A, completely devoid of CpGs) and ROBO1-B (containing a CpG island). Several notable differences were observed between these two, such as lack of Ub-H2A at ROBO1-B and an absence of DNMT3B at ROBO1-A (Fig. S5B). In 3BKO cells, activating H3 acetylation and trimethylated H3K4 marks were elevated at HAND1, while repressive H3K9 trimethylation and H3K27 dimethylation were reduced (Fig. 5C). Interestingly, however, H2AK119 monoubiquitination at HAND1 was completely lost in 3BKO cells. PRC2 binding, although reduced, was still clearly present. Our findings with HAND1 are consistent with it being a well-established target of PRC1 repression (34). Similar results for PRC2 and H2AK119 ubiquitination were observed at the IL1R2 and ROBO1-A promoters (Fig. 5D and Fig. S5B).
Given that loss of DNMT3B function in B cells resulted in not only gene-specific, but also global changes in the histone code (21), we examined total levels of select histone marks and PRC1/PRC2 subunits by western blotting in HCT116 cells. Notable changes include reduced H3K4 dimethylation and elevated H3K9 trimethylation and acetylation in 1KO and 3BKO cells (Fig. 6A). Levels of PRC2 subunits were increased in DNMT KO cells. Interestingly, some PRC1 subunits showed elevated expression (RING1/2 H2AK119 ubiquitin E3 ligases in all KOs and MEL-18 in 3BKO and DKO), while BMI1 expression was markedly reduced in KO cells (Fig. 6A). Increased expression of EZH2, SUZ12, RING1, RING2, and MEL-18 at the protein level could be accounted for, at least in part, by elevated mRNA levels (Fig. S6). Interestingly, however, BMI1 mRNA levels did not parallel their respective protein levels (Fig. 6A and Fig. S6). Taken together, these results indicate that DNMT1 and DNMT3B directly or indirectly regulate certain facets of the histone code globally (particularly H3K9 methylation and acetylation) and levels of both PRC1 and PRC2 subunits at the mRNA and/or protein level.
Fig. 6.
Relationships between DNMT status and levels of global and gene-specific epigenetic marks and their regulators. (A) Global levels of histone marks and PRC1/PRC2 complex proteins determined by western blotting. The antibody used is indicated at the left of the western panels. Staining of total core histones with coomassie blue (bottom) is used as a loading control. (B) ChIP analysis of the 14 indicated gene promoters for H2AK119 monoubiquitination, (C) trimethylated H3K27, (D) SUZ12 (PRC2 complex), (E) acetylated H3K9/K18, and (F) trimethylated H3K4. Following the gene symbol is the region analyzed by ChIP (relative to the transcription start site). The input is from DLGAP1.
We were intrigued by the marked loss of H2AK119 monoubiquitination in 3BKO cells at the three promoters examined thus far, therefore we examined this mark in greater detail at 14 other genes from Fig. 1B. Ubiquitinated H2A is a repressive mark that blocks transcriptional elongation, but not assembly, of RNA Pol II at promoters (35). For 11 out of 14 promoters, H2AK119 monoubiquitination was markedly or completely lost in 3BKO cells (Fig. 6B) despite global levels of this mark being relatively unchanged (Fig. 6A). This association was observed even if the gene did not become transcriptionally reactivated in 3BKO cells (e.g. DLGAP1 and JPH4, Fig. 1B, Fig. 6B, Fig. S7) strongly suggesting that DNMT3B inactivation causes Ub-H2A loss rather than Ub-H2A loss being an indirect consequence of gene reactivation. For the remaining three promoters, no Ub-H2A was observed in any of the examined samples indicating that they are targets of PRC2 but not of PRC1 (Fig. 6B–6D and Fig. S7). We also compared levels of PRC2 (SUZ12) binding and its mark, H3K27 trimethylation (Figs. 6C–6D), and two marks of transcriptional activity/permissiveness, H3K9/K18 acetylation and H3K4 trimethylation (Figs. 6E–6F). H3K27 trimethylation was reduced at 4/14 genes in 1KO cells and 2/14 genes in 3BKO cells, but was lost at almost all genes in DKO cells. Similar results were observed for SUZ12, although there were clearly cases where SUZ12 remained bound despite reduced H3K27 methylation. Levels of H3 acetylation and H3K4 trimethylation generally paralleled transcriptional activity and each other (Figs. 6E–6F). Analysis of a more complete set of histone marks for genes in Fig. 6 is presented in Fig. S7. Another interesting observation for several genes (e.g. HAND1, Fig. 5, Fig. 6, and Fig. S7) was that loss of one DNMT reduced binding of the other, suggesting that interactions among DNMTs (36) regulate their targeting. We corroborated our results from HCT116 KO cells by treating parental HCT116 cells with 5-azadC, which inactivates DNMTs. 5-azadC treatment resulted in changes in histone marks that largely paralleled those that we observed in DKO cells (Fig. S8). In summary, these findings demonstrate that most of the DNMT3B+PRC1/PRC2 target genes we identified based on in silico analyses are indeed bound by DNMT3B, PRC2, and are PRC1-marked in normal colon. In addition, our results suggest that DNMT3B is specifically involved in recruiting and/or maintaining ubiquitinated H2A at certain gene promoters.
DISCUSSION
In the present manuscript, we made use of our previously identified DNMT3B-regulated genes derived from an ICF syndrome model system, combined with published PRC1 and PRC2 targets derived from ChIP-chip and ChIP-seq studies, to examine the relationships between DNA methyltransferase and PcG complex binding, genes methylated in colon cancer, and the histone code. Nearly 50% of DNMT3B target genes in B cells are bound by PRC1 or PRC2 in ES cells or embryonic fibroblasts. Using DNMT3B-regulation and PRC1 and/or PRC2 binding as our sole criteria, we chose 44 genes and examined their expression and DNA methylation status. Nearly 80% were indeed hypermethylated and silenced in a DNMT-specific manner. Closer examination revealed discrete regions of hypomethylation in 3BKO cells flanking the transcription start sites and concomitant changes in the histone code. Interestingly, while loss of DNMT1 or DNMT3B resulted in marked changes in certain histone marks globally and in the levels of PRC1 and PRC2 complex subunits, especially BMI1 and MEL-18, the most consistent alteration at specific gene promoters in 3BKO cells was loss of H2AK119 monoubiquitination, a PRC1 complex-mediated mark. Taken together, these results show that the complement of epigenetic marks and DNMT3B binding in normal cells may serve as a good predictor of genes susceptible to acquiring aberrant DNA hypermethylation in cancer and that DNMT3B in particular plays a role in the activity and/or recruitment of PRC1 in differentiated cells.
Our findings show that use of DNMT3B regulation, in addition to PRC1 and/or PRC2 binding as criteria for predicting genes that become hypermethylated in colon cancer yields a high rate of positive hits. Further analysis of DNMT binding throughout the genome is therefore clearly warranted. Most of the genes we characterized have not been well studied in the context of colon cancer. Those acquiring aberrant methylation, such as the basic helix-loop-helix transcription factors ASCL1 and HAND1, may play important roles in regulating transcription of other genes that suppress growth or maintain the differentiated state. HAND1, which is expressed in developing and mature gut, may regulate expression of sonic hedgehog and the homeobox gene IRX4 (37). TBX4 is an interesting case where DNA methylation is involved in suppressing its expression in normal colon. While little is known about TBX4 in the context of cancer, TBX3 is highly overexpressed in breast cancer and represses the ARF tumor suppressor (38), supporting our hypothesis that TBX4 acts as an oncogene. CHRDL1 encodes a secreted BMP antagonist selectively expressed in colon crypts. Its epigenetic silencing may therefore negatively impact intestinal stem cell differentiation (39). Another group has also reported HAND1 methylation in colon cancer, consistent with our findings (40). Homeobox genes HOXD10, SIX3, and PROX1 were recently shown to be hypermethylated in lung cancer (41). Clearly, further study of how these genes regulate growth and differentiation of normal and transformed colon cells is warranted.
Here we have focused on how the activities of DNMT1 and DNMT3B are involved in regulating the histone code and PcG protein binding in tumor cells once methylation has been established. While histone methylation may initially recruit DNA methylation (11), once the densely hypermethylated state characteristic of tumor suppressor genes in cancer cells is established, our data suggest that DNMT3B is required for maintenance of H2AK119 monoubiquitination at a subset of polycomb targets. This may be mediated by direct physical contacts between the two complexes or indirectly via the DNA methylation mark itself. As we were unable to reliably ChIP for PRC1 with commercially available antibodies, we cannot yet determine if lack of DNMT3B interferes with PRC1 recruitment or with PRC1 enzymatic activity once recruited. DNMTs may bind to promoter regions in normal cells even if they are unmethylated, consistent with our previous ICF syndrome study (21). PcG complexes also bind to transcriptionally active or competent genes (28). DNMT3B may be kept catalytically inactive by PRC1 perhaps to have it poised for later de novo methylation-mediated repression, or DNMT3B participates in repression independent of methylation in this state. A shift to de novo methylation could occur due to changes in PRC1 complex subunit composition or post-translational modifications of PcG complex subunits or of DNMT3B itself. We observed reduced BMI1 and elevated MEL-18 levels in 3BKO cells, which may alter the overall composition and activity of PRC1. If this change occurred in a tissue-specific stem cell (e.g. intestinal crypt stem cell) it might affect downstream differentiation, leading, for example, to a cell blocked from terminally differentiating.
It remains a fundamental unanswered question why certain genes become hypermethylated in cancer. Recent findings demonstrating that PcG target genes in normal cells are more likely to acquire aberrant promoter hypermethylation in cancers (17, 18, 24), along with evidence of interactions between DNMT1/DNMT3B and several PcG complex subunits (EZH2 and BMI1 (12, 42)) are beginning to shed light on how this process may occur. Genes hypermethylated in cancer cells adopt a bivalent pattern of histone marks and low-level expression (characteristic of developmental genes in ES cells), when treated with DNA methylation inhibitors (43) lending further support for a mechanistic connection. Our results indicated that 5-azadC treatment altered histone marks in much the same way as DNMT1 and DNMT3B double knockout. Other data demonstrating that the EZH2-DNMT targeting process may not be universal (44) and that the association between tumor-specific DNA hypermethylation events and PcG binding may be cell-type specific (20, 45), demonstrates that we still have much to learn. It was striking that most PcG targets we examined lost H2AK119 monoubiquitination upon disruption of DNMT3B but not DNMT1. This despite the observation that DNMT1 loss results in marked reduction in BMI1 levels. Therefore DNMT1 and DNMT3B may play distinct roles in modulating PRC1 function. Interestingly, DNMT1 is essential for maintenance of and recruitment of BMI1 to PcG bodies (46). DNMT3B was not examined in this study although PcG bodies are known to co-localize with pericentromeric satellite DNA (47), an established DNMT3B target region. Understanding the full spectrum of connections between DNMT3B, H2A ubiquitination, and PRC1 will likely require the use of genome-wide approaches as part of future studies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R01CA114229 (KDR). We thank Drs. Hengbin Wang (University of Alabama, Birmingham) and Haruhiko Koseki (RIKEN Yokohama Institute, Japan) for providing antibodies.
REFERENCES
- 1.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Leonhardt H, Page AW, Weier H, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 3.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 4.Xu G-L, Bestor TH, Bourc'his D, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 5.Hansen RS, Wijmenga C, Luo P, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chedin F, Lieber MR, Hsieh C-L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ooi SKT, Qiu C, Bernstein E, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 9.Linhart HG, Lin H, Yamada Y, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 11.Lehnertz B, Ueda Y, Derijck AAHA, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 12.Vire E, Brenner C, Delpus R, et al. The polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 13.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature Rev Canc. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein BE, Mikkelesen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs JJL, Kieboom K, Marino S, De Pinho RA, van Lohuizen M. The oncogene and polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 17.Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nature Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 18.Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nature Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 19.McGarvey KM, Van Neste L, Cope L, et al. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–5759. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gal-Yam EN, Egger G, Iniguez L, et al. Frequent switching of polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci USA. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin B, Tao Q, Peng J, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet. 2008;17:690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- 22.Kim T-Y, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–7501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- 23.Qiu J, Ai L, Ramachandran C, et al. Invasion suppressor CST6 (cystatin E/M): High-level cell type-specific expression in normal brain and epigenetic silencing in gliomas. Lab Invest. 2008;88:910–925. doi: 10.1038/labinvest.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nature Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 26.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data -mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Endoh M, Endo TA, Endoh T, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 28.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 31.van der Stoop P, Boutsma EA, Hulsman D, et al. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLos ONE. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Squazzo SL, O' Geen H, Komashko VM, et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Gen Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roh Y-Y, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leeb M, Wutz A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol. 2007;178:219–229. doi: 10.1083/jcb.200612127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stock JK, Giadrossi S, Casanova M, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nature Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 36.Margot JB, Ehrenhofer-Murray AE, Leonhardt H. Interactions within the mammalian DNA methyltransferase family. BMC Mol Bio. 2003;4:7. doi: 10.1186/1471-2199-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Firulli AB. A HANDful of questions: the molecular biology of the heart and neural crest derivatives (HAND)-subclass of basic helix-loop-helix transcription factors. Gene. 2003;312:27–40. doi: 10.1016/s0378-1119(03)00669-3. [DOI] [PubMed] [Google Scholar]
- 38.Yarosh W, Barrientos T, Esmailpour T, et al. TBX3 is overexpressed in breast cancer and represses p14ARF by interacting with histone deacetylases. Cancer Res. 2008;68:693–699. doi: 10.1158/0008-5472.CAN-07-5012. [DOI] [PubMed] [Google Scholar]
- 39.Kosinski C, Li VSW, Chan ASY, et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci USA. 2007;104:15418–15423. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estecio MRH, Yan PS, Ibrahim AEK, et al. High-throughput methylation profiling by MCA coupled to CpG island microarray. Gen Res. 2007;17:1529–1536. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rauch T, Wang Z, Zhang X, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci USA. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xi S, Zhu H, Xu H, Schmidtmann A, Geiman TM, Muegge K. Lsh controls Hox gene silencing during development. Proc Natl Acad Sci USA. 2007;104:14366–14371. doi: 10.1073/pnas.0703669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatin chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 44.McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persists after depletion of EZH2. Cancer Res. 2007;67:5097–5102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- 45.Kondo Y, Shen L, Cheng AS, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nature Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 46.Hernandez-Munoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol Cell Biol. 2005;25:11047–11058. doi: 10.1128/MCB.25.24.11047-11058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saurin AJ, Shiels C, Williamson J, et al. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.