Abstract
We present a new version of the 3D TROSY HNCO pulse scheme, referred to as HR-TROSY HNCO, with comparable resolution in the 15N dimension to a 2D 1H-15N HSQC experiment. In the conventional 3D TROSY HNCO, the constant time period (1/2JNC′ ∼ 32 ms) severely limits the maximum resolution in the 15N dimension. In the HR-TROSY HNCO experiment presented here, both constant time periods (∼ 32 ms each) for coherence forward and backward transfer between 15N and 13C′ are utilized to double the 15N evolution time. This leads to a dramatic enhancement in peak separation along the 15N dimension, making the HR-TROSY HNCO an ideal pulse scheme for accurate paramagnetic relaxation enhancement and residual dipolar coupling measurements.
Keywords: TROSY HNCO, high resolution, RDC, PRE
Introduction
In recent years, several NMR methods, which complement traditional NOE-based approaches, have been developed to facilitate three-dimensional structure determination, to characterize molecular motions, and to visualize lowly-populated transient species. In particular, residual dipolar couplings (RDC) yield bond vector orientations relative to an external alignment tensor that are particularly useful for structure-determination of multidomain proteins, protein complexes and nucleic acids [1; 2; 3; 4]. In addition, accurate RDC data can potentially be used to study protein dynamics [5; 6; 7; 8; 9; 10; 11; 12; 13]. Paramagnetic relaxation enhancement (PRE) provides long-range distance information (up to ∼35 Å in suitable cases) that is particularly useful in the study of macromolecular complexes [14; 15; 16] and has been recently exploited to characterize highly transient, lowly populated species [17; 18; 19; 20; 21; 22; 23; 24], as well as unfolded and disordered states of proteins [25; 26; 27].
In cases where resolution permits, conventional backbone amide RDCs and PREs can be measured using 2D 1H-15N HSQC- or TROSY- [28] based experiments [29; 20], allowing the rapid measurement of a large number of RDCs and PREs. However, for larger proteins, or moderately-sized proteins with high α helical or unstructured coil content, cross-peak overlap greatly decreases the accuracy of the measured peak splittings and intensities.
Yang et al. extended the 2D 1H-15N correlation experiment to a 3D TROSY-based HNCO to separate overlapped peaks along an additional 13C′ dimension for the measurement of 1DNH RDCs [30]. In principle, a similar HNCO scheme can also be implemented for PRE Γ2 measurements by simply replacing the first INEPT element by a PRE measuring block [20]. However, the conventional TROSY-based HNCO [31] suffers from extremely low resolution along the 15N dimension because the maximum 15N evolution time is limited by the constant time period (1/2JNC′ ∼ 32ms), for coherence back transfer from 13C′ to 15N, and resolution is inversely proportional to this evolution time.
As a result, RDCs and PREs measured along the 15N dimension in a conventional TROSY HNCO would result in lower precision compared to the 2D 1H-15N HSQC counterpart.
Here, we present a high-resolution version of the TROSY HNCO, which we refer to as HR-TROSY HNCO, in which both constant time periods (∼ 32 ms each) for coherence forward and backward transfer between 15N and 13C′ are utilized, thereby permitting a maximum 15N evolution time of 64 ms. to maximize the 15N evolution time. This experiment separates cross-peaks along the 13C′ dimension while maintaining high resolution along the 15N dimension. An HNCO pulse scheme with improved resolution along the 15N dimension was previously proposed by Madsen & Sorensen [32]. In the latter implementation only the 15N to 13C′ (forward) INEPT transfer period (∼32 ms) is utilized for 15N chemical shift evolution, and improved resolution along the 15N dimension is realized by varying the evolution period from -32 ms to 32 ms by shifting the 180° pulses from one end of the constant time period to the other. This concept can also be easily incorporated into our HR-TROSY HNCO pulse sequence which would further extend the evolution time from -64 ms to 64 ms. We demonstrate the utility of the 3D HR-TROSY-HNCO experiment on a 1:1 complex of U-[15N, 13C, 2H]-labeled mouse KIX with phosphorylated KID (pKID) [33].
Experimental
Materials
A 29 residue peptide (pKID) comprising the kinase-inducible domain of the cAMP response-element binding protein (CREB, residues 119-146) was synthesized by solid state methods with Ser133 phosphorylated, the N-terminus acetylated, and the C-terminus amidated (Biopeptide Co., San Diego). U-[15N, 13C, 2H]-labeled mouse KIX domain (residues 586-672 of the CREB binding protein) was expressed and purified essentially as described previously [33].
NMR experiments
All NMR spectra were recorded at 27 °C on a Bruker 600 MHz spectrometer equipped with a z-gradient triple resonance cryoprobe. A 2D 1H-15N HSQC spectrum (non-constant time) was recorded as reference with 128(t1) × 512(t2) complex points along the 15N and 1H dimensions corresponding to acquisition times of 64 ms and 63.9 ms, respectively. The data matrix for the HR-TROSY HNCO comprises 100(t1) × 10(t2) × 512(t3) complex points along the 15N, 13C′ and 1H dimensions corresponding to acquisition times of 63.24 ms, 8.28 ms and 77.4 ms, respectively. The HR-TROSY HNCO spectrum was recorded with 16 scans per increment and an interscan delay of 1.1 s, resulting in ∼18 hours of total measurement time. For comparison, a conventional TROSY HNCO was recorded with 50(t1) × 10(t2) × 512(t3) complex points along the 15N, 13C′ and 1H dimensions corresponding to acquisition times of 31.62 ms, 8.28 ms and 77.4 ms, respectively, using 32 scans per increment, resulting in the same total measurement time as the HR-TROSY HNCO. For both the HR-TROSY HNCO and conventional TROSY experiments the 15N, 13C′ and 1H carrier frequencies were placed at 117, 176 and 4.75 ppm, respectively, and the sweep widths in the corresponding dimensions were 26, 8 and 11.02 ppm, respectively. (The same carrier frequencies and sweep widths for 1H and 15N were also used for the 1H-15N HSQC). The conventional TROSY and HR-TROSY HNCO spectra were processed identically using linear prediction and zero-filling along the 15N dimension, giving final spectra with 256 and 512 frequency data points, respectively. Replacing the first INEPT element (dashed block in Fig. 1A) with the PRE measuring block (Fig. 1B) adapts the HR-TROSY HNCO pulse sequence to one suitable for PRE Γ2 measurements. By setting the transverse relaxation delay T1 to be 0 and 14 ms and acquiring the data for both delays in an interleaved manner [20], the total experimental time is ∼39 hours. All data sets were processed using the NMRPipe package [34].
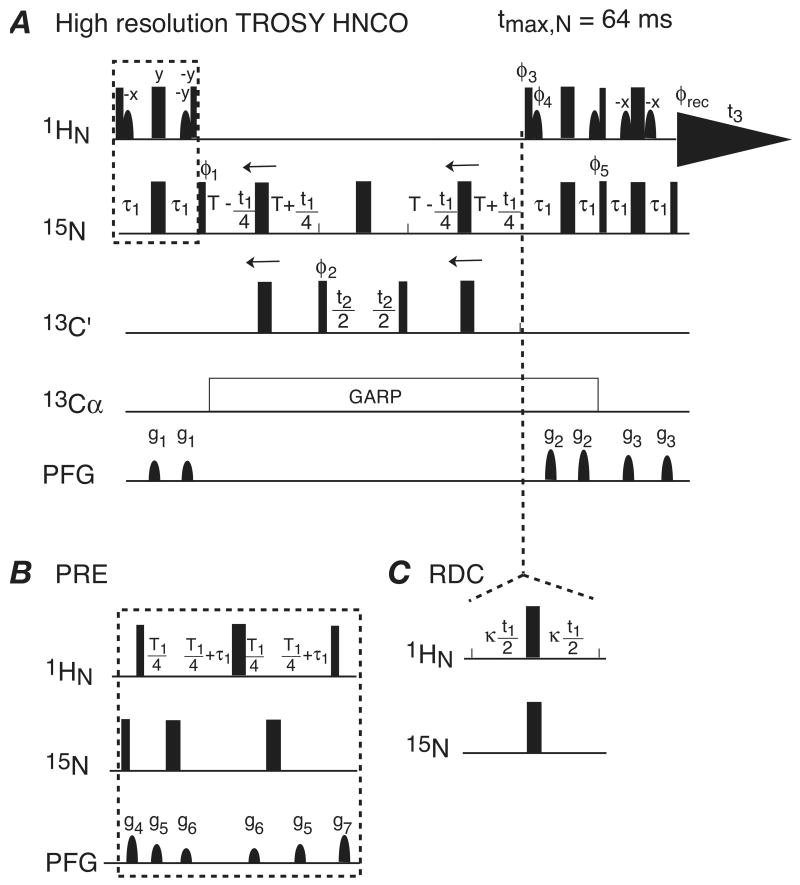
Figure 1.
(A) Pulse sequence of the 3D HR-TROSY HNCO with high resolution along the 15N dimension. The radio-frequency pulses on 1H, 15N, 13C′ and 13Cα are applied at 4.75, 117, 176 and 56 ppm, respectively. Narrow and wide black bars indicate nonselective 90° and 180° pulses, respectively. Water suppression is achieved using the Watergate pulse train. 13Cα decoupling is carried out using GARP with a field strength of γB2 = 0.625 kHz. Sine bell shape 1H pulses are water selective 90° pulses. The duration and strength of the pulsed field gradients applied along the z-axis are as follows: g1: 0.7 ms, 25 G/cm; g2: 1.0 ms, 60 G/cm; g3: 0.7 ms, 50 G/cm; g4: 0.45 ms, 50 G/cm; g5: 0.3 ms, 21 G/cm; g6: 0.3 ms, 19 G/cm. The delays are T = 16 ms, τ1 = 2.72 ms. The phase cycling is as follows: φ1 = y, -y, x, -x; φ2 4x, 4(-x); φ3 = -y; φ4 = y; φ5 = -y; φrec = y, -y, -x, x, -y, y, x, -x. All other radio-frequency pulses are applied with phase x except indicated. A phase-sensitive spectrum in the 15N (t1) dimension is obtained by recording a second FID for each t1 value, with φ1 = y, -y, -x, x; φ3 = y; φ4 = -y; and φ5 = y. Quadrature detection in the 13C′ (t2) dimension is achieved using States-TPPI applied to the phase φ2. The resolution can be further improved [32] by simultaneously shifting the 15N and 13C′ 180° pulses from one end of the constant time period 2T to the other for the coherence forward and backward transfer between 15N and 13C′; that is by simply replacing the two T - t1/4 periods by 2T - t1/4 periods, and the two T + t1/4 periods by t1/4 periods, thereby extending the 15N evolution period from -64 ms to 64 ms. (B) Modification of the 3D HR-TROSY HNCO pulse sequence for measurement of PRE Γ2 rates [20]. The INEPT element (dashed block in panel A) is replaced by the PRE measuring block (panel B). A two-time point measurement with different values of the relaxation delay T1 is carried out in an interleaved mode. (C) Modification of the 3D HR-TROSY HNCO pulse sequence for measurement of 1HN-15N dipolar couplings along the 15N dimension. In the first instance (not shown), the anti-TROSY component of 15N is simply selected by swapping the phases of φ3 and φ4 in panel (A): i.e φ3= y; φ4 = -y. In the second instance, a (J+D) coupling scaling element κt1 is inserted without changing anything else [35] as indicated by the dashed lines. Usually the value of the scaling factor κ can be set to 1.
Results and Discussion
Fig. 1A provides a schematic of the 3D HR-TROSY HNCO pulse scheme. After the first INEPT, magnetization on 15N is transferred to 13C′ and MQ coherence between 15N and 13C′ is generated by the 90° 13C′ pulse (φ2). By synchronously shifting the 180° 15N and 13C′ pulses (as indicated by the arrows), the chemical shift of 15N is encoded during both constant time periods for the coherence forward and backward transfer between 15N and 13C′, thereby extending the maximum 15N evolution time to as long as 1/JNC′ ∼ 64 ms. This allows the resolution along the 15N dimension of the 3D HR-TROSY experiment to be comparable to that of the 2D 1H-15N HSQC counterpart. A similar approach has been previously reported for the 3D HNCA TROSY used for sequence specific backbone assignments [31], in which the maximum 15N evolution time was 1/(1JNCα +2JNCα(i-1)) ∼ 44 ms. The 3D HR-TROSY HNCO offers even higher resolution along the 15N dimension, which results in better separation of amide cross-peaks and yields clearer 2D planes and strips.
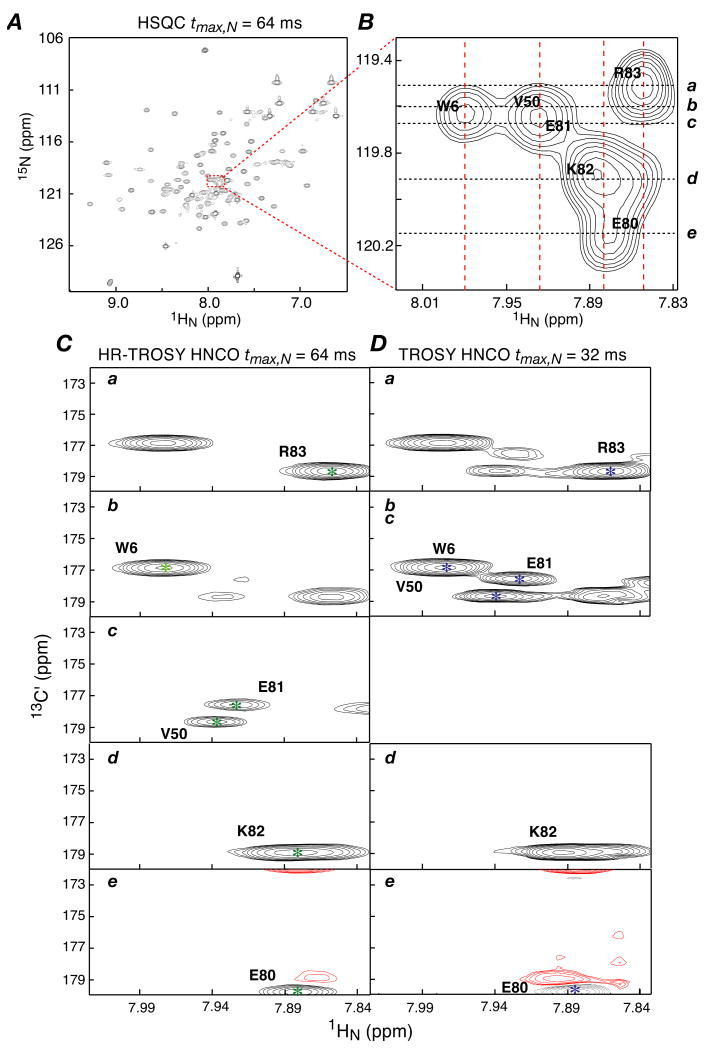
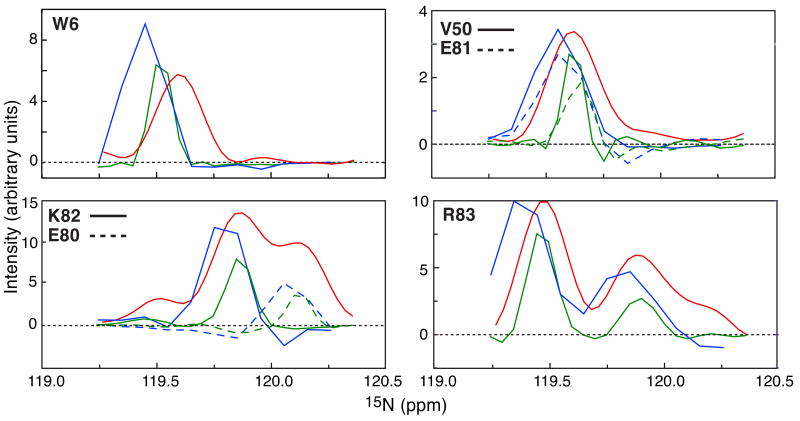
Fig. 2A shows the overall 2D 1H-15N HSQC spectrum for the KIX/pKID complex, and an expansion of the most crowded region is provided in Fig. 2. 1HN-13C′ planes of the HR-TROSY HNCO and conventional TROSY HNCO spectra are displayed in Figs. 2C and D, respectively, taken at the 15N chemical shifts corresponding to the dashed lines a to e in Fig. 2B. A comparison of Figs. 2C and D clearly demonstrates that the HR-TROSY HNCO spectrum is cleaner and better resolved than the conventional TROSY-HNCO, which exhibits significant peak leakage between different 1HN-13C′ planes due to the lower 15N resolution. Indeed, the 1HN-13C′ planes taken from the conventional TROSY-HNCO at the 15N chemical shifts b and c are completely overlappped (identical). Fig. 3 provides a comparison of 1D traces along the 15N dimension for the reference 2D 1H-15N HSQC (taken at the 1H chemical shifts indicated by the red dashed lines in Fig. 2), the 3D HR-TROSY HNCO (taken at the cross-peak positions indicated by the green stars in Fig. 2C) and the conventional 3D TROSY HNCO (taken at the cross-peak positions indicated by the blue stars in Fig. 2D). These traces correspond to peaks for W6, V50, E80, E81, K82 and R83 (all peaks in the green traces have narrower linewidths than those in the blue traces). The increased resolution along the 15N dimension makes the HR-TROSY HNCO particularly well suited for RDC and PRE measurements because peak positions and intensities can be measured more accurately in the 3D HR-TROSY HNCO than in the conventional 3D TROSY HNCO or 2D 1H-15N HSQC experiments.
Figure 2.
Demonstration of increased 15N resolution in the HR-TROSY HNCO spectrum. (A) 600MHz 2D 1H-15N HSQC spectrum of 0.25 mM U-[15N,13C,2H]-labeled mouse KIX complexed with pKID (1:1 stoichiometry) at 27 °C. (B) Expansion of a region with a high degree of spectral overlap to illustrate the high 15N resolution afforded by the 3D HR-TROSY HNCO. Peaks assignments are annotated with the residue numbering for the KIX domain of CBP. (C) 1HN-13C′ slices taken from the 3D HR-TROSY HNCO spectrum at the 15N chemical shifts indicated by the dashed lines a to e in (B), showing a very clean spectrum with well-resolved cross-peaks. (D) The corresponding 1HN-13C′ slices taken from the conventional 3D TROSY HNCO spectrum are shown for comparison. In the conventional TROSY HNCO 1HN-13C′ slices taken at the 15N chemical shifts of dashed lines b to c are identical owing to complete overlap due to the low resolution along the 15N dimension. In (C) and (D), peaks with red contours have negative signs arising from folding in the 13C′ dimension. The red dashed lines in (B) and the green and blue stars in (C) and (D), respectively, indicate the positions where the 1D slices along the 15N dimension shown in Fig. 3 were taken from the reference 2D 1H-15N HSQC, the 3D HR-TROSY HNCO and the conventional 3D TROSY HNCO, respectively.
Figure 3.
1D slices along the 15N dimension from the 2D 1H-15N HSQC (red), the 3D HR-TROSY HNCO (green) and the conventional 3D TROSY HNCO (blue) for residues W6, V50, E80, E81, K82 and R83 of KIX in the U-[15N,13C,2H]-KIX/pKID complex. Directly overlapping 1D 15N peak profiles for these residues clearly illustrate the high resolution along the 15N dimension afforded by the HR-TROSY HNCO (all peaks in green have narrower peak widths). The peak heights and noise levels from the different spectra are not normalized.
During the constant time periods of the HR-TROSY HNCO, no decoupling or 180° pulse is applied on 1H to maintain the 15N coherence under the TROSY state. To minimize intensity attenuation on MQ coherence due to passive J couplings between 1H and 15N (e.g., 2JHαN and 3JHα(i-1)N) during the long 15N evolution period (64 ms) and between 1H and 13C′ (e.g., 2JHαC′ and 3JHβC′) during the 13C′ evolution period (t2), we recommend perdeuterated samples for this type of experiment even for proteins of moderate molecular size. This is especially important when measuring peak intensities. Clear recognition of resolved cross-peaks with nearly undistorted peak shape is critical for measuring both peak position and intensity, which are important for accurate measurement of 1DNH RDCs and 1HN PRE relaxation rates, respectively.
Indeed, this basic 3D HR-TROSY HNCO pulse scheme is easily adapted to pulse sequences for the measurement of 1HN-15N PRE Γ2 rates [20] (Fig. 1B) and 1DNH RDCs (Fig. 1C). For PRE measurement, the INEPT element (delineated by the dashed block in Fig. 1A) is replaced by the PRE measuring block (Fig. 1B). For RDC measurements, two alternative schemes are available. For proteins of moderate molecular size, where the anti-TROSY component of 15N is not extremely broad and the peak shape remains undistorted, the anti-TROSY component can be directly selected by simply swapping the phases of φ3 and φ4 in the pulse scheme shown in Fig. 1A; selection of the TROSY and anti-TROSY components of 15N can be run in an interleaved mode to obtain a pair of peaks, from which the splitting J or (J+D) is measured. When the relaxation of the anti-TROSY component of 15N is too fast or the peak shape is severely distorted, a (J+D) coupling scaling element κt1 is inserted [35] as indicated by the dashed lines in Fig. 1, and no other changes are necessary. In this case, the relevant coherence is under the TROSY state during the two constant time periods (total as long as 64 ms), which better optimizes the relaxation properties of the pulse scheme. Insertion of the κt1 element allows the relevant coherence to evolve under the Hamiltonian (J+D). The measured splitting between the peak of this scheme and that of the original HR-TROSY-HNCO along the 15N dimension is (J+D) scaled by a factor of κ/2. In most cases, the optimal value of the scaling factor κ is 1. Lerche et al. [36] previously suggested a scheme for measuring 1DNH RDCs along the 1HN dimension in which the anti-TROSY and TROSY components of 1HN or the neutral (decoupled or refocused) 1HN chemical shift are chosen for the measurement of the couplings (J or J+D). To avoid potential linewidth broadening along the 1HN dimension due to passive 1H-1H RDCs, we made use of an ST2-PT element [37] in our pulse sequence to select the TROSY or anti-TROSY component for measuring 1DNH RDCs along the 15N dimension while detecting 1HN under the TROSY state.
Acknowledgments
This work was supported by the intramural program of NIDDK, National Institutes of Health, and the Intramural AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health (to G.M.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium Science. 1997;278:1111–4. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 2.Clore GM. Accurate and rapid docking of protein-protein complexes on the basis of intermolecular nuclear overhauser enhancement data and dipolar couplings by rigid body minimization. Proc Natl Acad Sci U S A. 2000;97:9021–5. doi: 10.1073/pnas.97.16.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prestegard JH, al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q Rev Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 4.Bax A, Kontaxis G, Tjandra N. Dipolar couplings in macromolecular structure determination. Methods Enzymol. 2001;339:127–74. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 5.Tolman JR. Dipolar couplings as a probe of molecular dynamics and structure in solution. Curr Opin Struct Biol. 2001;11:532–9. doi: 10.1016/s0959-440x(00)00245-1. [DOI] [PubMed] [Google Scholar]
- 6.Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH. Structural and dynamic analysis of residual dipolar coupling data for proteins. J Am Chem Soc. 2001;123:1416–24. doi: 10.1021/ja002500y. [DOI] [PubMed] [Google Scholar]
- 7.Briggman KB, Tolman JR. De novo determination of bond orientations and order parameters from residual dipolar couplings with high accuracy. J Am Chem Soc. 2003;125:10164–5. doi: 10.1021/ja035904+. [DOI] [PubMed] [Google Scholar]
- 8.Clore GM, Schwieters CD. Amplitudes of protein backbone dynamics and correlated motions in a small α/β protein: correspondence of dipolar coupling and heteronuclear relaxation measurements. Biochemistry. 2004;43:10678–91. doi: 10.1021/bi049357w. [DOI] [PubMed] [Google Scholar]
- 9.Clore GM, Schwieters CD. How much backbone motion in ubiquitin is required to account for dipolar coupling data measured in multiple alignment media as assessed by independent cross-validation? J Am Chem Soc. 2004;126:2923–38. doi: 10.1021/ja0386804. [DOI] [PubMed] [Google Scholar]
- 10.Bouvignies G, Bernado P, Meier S, Cho K, Grzesiek S, Bruschweiler R, Blackledge M. Identification of slow correlated motions in proteins using residual dipolar and hydrogen-bond scalar couplings. Proc Natl Acad Sci U S A. 2005;102:13885–90. doi: 10.1073/pnas.0505129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clore GM, Schwieters CD. Concordance of residual dipolar couplings, backbone order parameters and crystallographic B-factors for a small alpha/beta protein: a unified picture of high probability, fast atomic motions in proteins. J Mol Biol. 2006;355:879–86. doi: 10.1016/j.jmb.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 12.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–5. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 13.Vogeli B, Yao LS, Bax A. Protein backbone motions viewed by intraresidue and sequential H-N-H-alpha residual dipolar couplings. J Biomol NMR. 2008;41:17–28. doi: 10.1007/s10858-008-9237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–65. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 15.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–50. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- 16.Iwahara J, Schwieters CD, Clore GM. Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J Am Chem Soc. 2004;126:5879–96. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 17.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 18.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 19.Volkov AN, Worrall JA, Holtzmann E, Ubbink M. Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc Natl Acad Sci U S A. 2006;103:18945–50. doi: 10.1073/pnas.0603551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwahara J, Tang C, Clore GM. Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449:1078–82. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 22.Clore GM. Visualizing lowly-populated regions of the free energy landscape of macromolecular complexes by paramagnetic relaxation enhancement. Mol Biosyst. 2008;4:1058–69. doi: 10.1039/b810232e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang C, Louis JM, Aniana A, Suh JY, Clore GM. Visualizing transient events in amino-terminal autoprocessing of HIV-1 protease. Nature. 2008;455:693–U92. doi: 10.1038/nature07342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X, Reinle W, Hannemann F, Konarev PV, Svergun DI, Bernhardt R, Ubbink M. Dynamics in a pure encounter complex of two proteins studied by solution scattering and paramagnetic NMR spectroscopy. J Am Chem Soc. 2008;130:6395–403. doi: 10.1021/ja7101357. [DOI] [PubMed] [Google Scholar]
- 25.Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured α-synuclein. Proc Natl Acad Sci U S A. 2005;102:1430–5. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dedmon MM, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson CM. Mapping long-range interactions in α-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J Am Chem Soc. 2005;127:476–7. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 27.Felitsky DJ, Lietzow MA, Dyson HJ, Wright PE. Modeling transient collapsed states of an unfolded protein to provide insights into early folding events. Proc Natl Acad Sci U S A. 2008;105:6278–83. doi: 10.1073/pnas.0710641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–71. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ottiger M, Delaglio F, Bax A. Measurement of J and Dipolar Couplings from Simplified Two-Dimensional NMR Spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 30.Yang DW, Venters RA, Mueller GA, Choy WY, Kay LE. TROSY-based HNCO pulse sequences for the measurement of 1HN-15N,15N-13CO, 1HN-13CO, 13CO-13Cα and 1HN-13Cα dipolar couplings in 15N, 13C, 2H-labeled proteins. J Biomol NMR. 1999;14:333–343. [Google Scholar]
- 31.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U S A. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madsen JC, Sørensen OW. Multidimensional NMR experiments with improved resolution. J Magn Reson. 1992;100:431–436. [Google Scholar]
- 33.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based On Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Rexroth A, Schmidt P, Szalma S, Geppert T, Schwalbe H, Griesinger C. New Principle For the Determination of Coupling-Constants That Largely Suppresses Differential Relaxation Effects. J Am Chem Soc. 1995;117:10389–10390. [Google Scholar]
- 36.Lerche MH, Meissner A, Poulsen FM, Sørensen OW. Pulse sequences for measurement of one-bond 15N-1H coupling constants in the protein backbone. J Magn Reson. 1999;140:259–263. doi: 10.1006/jmre.1999.1820. [DOI] [PubMed] [Google Scholar]
- 37.Peruvshin KV, Wider G, Wüthrich K. Single transition-to-single transition polarization transfer (ST2-PT) in [15N,1H]-TROSY. J Biomol NMR. 1998;12:345–348. doi: 10.1023/A:1008268930690. [DOI] [PubMed] [Google Scholar]