Abstract
The base excision repair (BER) pathway is required to repair endogenous and exogenous oxidative DNA damage. Multiple DNA repair pathways have been shown to be down-regulated in the tumor microenvironment, whereas APE1/Ref1, a central protein in BER, is overexpressed in many types of solid tumors. APE1/Ref1 has dual functions, participating both in BER and in redox regulation of oxidized transcription factors. Here, we show that inhibition of the BER pathway in an acidic tumor microenvironment increases oxidative DNA damage temporally related to increased intracellular reactive oxygen species. Unrepaired oxidative DNA damage results in cell cycle arrests and increased DNA double strand breaks, leading to cell death. Therefore, up-regulation of BER in solid cancers may represent an adaptive survival response. Consequently, BER inhibition may confer tumor microenvironment targeted cytotoxicity in human cancers. Our data suggest that BER inhibition is a rational basis for cancer therapy with or without other cytotoxic therapy. Additionally, our results offer insight as to why APE1/Ref1 retains it’s unique dual functionality, both of which counteract environmental oxidative stress.
Keywords: base excision repair, APE1/Ref1, XRCC1, tumor microenvironment, DNA damage
INTRODUCTION
Oxidative stress generates various types of DNA damage including base damage, apurinic/apyrimidinic (AP) sites, single strand breaks (SSBs), and double strand breaks (DSBs). In addition to exogenous stress such as ionizing radiation (IR), endogenous oxidative stress is thought to be a significant threat to cells with aerobic respiration. The base excision repair (BER) system is essential to repair some forms of oxidative DNA damage (1). Spontaneous DNA damage induced by endogenous oxidative stress may be enough to reduce cell proliferation and induce apoptosis if BER is strongly inhibited (2).
APE1/Ref1 is a bi-functional protein serving as a key enzyme in the BER pathway (apurinic/apyrimidinic endonuclease 1 [APE1]) as well as in redox regulation (redox factor 1 [Ref1]). Ref1 was independently identified and shown to reductively activate several key transcription factors such as c-Fos, c-Jun, and NF-κB (3). As such, APE1/Ref1 has a unique and critical role in both genomic protection and cellular maintenance against an environmental oxidative stress. Transfection of another BER-related gene, XRCC1, into the Chinese hamster ovary (CHO) mutant EM9 cell line was found to correct defective DNA strand break repair and reduce cytotoxicity to a variety of DNA damages (4). XRCC1 coordinates multiple steps in the BER pathway through protein-protein interactions with DNA ligase III (LIGIII) (5), poly(ADP-ribose) polymerase 1 (PARP-1) (6, 7), DNA polymerase β (Polβ) (6, 8), and polynucleotide kinase (PNK) (9). XRCC1 also interacts with APE1 and is required for efficient APE1 function (10).
Solid tumors are often found to have an altered extracellular condition called the tumor microenvironment. The tumor microenvironment may be characterized by acute/chronic hypoxia, low extracellular pH (pHe), and low nutrition. The tumor microenvironment has been shown to impact on local disease progression, metastatic potential, and response to radiation and chemotherapy (11). The tumor microenvironment is also associated with genetic instability (12, 13). Multiple DNA repair systems are inhibited under hypoxic and/or low pH extracellular conditions including mismatch repair (14, 15), nucleotide excision repair (16), and homologous recombination (17, 18). In contrast to these DNA repair systems, APE1 was found to have greater activity in various types of tumors compared to normal tissues (19–21). Additionally, certain polymorphisms of XRCC1, particularly single amino acid changes at codons 194, 280 and 399 are associated with a risk of several types of gastrointestinal cancers, breast cancer and lung cancer (22). The indispensable role of BER in mediating oxidative stress and the observation of overexpression of BER in various types of cancer raises the question of whether BER can counteract increased genotoxic stress in an acidic tumor microenvironment compared to a normal pH extracellular environment. Therefore, we hypothesize that the BER plays an essential role in mediating cytotoxicity in an acidic tumor microenvironment.
MATERIALS AND METHODS
Cell lines and chemicals
The cell lines used were the isogenic Chinese hamster ovary cell lines, CHO AA8 (parental wild type), CHO EM9 (XRCC1 deficient), and CHO H9T3 (XRCC1 cDNA-complemented in EM9 cells), and the human colon cancer cell lines, HCT116 and RKO. RKO cells conditionally expressing shRNA for the APE/Ref1 sequence and for a control sequence were kind gifts from Dr. Y-C Cheng of Yale University (23). Expression of shRNA was induced by incubation with 1µg/ml doxycycline. The cells were maintained in DMEM, supplemented with 10% fetal bovine serum, L-glutamine, and penicillin/streptomycin at 37°C in a humidified 10% CO2 atmosphere. The pH of culture media was adjusted by phosphoric acid at ambient conditions before adding supplemental factors to the media. All chemicals were purchased from Sigma (St. Louis, MO), unless otherwise specified. A potent inhibitor of APE1, 7-nitroindole-2-carboxylic acid (CRT0044876) (24), was obtained from Calbiochem (San Diego, CA). CRT0044876 was dissolved in DMSO at a concentration of 200mM and stored at −0°C. For experiments using CRT0044876 treatments, the same dose of DMSO was added to control groups.
Survival assay
Clonogenic cell survival was determined by a standard colony-forming assay as described previously (25). Survival fractions were calculated by a formula:
Multicellular spheroid
Ten-thousand exponentially growing cells were seeded in each well of a 96 U-bottom culture plate pre-coated with 1% agarose in DMEM. The plates were placed on a plate shaker for up to one hour at ambient conditions to facilitate cell aggregation in the center of the wells. The cells were incubated at 37°C in a humidified 10% CO2 atmosphere. The cells started to grow as one spheroid in each well, typically 5 to 10 days later. Following specified treatments, spheroids were fixed in 3.7% paraformaldehyde for immunohistochemical analyses. The spheroids were treated with pimonidazole for 24 hours prior to harvest to label hypoxic regions, according to the manufacturer’s instructions (NPI, Burlington, MA). The fixed spheroids were aggregated in Histogel (Thermo Fisher Scientific, Waltham, MA), and then paraffin-embedded. For a spheroid growth assay, images of spheroids were taken under phase-contrast microscopy. The volume of spheroids was estimated by the equation:
Intracellular ROS measurement
At subconfluent conditions, HCT116 cells were counterstained with Hoechst33342. Then, a ROS responsive fluorochrome, CM-H2DCFDA, was loaded into the cells, according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). The cells were next incubated at 37°C in pH7.4 PBS, pH6.2 PBS, or pH7.4 PBS with 25µM H2O2. After 20 min, the cells were observed by reverse fluorescent microscopy. In a parallel experiment, cells were harvested by trypsinization. CM-H2DCFDA was then loaded into the resultant single cell suspensions. The cells were incubated under similar conditions to the attached cells, and then assayed by flow cytometry.
Western blotting
Western blot analyses were conducted using standard procedures (SDS-PAGE followed by an electrotransfer onto a membrane). The antibodies used were γH2AX and PARP-1 (Calbiochem, San Diego, CA), APE1/Ref1 (Novus Biologicals, Littleton, CO), and β-actin. Relative protein levels of APE1/Ref1 were determined by an optical density measurement using a software ImageJ-1.39u against control bands with 100% and 10% protein loading.
Immunochemical staining
Paraffin-embedded slides were processed using standard procedures with de-paraffinization and rehydration. For tissue culture samples, cells grown on cover glass were fixed and permeabilized using ice-cold methanol. The primary antibodies used were γH2AX (1:500 dilution) (Calbiochem, San Diego, CA) and pimonidazole (1:250 dilution) (NPI, Burlington, MA). Texus-Red conjugated or AlexaFluor488 secondary antibody (Invitrogen, Carlsbad, CA) were used for visualization.
Immunocytochemistry and flow cytometry of spheroids
Following the indicated treatments, cellular debris-like materials adjacent to spheroids were carefully harvested with medium separately from the adjacent spheroids. The debris-like materials were recovered by centrifugation. Separately, a single cell suspension of spheroids was obtained by trypsinization. Following fixation in 70% ethanol at −20°C, the samples were dropped onto a microscope slide and air-dried. The slides were immuno-stained for pimonidazole, as described above. The remaining ethanol-fixed single cell suspension was similarly immuno-stained for pimonidazole and counter-stained with propidium iodide. The samples were then subjected to flow cytometry.
Comet assay
An alkaline comet assay was performed as previously described (25). Briefly, a single cell suspension was embedded in 1% low-melting-point agarose in PBS, and then spread onto a microscope slide. Following a cell lysing process and immersion in alkaline solution, the slide was subjected to electrophoresis using 1V/cm in a neutral buffer for 10 min. Following fixation, DNA was visualized with SYBR-green I (Invitrogen, Carlsbad, CA) and tail moments were measured.
Cell cycle analysis
Following the indicated treatments, cells were harvested by trypsinization and fixed with 70% ethanol at −20°C. Following propidium iodide staining, the cells were subjected to flow cytometry.
RESULTS
To test our hypothesis that up-regulation of BER provides a greater tolerance to cancer cells in an acidic tumor microenvironment, we examined the impact of BER inhibition on cell survival. APE1 and XRCC1 were selected as targets of inhibition since: 1) APE1 is a rate-limiting enzymatic step in BER and is frequently overexpressed in solid tumor tissues; and 2) XRCC1 has a crucial role in efficient BER and is involved in multiple steps of the BER pathway including binding to APE1.
We first compared growth of the XRCC1-proficient and -deficient isogenic pair of CHO cells at pHe7.4 for 5 days. By counting live cells (i.e. with consideration of cell loss factor), no difference was found in cell doubling times between CHO AA8 cells and CHO EM9 cells. The doubling times of CHO AA8 cells and CHO EM9 cells were 20.1 [18.3–22.2] hours and 20.5 [18.6–22.5] hours, respectively [95%CI]. However, incubation in an acidic medium (pHe6.0) showed a differential effect within 3–5 days of incubation based on microscopic evaluation of cell density and morphology in XRCC1-deficient CHO EM9 cells compared to wild type CHO AA8 cells (Fig.1A). Subsequent comparison of clonogenic survival at day 14 using a colony forming assay demonstrated that incubation at pHe6.0 for 5 days significantly reduced survival in CHO EM9 cells compared to CHO AA8 cells (Fig.1B). The reduced survival was partially reversed in CHO H9T3 cells, in which XRCC1 function was complemented by stable transfection of XRCC1 cDNA into CHO EM9 cells.
Fig. 1.
(A) Cell density and morphology at pHe6.0 were examined under phase-contrast microscopy (×100) for up to 5 days. CHO EM9 cells appeared to have reduced density and viability at pHe6.0 compared to CHO AA8 cells. (B) Clonogenic survival of isogenic CHO cells. (C) Western blot analysis showed down-regulation of APE1 using stable transfection of shRNA in RKO cells. (D) Following a 48-hour incubation at pHe7.4 or 6.0, clonogenic survival was measured in APE1-inhibited RKO cells (APE1-shRNA with DOX [grey bar]) compared to control RKO cells (control-shRNA with DOX [black bar] and APE1-shRNA without DOX [white bar]). Significance of difference between the two groups was examined by t-test (*p<0.01, **p<0.001).
We next tested whether APE1 inhibition alters cell survival under acidic conditions. APE1 was down-regulated in RKO cells using shRNA (Fig.1C). The APE1 protein levels were gradually reduced to approximately 10% following a 12-day incubation with 1µg/ml doxycycline. For a clonogenic survival assay, we used RKO cells pre-treated with doxycycline for greater than 12 days and then continued to incubate the cells with doxycycline throughout the assay period. At pHe7.4, there was no difference in cell survival between APE1-down-regulated RKO cells and control RKO cells (shRNA for APE1 without doxycycline or shRNA for control sequence with doxycycline). However, following a 48-hour incubation at pHe6.0, cell survival was significantly reduced in the APE1-down-regulated RKO cells compared to control RKO cells (p<0.01) (Fig.1D).
Similarly, clonogenic survival was measured in HCT116 cells following a 72-hour period of chemical inhibition of APE1 using varying concentrations of CRT0044876 (50–2000µM). HCT116 cells were significantly more sensitive to CRT0044876 at pHe6.2 compared to pHe7.4 (p<0.001) (Fig.2A). The 50% lethal dose (LD50) and 90% lethal dose (LD90) were 189µM and 439µM at pHe6.2, and 1590µM and 2510µM at pHe7.4, respectively. Additionally, we measured the effect on HCT116 cell proliferation in monolayer cultures following up to a 4 day exposure to 600µM CRT0044876 (Fig.2B, upper panel). At pHe7.4, there was no effect on cell proliferation (relative cell number determined once daily × 4 days). However, at both pHe6.8 and 6.2, exposure to the drug significantly reduced the cell numbers compared to control (vehicle alone) cells. To generalize our observation to another human tumor cell line, we conducted a similar experiment using the human glioblastoma cell line U251. We found a qualitatively similar result in U251 cells (Fig.2B, lower panel). Since the APE1 inhibitor CRT0044876 has been shown to enhance cytotoxicity to other oxidative damaging agents (24), we tested whether CRT0044876 enhances cytotoxicity to exogenous oxidative stress using H2O2 in HCT116 cells. We found that there was a positive interaction between 600µM CRT0044876 and a range of H2O2 concentrations in HCT116 cells (Fig.2C), consistent with the hypothesis that CRT0044876 inhibits repair of oxidative DNA damages.
Fig. 2.
(A) Clonogenic survival following treatment with the APE1 inhibitor CRT0044876 in HCT116 cells at pHe7.4 versus 6.2. The cell survival was dependent on pHe (p<0.001) as well as a dose of CRT0044876 (p<0.001), assessed by ANOVA. (B) 600µM CRT0044876 did not affect cell proliferation of HCT116 cells (upper panel) in monolayer cultures at pHe7.4, whereas cell proliferation was significantly inhibited at the lower pHe values (mean±sem). A similar result was found using U251 cells (lower panel). (C) Clonogenic survival assay following a 48-hour exposure to various doses of H2O2 with or without co-incubation of 600µM CRT0044876 (mean±sem).
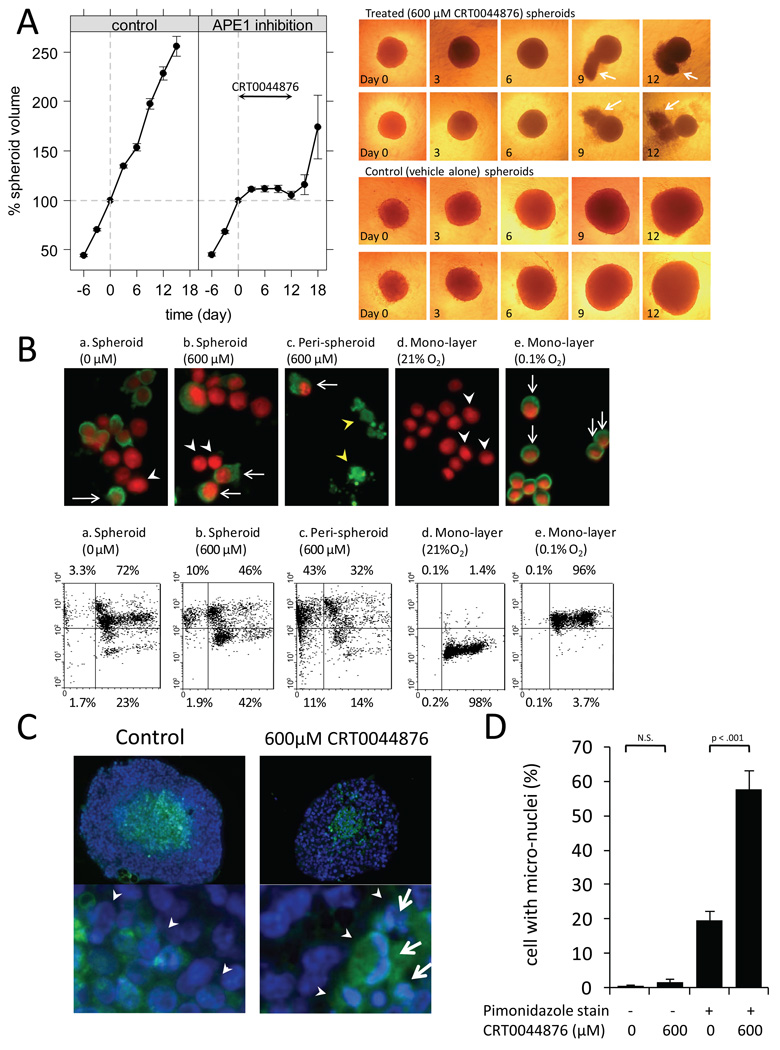
We also examined the impact of APE1 inhibition on the growth of HCT116 tumor spheroids. We chose 600µM CRT0044876 as a dose of the APE1 inhibitor for these experiments, since greater than 90% of cell killing occurred at pHe6.2 compared to less than 10% cell killing at pHe7.4 in monolayer cultures of HCT116 cells (Fig.2B). APE1 inhibition by incubation with 600µM CRT0044876 for up to 12 days resulted in substantial growth retardation of spheroids (Fig.3A). However, following removal of the chemical APE1 inhibitor, the spheroids started to re-grow.
Fig. 3.
(A) Growth of HCT116 multicellular spheroids treated with 0µM or 600µM CRT0044876 at pHe7.4 for 12 days (mean±sem of greater than 100 spheroids in each group from the 5 independent experiments). Representative images of the spheroids by phase-contrast microscopy (×40) are also shown. Note the debris-like material adjacent to the CRT0044876-treated spheroids (white arrows) at days 9 and 12. (B) Following one-week treatment with 0µM or 600µM CRT0044876, spheroids (subpanel a, b) and peri-spheroid stranding (debris-like materials shown in the panel A) (subpanel c) were separately collected. No detectable materials were recovered as a peri-spheroid stranding from the spheroids treated with 0µM CRT0044876. The upper panels are representative images (×400) of immunocytochemical staining for pimonidazole (green). Nuclei were counterstained with propidium iodide (red). (white arrows: pimonidazole-positive cells, white arrow heads: pimonidazole-negative cells, yellow arrow heads: pimonidazole-positive cellular remnants). The lower panels show flow cytometric analysis of the same samples (x-axis: DNA contents, y-axis: a log scale of pimonidazole stain). (C) Immunohistochemical analyses from paraffin-embedded sections of the HCT116 spheroids following a 4-day treatment with CRT0044876 (×200) (green: pimonidazole, blue: Hoechst33342). Arrow heads indicate an interface between hypoxic and normoxic regions. 600µM CRT0044876 resulted in micronuclei (white arrow) in the central hypoxic regions but not in the peripheral normoxic regions. (D) The frequency of micronuclei was quantitated in the 5 pairs of spheroid sections with 0 or 600µM CRT0044876 treatment (Bar: mean±sem).
To determine whether the effect of CRT0044876 treatment (600µM × 12 days) in HCT116 tumor spheroids represents a cytostatic versus a cytotoxic effect, we observed spheroids over the 12 day period by phase-contrast microscopy (Fig.3A). The treated spheroids (upper panel) were less compact with increased peri-spheroid stranding, typically occurring following one week of treatment (arrows at days 9 and 12). In separate experiments, the hypoxic regions of spheroids were labeled using pimonidazole (an exogenous hypoxic marker) for 24 hour prior to CRT0044876 or vehicle alone treatment. Following one week of treatment with CRT0044876 or vehicle alone, spheroids and the adjacent peri-spheroid stranding were separately harvested. The resultant cellular suspensions were analyzed by immunocytochemistry and flow cytometry with pimonidazole staining (Fig.3B). DNA contents were measured with propidium iodide staining. We found that a large portion of cellular materials from the peri-spheroid stranding were positive for pimonidazole staining with either sub-G1 (43%) or normal DNA contents (32%) (subpanel c). Additionally, single cell suspensions derived from spheroids treated with 600µM CRT0044876 (subpanel b) showed a larger proportion of pimonidazole-negative (oxic) cells as well as a larger pimonidazole-positive sub-G1 population than single cell suspensions from control spheroids (subpanel a). HCT116 cells in mono-layer cultures under normoxic (21% O2) or hypoxic (0.1% O2) conditions were used as negative and positive controls, respectively (subpanel d and e). These observations suggest that peri-spheroid stranding is derived from a hypoxic region of spheroids at the time of CRT0044876 treatment. Additionally, using immunohistochemical analysis of tumor spheroid sections following 4 days of treatment, there was enhanced micronuclei formation (by Hoechst33342 staining) corresponding to areas of enhanced pimonidazole staining in CRT0044876 treated spheroids compared to control spheroids (Fig.3C, D).
Collectively, these data suggest a cytotoxic effect to APE1 inhibition by CRT0044876 treatment in HCT116 spheroids, particularly in hypoxic regions. The observed re-growth of spheroids following a removal of CRT0044876 is consistent with the hypothesis that the peripheral (less hypoxic/acidic) area of spheroids is spared from cytotoxicity induced by the APE1 inhibition.
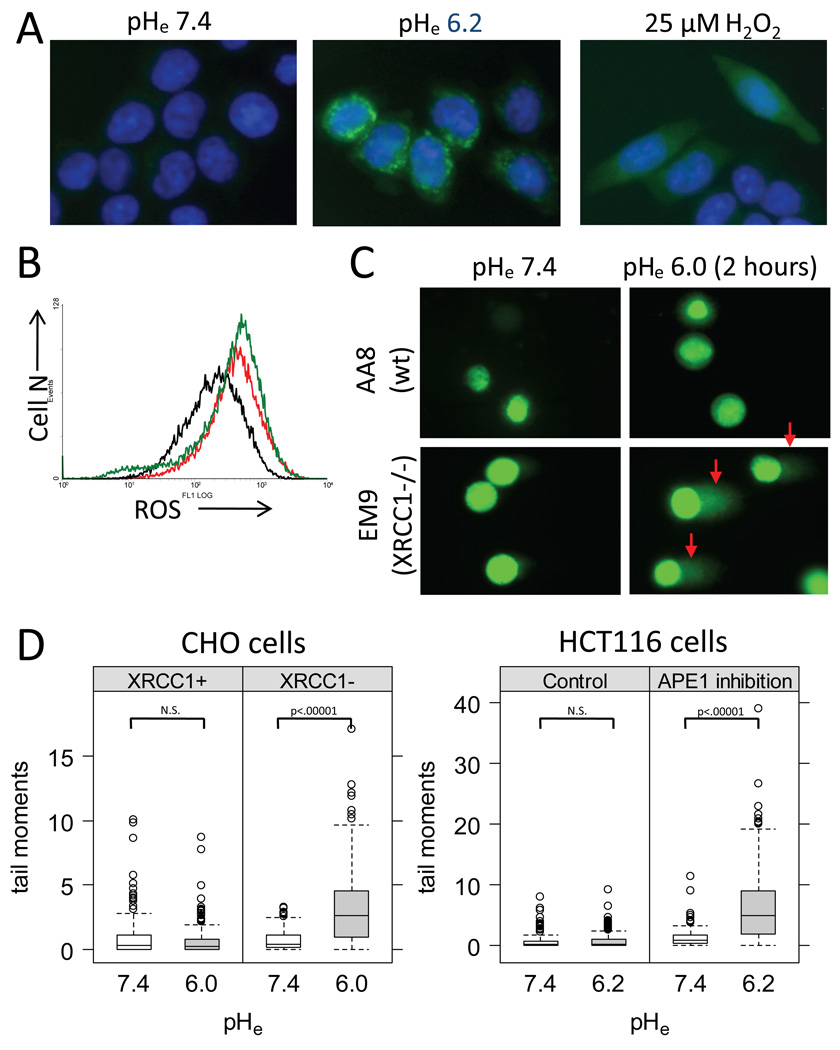
To explore possible mechanisms of enhanced cytotoxicity to BER inhibition in an acidic microenvironment, we examined whether low pHe increases intracellular reactive oxygen species (ROSs). A ROS-responsive dye, CM-H2DCFDA, was used to measure intracellular ROS levels in HCT116 cells grown in monolayer culture. Incubation with 25µM H2O2 was used as a positive control. We observed increased intracellular ROSs levels following a 20-min incubation at pHe6.2, similar to treatment with 25µM H2O2 (Fig.4A, B). However, the distribution of the fluorescent signals representing intracellular ROSs appeared different following an incubation at low pHe (focal distribution) compared to exogenous H2O2 treatment (diffuse distribution), which may implicate the existence of a localized source of intracellular ROS generation upon an exposure to an acidic environment (Fig.4A).
Fig. 4.
(A) Intracellular ROS levels measured in HCT116 cells using CM-H2DCFDA (green) (×400). Nuclei were counterstained using Hoechst33342 (blue). A 20 min incubation at pHe 6.2 increased intracellular ROS levels compared to pHe7.4. (B) CM-H2DCFDA fluorescent intensity was measured by flow cytometry under similar conditions as Fig.3A (Black: control at pHe7.4, Green: pHe6.2, Red: H2O2). (C) DNA damages were measured using an alkaline comet assay in the CHO cell pair following a 2-hour incubation at pHe6.0 or 7.4. (D) The distributions of the comet tail moments from >100 cells in the CHO cells pair (left) and in HCT116 cells with 0µM or 600µM CRT0044876 (right) are graphed. Significance of difference between two groups was tested using Wilcoxon rank-sum test (N.S.; not significant).
To further evaluate the mechanism of cytotoxicity following BER inhibition at low pHe, we evaluated the extent and time-course of DNA damages. Using the alkaline comet assay, we measured the amount of DNA damages following a 2-hour exposure to low pHe (6.0–6.2) in the isogenic CHO cell pair and in HCT116 cells with and without 600µM CRT0044876 treatment (Fig.4C, D). The alkaline comet assay can detect a variety of DNA damages including AP sites, SSBs, and DSBs. Increased DNA damages, as measured by increased comet tail moments, were found in both CHO EM9 cells and in CRT0044876-treated HCT116 cells at low pHe compared to normal pHe (Fig.4D). CHO AA8 cells and control HCT116 cells showed no increased DNA damages following the 2-hour exposure to low pHe.
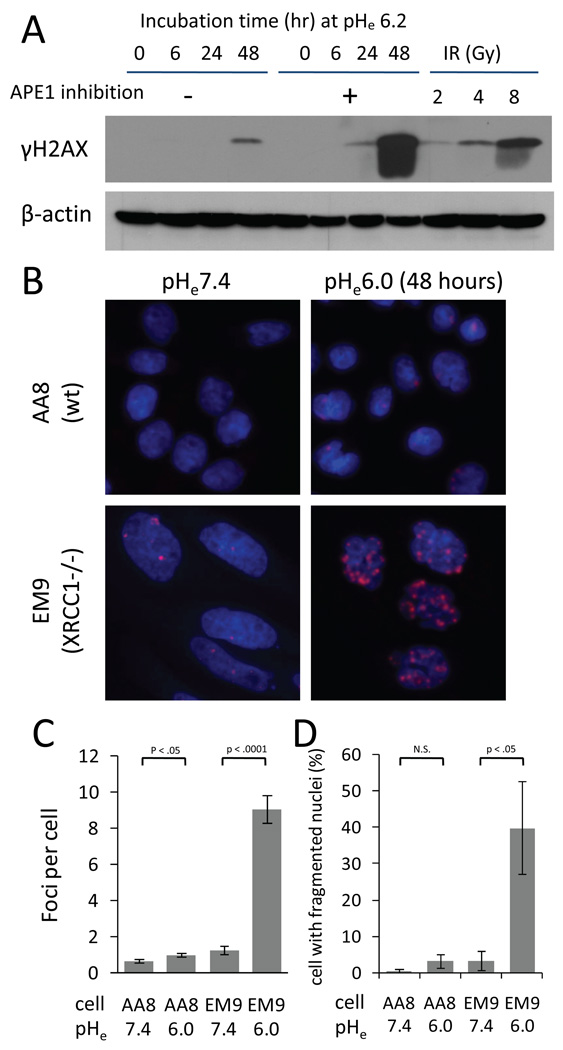
Unlike other types of DNA damage, DSBs are an imminent threat to cells. To access the extent and time-course of DSB formation, we measured cellular γH2AX levels as a marker of DNA DSB (26, 27) in HCT116 cells treated with 0µM or 600µM CRT0044876 while incubated at pHe6.2. A time course experiment revealed that phosphorylation of histone H2AX increased over 24 hours with APE1 inhibition. Following 48 hours of APE1 inhibition, γH2AX levels were greater than that at 30 min following 8Gy IR (Fig.5A). However, we did not detect an increase of γH2AX levels at 48 hours with APE1 inhibition at pHe7.4 (0 hour at pHe6.0; lane5 in Fig.5A). Similarly, we examined γH2AX foci formation using the CHO cell pair incubated at pHe6.0 for 48 hours (Fig.5B, C). Under normal pHe, CHO EM9 cells showed a slightly greater number of γH2AX foci per cell than CHO AA8 cells. Incubation at pHe6.0 significantly increased the number of γH2AX foci in both CHO AA8 and CHO EM9 cells. However, the increase in γH2AX foci number was substantially greater in CHO EM9 cells (mean 7.2 [95%CI 6.4–8.1]) than CHO AA8 cells (0.31 [0.13–0.49]). A comparison of the extent of micro/fragmented nuclei revealed a similar finding (Fig.5D). A significant enhancement in the number of micro/fragmented nuclei was found in CHO EM9 cells following incubation at pHe6.0.
Fig. 5.
(A) The time-course of phosphorylation of histone H2AX by Western blotting in HCT116 cells treated with 0µM or 600µM CRT0044876 at pHe6.2. For comparison, the extent of H2AX phosphorylation at 30 min following ionizing radiation (IR) is also shown. (B) Immunocytochemical analyses for γH2AX foci formation (red) (×640). Nuclei were counterstained with Hoechst33342 (blue). A 48-hour incubation at pHe6.0 induced greater γH2AX foci formation in CHO EM9 cells compared to CHO AA8 cells. (C) Quantitative analysis of γH2AX foci formation in >500 nuclei from 5 independent experiments (mean±sem). (D) The number of cells with micronuclei was counted in the same experiments shown in the panel B and C (mean±sem). Significance of difference between the two groups was tested using t-test.
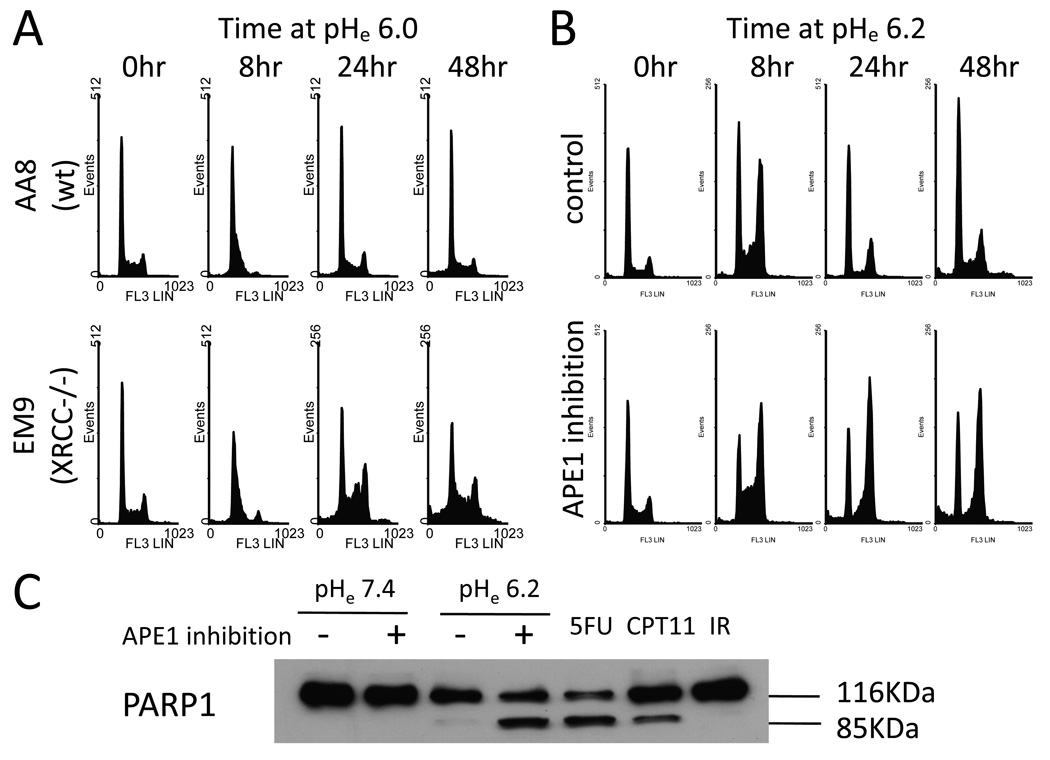
BER pathway inhibition also significantly impacted cell cycle progression under acidic conditions (Fig.6A, B). Both CHO AA8 cells and CHO EM9 cells showed an intra S-phase arrest following a 8-hour incubation at pHe6.0. CHO EM9 cells continued to show prolonged intra S-phase as well as G2/M phase arrests at the later time points (24–48 hours), whereas the intra S-phase arrest appeared to be transient and the cell cycle recovered to the baseline in CHO AA8 cells. A sub-G1 population was apparent at 48 hours and also significantly greater in CHO EM9 cells. Similar effects on the cell cycle progression were found in HCT116 cells following chemical APE1 inhibition (Fig.6B). Consistent with the increased sub-G1 population by flow cytometry, cleavage of PARP-1 was found in HCT116 cells following chemical APE1 inhibition at low pHe (Fig.6C). Similar PARP-1 cleavage was found in HCT116 cells treated for 48 hours with 5-fluorouracil or camptothecin (both known to induce apoptosis in colorectal cancers), but not in HCT116 cells at 48 hours following treatment with 10 Gy ionizing radiation (which induced >99% cell killing primarily through non-apoptotic mechanism, data not shown). These data suggest that an apoptotic pathway is, at least partially, involved in cell death induced by APE1 inhibition at low pHe.
Fig. 6.
(A) Cell cycle progression in an acidic extracellular environment in CHO AA8 cells versus CHO EM9 cells. (B) Using HCT116 cells with chemical APE1 inhibition, a differential cell cycle progression was found similarly to the CHO cell pair. (C) A substantial amount of the cleaved PARP-1 85kDa fragments was detected in HCT116 cells following a 48-hour incubation at pHe6.2 when APE16.2 when APE1 was inhibited. A 48-hour incubation with 150µM 5-fluorouracil or 5µM camptothecin was used as a positive control for apoptotic activity while a 48-hour measurement after 10Gy ionizing radiation was used as a negative control.
DISCUSSION
The extracellular environment is often acidic in solid cancers. Impaired microcirculation and increased acid production can be sources of acidity (28). Metabolically transformed cancer cells also produce a greater amount of acid through aerobic glycolysis regardless of the level of oxygenation. This hypermetabolic phenomenon is often referred to as the Warburg effect and is the mechanistic basis of 18F-fluoro-deoxy-glucose positron emission tomography for cancer imaging.
In our study, we use pHe levels of 6.0 and 6.2 to assess the effects of BER inhibition in our CHO and RKO cell models and in the HCT116 cell model, respectively, based on preliminary growth experiments in these cell lines. This pHe range is comparable to a study of a mouse tumor model using a fluorescent imaging technique (29). However, other studies using micro-electrode pH measurements show a wide range of values, although this technique records a composite of both extracellular pH and intravascular pH. For example, two studies in typically large human tumors showed significant intra-and inter-tumor variations in pH with a range of 5.5–7.8 (30, 31). Using magnetic resonance spectroscopy, a pH range of 5.9–7.1 (mean 6.6–6.9) was found in several different mouse tumors (32). As such, the pHe of 6.0–6.2 used in our study is in a low but clinically relevant pHe range.
The association between ischemia/acidosis and oxidative stress has been more intensively investigated in the field of the nervous and cardiovascular systems (33). Others have demonstrated how extracellular acidity increases intracellular ROS production. In brain tissue, hyperglycemia-induced lactic acidosis is well known to strongly enhance cell death following an ischemic event. Ying et al. revealed that acidosis enhances oxidative neuronal injury by impairing antioxidant enzyme function and increasing intracellular free iron levels (34). In cardiac myocytes, an acidic medium induced by chronic hypoxia or exogenous addition of lactic acid caused apoptosis irrespective of aerobic or anaerobic conditions (35). In their study, chronic hypoxia alone was not a significant factor for inducing apoptosis, but low extracellular pH or reoxygenation resulted in apoptosis in cardiac myocytes. In vascular endothelial cells, ketoacidosis has been shown to generate oxygen radicals which result in lipid peroxidation and possibly atherosclerosis (36). These findings are consistent with our observation showing that an acidic extracellular environment increased oxidative stress in the human cancer cell line, HCT116. Interestingly, the intracellular spatial distribution of ROS-responsive fluorescent dye following the incubation in the acidic environment was different from the exogenous H2O2 treatment, while similar enhanced signal intensity per cell was found using the flow cytometric analysis (Fig.4A, B). The significance of this observation is not yet established, although a focal increase of signal intensity implies a presence of an intracellular localized ROS source in response to lowered pHe.
Oxidative attacks to DNA can generate strand breaks directly or indirectly through the BER processing. An oxidized base can be excised by bi-functional DNA glycosylases/AP-lyases (e.g. OGG1, NTH1, and NEIL1/2), which leaves a SSB with damaged 5’ or 3’ termini (37). For both the direct and indirect formation of SSBs, a DNA end processing step is necessary to proceed to the next gap-filling step in both the short patch and long patch BER pathways. The end processing is extremely important for cellular viability because the repair intermediates may be more toxic than the original base damages. An imbalanced increase of glycosylase activity can paradoxically increase cell death because of accumulated repair intermediates. Depending on types of damaged termini, APE1 or PNK is the major enzyme to process the termini. XRCC1 has a critical role in coordinating the end processing and the downstream steps, gap filling and ligation. Accordingly, inhibition of APE1 and/or XRCC1 can cause excess SSBs. A consequence of unrepaired SSBs in proliferating cells is likely a blockage or collapse of replication forks during the S-phase, which results in DNA DSB formation (38). Consistent with this model, we observed prolonged intra-S-phase and G2/M phase cell cycle arrests in an acidic environment with APE1 or XRCC1 inhibition. In non-cycling cells, SSBs located in the opposed strands may form a DSB if they are close spatially. Although cells are equipped to repair DSBs efficiently, the tumor microenvironment inhibits homologous recombination repair which works primarily in S-phase (17). Reduced DSB repair may contribute, to some extent, to our observation that APE1 or XRCC1 inhibition caused greater cell death in an acidic environment.
DNA repair systems are under active investigations as targets for cancer therapeutics (39). APE1 is considered to be one of promising targets, and multiple pharmacologic inhibitors are being evaluated in addition to CRT0044876 (40). Methoxyamine is a potential APE1 inhibitor that physically binds to DNA AP-sites, inhibiting further processing by APE1 (41). Methoxyamine is currently under clinical investigation as a modulator of cytotoxicity to the DNA methylating agent, temozolomide. Recently, another more potent APE1 inhibitor has been identified (42). Although these APE1 inhibitors are being developed to sensitize other DNA damaging agents (43), our data suggests that APE1 inhibitors alone may induce cell death in an acidic environment independent of exogenous DNA damaging agents.
In this study, we focused on enhanced DNA damages as a mechanism of the differential cytotoxicity induced by the BER inhibition in an acidic versus normal pH environment. A further investigation of other potential mechanisms, including Ref1 function, may be necessary to consider, since Ref1 function is also essential for cell survival (44). Ref1 is necessary to reductively activate oxidized transcription factors that play essential roles in cellular viability and proliferation in the tumor microenvironment, such as c-Jun, c-Fos, NF-κB, and HIF-1α. Inhibition of Ref1 using a small molecule inhibitor has recently been shown to reduce proliferation and migration in pancreatic cancer cell lines (45). The reduced proliferation was more pronounced under hypoxic conditions than normoxic conditions. Taken together with our data in this study, APE1/Ref1 appears to possess dual functions which at first glance appear distinct and unrelated but both counteract against environmental oxidative stress.
In conclusion, inhibition of APE1 and/or XRCC1 induces acidic tumor microenvironment targeted cytotoxicity. Therefore, it seems logical to target the BER pathway for solid cancer therapeutics as monotherapy or combined with other standard cytotoxic therapies.
Acknowledgments
This work was supported, in part, by NIH grant U56 CA112963, the DBJ Foundation, and the University Radiation Medicine Foundation. There are no conflicts of interest cited.
REFERENCES
- 1.Horton JK, Watson M, Stefanick DF, et al. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 3.Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. Embo J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol Cell Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular 'nick-sensor' in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masson M, Niedergang C, Schreiber V, et al. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubota Y, Nash RA, Klungland A, et al. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. Embo J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 9.Whitehouse CJ, Taylor RM, Thistlethwaite A, et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 10.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. Embo J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overgaard J. Hypoxic radiosensitization: adored and ignored. J Clin Oncol. 2007;25:4066–4074. doi: 10.1200/JCO.2007.12.7878. [DOI] [PubMed] [Google Scholar]
- 12.Reynolds TY, Rockwell S, Glazer PM. Genetic instability induced by the tumor microenvironment. Cancer Res. 1996;56:5754–5757. [PubMed] [Google Scholar]
- 13.Bindra RS, Crosby ME, Glazer PM. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev. 2007;26:249–260. doi: 10.1007/s10555-007-9061-3. [DOI] [PubMed] [Google Scholar]
- 14.Mihaylova VT, Bindra RS, Yuan J, et al. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koshiji M, To KK, Hammer S, et al. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 16.Yuan J, Narayanan L, Rockwell S, Glazer PM. Diminished DNA repair and elevated mutagenesis in mammalian cells exposed to hypoxia and low pH. Cancer Res. 2000;60:4372–4376. [PubMed] [Google Scholar]
- 17.Bindra RS, Schaffer PJ, Meng A, et al. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan N, Koritzinsky M, Zhao H, et al. Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res. 2008;68:605–614. doi: 10.1158/0008-5472.CAN-07-5472. [DOI] [PubMed] [Google Scholar]
- 19.Kelley MR, Cheng L, Foster R, et al. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin Cancer Res. 2001;7:824–830. [PubMed] [Google Scholar]
- 20.Robertson KA, Bullock HA, Xu Y, et al. Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res. 2001;61:2220–2225. [PubMed] [Google Scholar]
- 21.Yoo DG, Song YJ, Cho EJ, et al. Alteration of APE1/ref-1 expression in non-small cell lung cancer: the implications of impaired extracellular superoxide dismutase and catalase antioxidant systems. Lung Cancer. 2008;60:277–284. doi: 10.1016/j.lungcan.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Ladiges WC. Mouse models of XRCC1 DNA repair polymorphisms and cancer. Oncogene. 2006;25:1612–1619. doi: 10.1038/sj.onc.1209370. [DOI] [PubMed] [Google Scholar]
- 23.Lam W, Park SY, Leung CH, Cheng YC. Apurinic/apyrimidinic endonuclease-1 protein level is associated with the cytotoxicity of L-configuration deoxycytidine analogs (troxacitabine and beta-L-2',3'-dideoxy-2',3'-didehydro-5-fluorocytidine) but not D-configuration deoxycytidine analogs (gemcitabine and beta-D-arabinofuranosylcytosine) Mol Pharmacol. 2006;69:1607–1614. doi: 10.1124/mol.105.021527. [DOI] [PubMed] [Google Scholar]
- 24.Madhusudan S, Smart F, Shrimpton P, et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo Y, Yan T, Schupp JE, et al. The Interaction between Two Radiosensitizers: 5-Iododeoxyuridine and Caffeine. Cancer Res. 2006;66:490–498. doi: 10.1158/0008-5472.CAN-05-2766. [DOI] [PubMed] [Google Scholar]
- 26.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 27.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raghunand N, Gatenby RA, Gillies RJ. Microenvironmental and cellular consequences of altered blood flow in tumours. Br J Radiol. 2003;76:S11–S22. doi: 10.1259/bjr/12913493. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 29.Mordon S, Devoisselle JM, Maunoury V. In vivo pH measurement and imaging of tumor tissue using a pH-sensitive fluorescent probe (5,6-carboxyfluorescein): instrumental and experimental studies. Photochem Photobiol. 1994;60:274–279. doi: 10.1111/j.1751-1097.1994.tb05104.x. [DOI] [PubMed] [Google Scholar]
- 30.Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- 31.Engin K, Leeper DB, Cater JR, et al. Extracellular pH distribution in human tumours. Int J Hyperthermia. 1995;11:211–216. doi: 10.3109/02656739509022457. [DOI] [PubMed] [Google Scholar]
- 32.Ojugo AS, McSheehy PM, McIntyre DJ, et al. Measurement of the extracellular pH of solid tumours in mice by magnetic resonance spectroscopy: a comparison of exogenous (19)F and (31)P probes. NMR Biomed. 1999;12:495–504. doi: 10.1002/(sici)1099-1492(199912)12:8<495::aid-nbm594>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 33.Leake DS. Does an acidic pH explain why low density lipoprotein is oxidised in atherosclerotic lesions? Atherosclerosis. 1997;129:149–157. doi: 10.1016/s0021-9150(96)06035-2. [DOI] [PubMed] [Google Scholar]
- 34.Ying W, Han SK, Miller JW, Swanson RA. Acidosis potentiates oxidative neuronal death by multiple mechanisms. J Neurochem. 1999;73:1549–1556. doi: 10.1046/j.1471-4159.1999.0731549.x. [DOI] [PubMed] [Google Scholar]
- 35.Webster KA, Discher DJ, Kaiser S, et al. Hypoxia-activated apoptosis of cardiac myocytes requires reoxygenation or a pH shift and is independent of p53. J Clin Invest. 1999;104:239–252. doi: 10.1172/JCI5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain SK, Kannan K, Lim G. Ketosis (acetoacetate) can generate oxygen radicals and cause increased lipid peroxidation and growth inhibition in human endothelial cells. Free Radic Biol Med. 1998;25:1083–1088. doi: 10.1016/s0891-5849(98)00140-3. [DOI] [PubMed] [Google Scholar]
- 37.Hazra TK, Das A, Das S, et al. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 39.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 40.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med. 2007;28:375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Liu L, Nakatsuru Y, Gerson SL. Base excision repair as a therapeutic target in colon cancer. Clin Cancer Res. 2002;8:2985–2991. [PubMed] [Google Scholar]
- 42.Seiple LA, Cardellina JH, 2nd, Akee R, Stivers JT. Potent inhibition of human apurinic/apyrimidinic endonuclease 1 by arylstibonic acids. Mol Pharmacol. 2008;73:669–677. doi: 10.1124/mol.107.042622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horton JK, Wilson SH. Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency. DNA Repair (Amst) 2007;6:530–543. doi: 10.1016/j.dnarep.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Izumi T, Brown DB, Naidu CV, et al. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci U S A. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou GM, Maitra A. Small-molecule inhibitor of the AP endonuclease 1/REF-1 E3330 inhibits pancreatic cancer cell growth and migration. Mol Cancer Ther. 2008;7:2012–2021. doi: 10.1158/1535-7163.MCT-08-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]