Abstract
IL-10 and TNF-α are cytokines that have complex and opposing roles in the inflammatory responses. G/A polymorphisms at position –1082 of IL10 and –308 of TNFA genes have been reported to influence the expression of IL-10 and TNF-α, respectively. The aim of this study was to investigate the association between the IL10 (-1082) and TNFA (- 308) gene polymorphisms with different clinical forms or severity of periodontitis in a sample of Brazilian individuals. DNA was obtained from oral swabs of 165 Brazilian individuals, which were divided into three groups: individuals with chronic periodontitis, aggressive periodontitis and individuals without clinical evidence of periodontitis. Evaluation of IL10 and TNFA polymorphisms was performed by RFLP analysis. Statistical analysis of data was performed using the χ2 likelihood ratio and Fisher`s exact test. No significant differences in the genotype and allele distribution of either IL10 or TNFA were observed among individuals with different clinical forms or with different degrees of severity of periodontitis. Moreover, combined analysis of IL10 and TNFA polymorphisms did not show any association with periodontal status. As conclusion, the IL10 and TNFA gene promoter polymorphisms investigated are not associated with different clinical forms of periodontitis or with severity of the disease in the Brazilian population polymorphisms.
Keywords: IL-10, TNF-alpha, periodontitis, polymorphism.
INTRODUCTION
Periodontitis is a multifactorial polymicrobial infection characterized by an inflammatory process that leads to destruction of teeth supporting tissues [1]. There is a complex network of pro- and anti-inflammatory cytokines acting in the inflamed periodontal tissues [2, 3]. Tumor necrosis factor-alpha (TNF-α), a pro-inflammatory cytokine, has been detected in gingival crevicular fluid and gingival tissues from individuals with periodontitis [4, 5]. This cytokine is a potent immunologic mediator that, in addition to its inflammatory effects, increases bone resorption and regulates fibroblast proliferation [6, 7]. TNF-α activity is regulated by interleukin-10 (IL-10) and other anti-inflammatory molecules. IL-10 has the ability to block cytokine synthesis, as well as several accessory cell functions [8] and also may contribute to the regulation of cell proliferation and differentiation [9]. The balance between these and other cytokines regulate the homeostasis of the immune system. Thus, in an inflammatory disease, the predominance of pro-inflammatory cytokines is expected, leading to an unbalanced response and further tissue destruction [10].
The presence of functional polymorphisms in cytokine genes affects cytokine expression and thus may have an important role in the genetic regulation of inflammatory response and of resistance or susceptibility to infections, such as periodontal disease [11-13]. The gene encoding TNF-α has been mapped to chromosome 6 and a polymorphism in the locus –308, causing a substitution from the guanine (G) to adenine (A), leads to two- to threefold higher transcriptional activity of TNF-α upon stimulation with bacterial lipopolysaccharide [14]. The gene encoding IL-10 is mapped to chromosome 1 and displays a polymorphism within the promoter region at position –1082 (also referred to as -1087), being composed of a G to A substitution [15]. This polymorphism lies within as ETS-like recognition site [16] and may affect the binding of this transcription factor producing a higher IL-10 protein levels in GG individuals [15].
Considering the importance of TNF-α and IL-10 in the dynamics of inflammatory responses and, thus, in the pathogenesis of periodontitis, and that the presence of functional polymorphisms in TNFA and IL10 genes affect the levels of expression of these cytokines, the aim of this study was to evaluate the association between the occurrence of the IL-10 (-1082) and TNFA (-889) gene polymorphisms in a sample of Brazilian patients with different clinical forms periodontitis, as well as with different degrees of disease severity.
MATERIALS AND METHODOLOGY
Patients
The study employed a cross-sectional design involving individuals from the State of Minas Gerais from the Southeastern region of Brazil. A total of 165 patients receiving treatment at the Dentistry School, Federal University of Minas Gerais, were included in this study. The patients were stratified into three groups: subjects with aggressive periodontitis (AP, n=55), subjects with chronic periodontitis (CP, n=67) and healthy volunteers, without clinical evidence of periodontitis, as the control group (C, n=43). All patients came from the same geographical area, had a similar socioeconomic status, and displayed no significant differences in the ratio of men to women, or age, between the groups. Patients in the AP group were 15-46 years old and exhibited highly destructive forms of periodontitis; in these patients, the amount of microbial deposits did not justify the severity of periodontal tissue destruction. Patients in the CP group were 25-67 years old and exhibited loss of clinical attachment and amount of destruction consistent with the presence of local factors. Individuals with more than three sites with a probing depth of >5mm and lesions distributed on more than two teeth in each quadrant, were included in this group. No case that produced doubt in classification was included in the study. Diagnosis of disease was made considering the patient’s medical and dental histories, radiographic findings and observation of clinical signs and parameters including probing depth, assessment of clinical attachment loss (CAL), observation of tooth mobility, bleeding on probing and presence of plaque/calculus. Clinical diagnosis of periodontitis was based on criteria established in 1999 at the International Workshop for a Classification of Periodontal Diseases and Conditions [17]. Measurements of probing depth and CAL were assessed at six locations around each tooth. The severity of disease was characterized on the basis of the mean of CAL, within each clinical form. Assessment of CAL was performed by insertion of a periodontal probe in the gingival sulcus and the measurement corresponding to the distance from the cemento-enamel junction to the location of a periodontal probe tip was defined as CAL. Results were expressed as mean CAL; that is, the average of CAL in all six sites of the affected teeth. Patients exhibiting CAL≥5mm were considered with severe and those exhibiting 3mm ≤ CAL>5mm were considered with moderate periodontitis, as previously used by us [18]. Healthy control individuals included in study were 20-70 years old and did not have, at the time of sample collection, periodontal disease; as determined by absence of sites with probing depth >3mm. Moreover, upon questionnaire and clinical evaluation control individuals did not have history of periodontal disease. Measurements of probing depth and clinical attachment loss were assessed at six locations around each tooth in all the individuals involved in present study.
A questionnaire was applied to all individuals enrolled in this study, in order to obtain information regarding dental history, family history of periodontal disease, smoking habit, as well as general health concerns. Use of orthodontic appliances; chronic use of anti-inflammatory drugs; history of diabetes, hepatitis or HIV infection; immunosupressive chemotherapy; bleeding disorders; severely compromised immune function; pregnancy or lactation were regarded as exclusion criteria. Except for the presence of periodontitis, the patients included in this study were systemically healthy. Because tobacco smoking is an important risk factor for periodontitis, we also analyzed our data taking the habit of smoking under consideration. “Smokers” were defined as current smokers/former smokers (more than ten cigarettes/day) and “non-smokers” included individuals that had never smoked. Table 1 summarizes the patient data, as well as their classification into different groups.
Table 1.
Characteristics of the Study Groups
| Clinical Forms | Control (C) | Aggressive Periodontitis (AP) |
Chronic Periodontitis (CP) |
|---|---|---|---|
| Number of individuals (n) | 43 | 55 | 67 |
| Age range (years) | 20-70 | 15-46 | 25-67 |
| Gender male (%) female (%) |
17 (39.5) 26 (60.5) |
20 (36.4) 35 (63.6) |
20 (29.8) 47 (70.2) |
| CAL (mm) mean (±SD) |
- |
6.17 ± 1.57 |
5.69 ± 1.22 |
| Smoking status Non-smokers (%) Smokers (%) |
39 (90.7) 4 (9.3) |
36 (65.5) 19 (34.5) |
38 (56.7) 29 (43.3) |
This study was approved by Universidade Federal de Minas Gerais’s Ethics Committee (nº 003/03) and a signed informed consent was obtained from all participants.
Sample Collection and DNA Extraction
Epithelial cells were obtained through an oral swab performed with a sterile plastic spatula and placed in 1500μl of Krebs buffer (NaCl 20%, KCl 2%, CaCl2 2%, H2O 2%, MgSO4, KH2PO4, C6H12O6). DNA extraction was performed as described previously by us [18]. A pellet of cells was obtained, the supernatant was removed, 20μl of silica (SiO2, Sigma, St. Louis-USA) and 450μl of lyses buffer (6,0M GuSCN, 65mM Tris-HCl pH=6,4, 25mM EDTA and 1,5% Triton X-100) were added to the microtubes. Samples were homogenized and incubated for 30 min at 56ºC. After another centrifugation, the pellet obtained was washed twice with 450μl washing buffer (6,0M GuSCN, 65mM Tris-HCl pH=6,4), twice with 450μl of 70% ethanol, once with 450μl acetone and dried at 56ºC for 20 min. Finally, 100μl of TE buffer (10mM Tris-HCl pH=8,0 and 1mM EDTA) was added and incubated at 56ºC for 12 h. After incubation, the solution was homogenized, centrifuged and the supernatant containing DNA obtained.
Polymerase Chain Reaction (PCR) and Restriction Endonuclease Digestion
IL-10 (-1082) and TNFA (-308) polymorphisms were assessed by standard polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay. The sequences of PCR primers used were 5’CCAAGACAACACTACTAAGGCTCCTTT3’and 5’GCTTCTTATATGCTAGTCAGGTA3’ with expected PCR product size of 377bp to IL10 polymorphism [19] and 5’AGGCAATAGGTTTT GAGGGCCAT3’ and 5’TCCTCCCTGCTCCGATTCCG3’ with expected PCR product size of 107bp to TNFA polymorphism [20]. PCR was carried out in a total volume of 50μl, containing 10μl of solution DNA, Pre-mix buffer (Phoneutria Biotecnologia-Brazil) and primers (20 pMol/ reaction). The amplification conditions consisted of 94ºC for 3 min followed by 40 cycles of 94ºC for 30s, 56ºC for 35s and 72ºC for 30s, and final elongation at 72ºC for 5 min for both primers used. The products were digested with 5 units of enzyme at 37ºC for 4 hours. For IL-10 polymorphism was used XagI enzyme (MBI Fermentas) and digestion products of 280+97 bp and 253+27 bp were obtained for A and G alleles, respectively. For TNFA polymorphism was used NCoI enzyme (Promega-USA) and digestion products of 107 bp and 87+20 bp were obtained for A and G alleles, respectively. The visualization was performed in a 10% acrylamide gel electrophoresis, stained with silver.
Statistical Analysis
Statistical analysis of data was performed using the JMP statistical software (SAS, Cary, NC, USA). The χ2 likelihood ratio was used to compare the genotypes distributions between C and CP groups, between C and AP groups, and between CP and AP groups (3 x 2 contingency table, degrees of freedom (d.f.)=2). The G/A allele and G+/G- or A+/A- genotype distribution between C and CP groups, between C and AP groups, and between CP and AP groups were assessed in 2 x 2 contingency table (df=1), as well as the comparisons regarding severity and “inflammatory/anti-inflammatory” genotypes. Fisher`s exact test was performed in analysis of the 2 x 2 contingency table. The study groups were tested for Hardy-Weinberg equilibrium comparing the expected with the observed genotypes frequencies for both polymorphisms. To exclude the possible confounding effect of smoking, in a second analysis we excluded smokers from all the different clinical groups. A p-value <0.05 was considered statistically significant.
RESULTS
The genotype and allele distributions of the TNFA (-308) and IL10 (-1082) polymorphisms are shown in Table 2 and 3, respectively. The evaluation of the genotype and allele distributions were performed comparing patients with different clinical forms of periodontitis and individuals without clinical evidence of disease, considering or not the habit of smoking. With regards to TNFA (-308) polymorphism, the frequency of homozigous individuals to allele A was very low (0.61%) in the studied population. Only one individual out of all displayed this genotype. No significant difference in the genotype and allele distributions was observed among groups, considering or not the habit of smoking (Tables 2 and 3). The presence of the TNF-α high-producer allele in the population, as evaluated by the frequency of A+ individuals, showed that the frequency of A+ versus A- individuals between the groups was not significantly different when analyzing non-smokers, nor when smokers were included (Table 2).
Table 2.
Distribution of the TNFA (-308) and IL10 (-1082) Genotypes in the Study Groups
| Genotype | Control (C) | Aggressive Periodontitis (AP) | Chronic Periodontitis (CP) |
|---|---|---|---|
| TNFA (-308) Non-smokers | |||
| GG (%) | 28 (71.8) | 24 (66.7) | 23 (60.5) |
| GA (%) | 11 (28.2) | 12 (33.3) | 14 (36.9) |
| AA (%) | - | - | 1 (2.6) |
| A+ (%) | 11 (28.2) | 12 (33.3) | 15 (39.5) |
| A - (%) | 28 (71.8) | 24 (66.7) | 23 (60.5) |
| TNFA (-308) Non-smokers +Smokers | |||
| GG (%) | 29 (67.4) | 40 (72.7) | 44 (65.7) |
| GA (%) | 14 (32.6) | 15 (27.3) | 22 (32.8) |
| AA (%) | - | - | 1 (1.5) |
| A+ (%) | 14 (32.6) | 15 (27.3) | 23 (34.3) |
| A- (%) | 29 (67.4) | 40 (72.7) | 44 (65.7) |
| IL10 (-1082) Non-smokers | |||
| GG (%) | 5 (12.8) | 3 (8.3) | 4 (10.6) |
| GA (%) | 19 (48.7) | 15 (41.7) | 17 (44.7) |
| AA (%) | 15 (38.5) | 18 (50) | 17 (44.7) |
| G+ (%) | 24 (61.5) | 18 (50) | 21 (55.3) |
| G - (%) | 15 (38.5) | 18 (50) | 17 (44.7) |
| IL10 (-1082) Non-smokers + Smokers | |||
| GG (%) | 7 (16.3) | 4 (7.3) | 8 (11.9) |
| GA (%) | 20 (46.5) | 25 (45.4) | 28 (41.8) |
| AA (%) | 16 (37.2) | 26 (47.3) | 31 (46.3) |
| G+ (%) | 27 (62.8) | 29 (52.7) | 36 (53.7) |
| G- (%) | 16 (37.2) | 26 (47.3) | 31 (46.3) |
* The differences among groups were not significant as analyzed with χ2 test and Fisher´s exact test, considering or not the habit of smoking.
Table 3.
Distribution of the TNFA (-308) and IL10 (-1082) Alleles in the Study Groups
| Allele | Control (C) | Aggressiv Periodontitis (AP) | Chronic Periodontitis (CP) |
|---|---|---|---|
| TNFA (-308) | |||
| Non-smokers | |||
| G (%) | 67 (85.9) | 60 (83.3) | 60 (78.9) |
| A (%) | 11 (14.1) | 12 (16.7) | 16 (21.1) |
| Non-smokers +Smokers | |||
| G (%) | 72 (83.7) | 95 (86.4) | 110 (82.1) |
| A (%) | 14 (16.3) | 15 (13.6) | 24 (17.9) |
| IL10 (-1082) | |||
| Non-smokers | |||
| G (%) | 29 (37.2) | 21 (29.2) | 25 (32.9) |
| A (%) | 49 (62.8) | 51 (70.8) | 51 (67.1) |
| Non-smokers + Smokers | |||
| G (%) | 34 (39.5) | 33 (30) | 44 (32.8) |
| A (%) | 52 (60.5) | 77 (70) | 90 (67.2) |
* The differences among groups were not significant as analyzed with χ2 test and Fisher’s exact test, considering or not the habit of smoking.
With regards to IL10 (-1082) polymorphism, no significant difference in the genotype and allele distributions was observed among groups, considering or not the habit of smoking (Tables 2 and 3). The presence of the IL-10 high-producer allele in the population, as evaluated by the frequency of G+ individuals, showed that the frequency of G+ versus G- individuals between the groups was not significantly different when analyzing non-smokers, nor when smokers were included (Table 2).
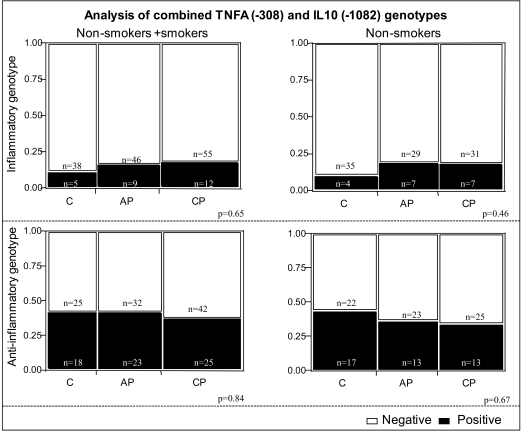
We also performed an additional analysis evaluating the expression of TNF-α and IL-10 polymorphisms, characterizing what we defined as “inflammatory” and “anti-inflammatory” genotypes. Inflammatory genotype was defined as individuals expressing the alleles consistent with high TNF-α and low IL-10 (A+ and G-, respectively) while anti-inflammatory was defined as individuals expressing the alleles consistent with low TNF-α and high IL-10 (A- and G+, respectively). No association was observed between these combined genotypes and the clinical forms of periodontitis analyzed (Fig. 1).
Fig. (1).
Analysis of combined genotypes concerning TNFA (-308) and IL10 (-1082) polymorphisms in the different study groups. Individuals with the A+ genotype of the TNFA (-308) polymorphism are classified as “high TNF- alpha producers” and individuals with the A- genotype are classified as “low TNF- alpha producers”. Individuals with the G+ genotype of the IL10 (-1082) polymorphism are classified as “high IL-10 producers” and individuals with G- genotype are classified as “low IL-10 producers”. The “anti-inflammatory genotype” contains individuals classified as “low TNF- alpha and high IL-10” producers (A-/G+ for TNF- alpha and IL-10, respectively) and the “inflammatory genotype” contains individuals classified as “high TNF-alpha and low IL-10” producers (A+/G- for TNF- α and IL-10, respectively). Black bars (positive) represent individuals with the “inflammatory” or “anti-inflammatory” genotypes in respective graphs. Clear bars (negative) contain individuals that do not bear the specific combinations defined as “inflammatory” or “anti-inflammatory” genotypes. The clear bars in “inflammatory” graphs contains individuals with the genotypes (A+/G+ and A-/G- for TNF-alpha/IL-10) and in the “anti-inflammatory” graph, individuals that express A+/G+ and A+/G- for TNF-alpha/IL-10. No statistical difference was observed among groups, considering or not the habit of smoking (χ2 test, 3 x 2 contingency tables). C= Control, AP= aggressive periodontitis, CP= chronic periodontitis.
With regards to severity of periodontitis evaluated by stratification of the groups according to CAL, no difference was observed between the groups and the occurrence of each polymorphism individually or of combined genotypes (data not shown).
DISCUSSION
Strong evidences supports that genes play a role in the pathogenesis of periodontal disease [12] and the observation that genetic polymorphisms may be associated with the establishment of distinct immune responses to microbial infections has enhanced the interest in identifying genetic polymorphisms associated with diseases [21].
Some studies evaluating the occurrence of polymorphisms in genes of IL-10 and TNF-α were performed in individuals with periodontal disease in different populations. With regards to IL-10 polymorphism, in general a lack of association between the IL-10 polymorphisms and periodontitis has been reported [22-24]. In our study, we also did not find an association of -1082 polymorphism of this gene with the susceptibility to chronic periodontitis or aggressive periodontitis, or with the severity of disease. In the literature, an association between the –1082 polymorphism and periodontal disease were found in Caucasian individuals of a northern European [25] and in Egyptian individuals [26]. These contradictory results can be explained on the basis of racial difference or association with other interactive types of cytokines or genetic markers predisposing for the disease in the population evaluated, as reported by Settin et al. (2006) [26].
In our study, the organization of the sample of Brazilian individuals into ethnic groups was not performed due to the strong miscegenation present among Brazilians, and based on the fact that classification into ethnic groups according to physical appearance is of little use in this population. It has been clearly demonstrated that Brazilian individuals classified as “white” or “black” have significantly overlapping genotypes, likely due to miscegenation [27]. Thus, performing additional analysis of these polymorphisms considering race is of questionable value. In our study, all the patients and controls were selected from the same geographic area and were at the same socio-economic status.
Considering possible haplotypes, each polymorphism may contribute to the production of IL-10 differently and specific haplotypes may influence in progression of periodontal disease [28, 29]. IL10 promoter SNP and haplotype frequencies in different populations have demonstrated that these polymorphisms exhibit a different distribution according to ethnicity [30-33]. The same disease may have different contributing factors in one ethnic group compared with another, as certain polymorphisms may exist in one ethnic group but not in others [34]. With regards to the Brazilian population, association was observed between specific haplotypes in the IL-10 gene and the susceptibility to chronic periodontitis in patients from Piracicaba/SP-Brazil [35]. However, the authors verified no association with the –1082 polymorphism; as observed in our study. Thus, although we did not find association of periodontitis with the polymorphism evaluate, other polymorphisms in IL10 gene may be present and the possible haplotypes may contribute to the production of IL-10 differently.
With regards to TNFA polymorphism, an association between -308 polymorphism and periodontitis was not observed in our study, in accordance other data [36-38]. However, a higher frequency of homozygous (G/G) genotype was observed in individuals with periodontitis when compared to controls in Egyptians [26]. Additionally, combined genotypes composed of TNF-α (-308) and lymphotoxin-α (+252) gene polymorphisms were reported to influence the susceptibility to chronic periodontitis in the Japanese population [39]. It has been reported that specific combinations of functional polymorphisms in different genes may significantly alter the individual risk to development of a phenotype in a disease [21].
Considering the anti-inflammatory and inflammatory profiles of IL-10 and TNF-α, respectively, we evaluated the frequency of specific combinations of functional polymorphisms. We determined the frequency of individuals with “anti-inflammatory genotype”, characterized as “low TNF-α and high IL-10” producers, and of individuals with “inflammatory genotype”, characterized as “high TNF-α and low IL-10” producers. No association was observed between these genotypes and the study groups. However, interpersonal differences in the regulation of IL-10 and TNF-α production may be critical to determine the final outcome of an inflammatory response and may be a consequence of occurrence of multiple polymorphisms in individuals.
We have previously shown that the IL-1B (+3954) and IL-1A (-889) genes polymorphisms are associated with chronic periodontitis [18, 40] and that the IL6 (-174) polymorphism is associated with the severity of periodontitis [41] in a sample of Brazilian individuals. These data suggests that altered levels these cytokines may exist in these individuals and thus may influence the expression of other cytokines, as TNF-α or IL-10.
CONCLUSION
The IL10 and TNFA gene promoter polymorphisms investigated are not associated with different clinical forms of periodontitis or with severity of the disease in the Brazilian population. Considering that little is known about the genetic basis of susceptibility to periodontitis in Brazilian population, the analysis of genetic polymorphisms in a sample this population represents important information concerning periodontal disease in Brazil. Thus, information regarding the presence as well as a lack of association between polymorphisms and disease are of great interest.
ACKNOWLEDGMENT
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)- Brazil.
REFERENCES
- 1.Bodet C, Chandad F, Grenier D. Porphyromonas gingivalis-induced inflammatory mediator profile in a ex vivo human whole blood model. Clin Exp Immunol. 2005;143:50–7. doi: 10.1111/j.1365-2249.2005.02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodet C, Chandad F, Grenier D. Inflammatory responses of a macrophage/epithelial cell co-culture model to mono and mixed infections with Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia. Microbes Infect. 2006;8:27–35. doi: 10.1016/j.micinf.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Okada H, Murakami S. Cytokine expression in periodontal health and disease. Crit Rev Oral Biol Med. 1998;9:248–266. doi: 10.1177/10454411980090030101. [DOI] [PubMed] [Google Scholar]
- 4.Birkedal-Hansen H. Role of cytokines and inflammatory mediators in tissue destruction. J Periodontal Res. 1993;28:500–10. doi: 10.1111/j.1600-0765.1993.tb02113.x. [DOI] [PubMed] [Google Scholar]
- 5.Rossomando EF, Kennedy JE, Hadjimichael J. Tumour necrosis factor alpha in gingival crevicular fluid as a possible indicator of periodontal disease in humans. Arch Oral Biol. 1990;35:431–4. doi: 10.1016/0003-9969(90)90205-o. [DOI] [PubMed] [Google Scholar]
- 6.Dufour A, Baran C, Langkamp HL, Piesco NP, Agarwal S. Regulation of differentiation of gingival fibroblast and periodontal ligament cells by rhIL-1 b and rhTNF-a. J Periodontal Res. 1993;28:566–8. doi: 10.1111/j.1600-0765.1993.tb02126.x. [DOI] [PubMed] [Google Scholar]
- 7.Manolagas SC. Role of cytokines in bone resorption. Bone. 1995;17:63S–7S. doi: 10.1016/8756-3282(95)00180-l. [DOI] [PubMed] [Google Scholar]
- 8.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O`Garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815–22. [PubMed] [Google Scholar]
- 9.Rousset F, Garcia E, Defrance T, et al. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc Natl Acad Sci USA. 1992;89:1890–3. doi: 10.1073/pnas.89.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kantarci A, Hasturk H, Van Dyke T. Host-mediated resolution of inflammation in periodontal diseases. Periodontol 2000. 2006;40:144–63. doi: 10.1111/j.1600-0757.2005.00145.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang XY, Hurme M, Jylha M, Hervonen A. Lack of associationbetween human logevity and polymorphisms of IL-1 cluster, Il-6, IL-10 and TNF-a genes in Finnish nonagenarians. Mech Ageing Dev. 2001;123:29–38. doi: 10.1016/s0047-6374(01)00338-4. [DOI] [PubMed] [Google Scholar]
- 12.Kinane DF, Hart TC. Genes and Gene polymorphisms associated with periodontal disease. Crit Rev Oral Biol Med. 2003;14:430–49. doi: 10.1177/154411130301400605. [DOI] [PubMed] [Google Scholar]
- 13.Greenstein G, Hart TC. A critical assessment of interleukin-1 (IL-1) genotyping when used in a genetic susceptibility test for severe chronic periodontitis. J Periodontol. 2002;73:231–47. doi: 10.1902/jop.2002.73.2.231. [DOI] [PubMed] [Google Scholar]
- 14.Wilson AG, Symons JA, Mcdowell TL, Mcdevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor-alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997;94:3195–99. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 16.Kube D, Platzer C, von Knethen A, et al. Isolation of the human interleukin-10 promoter. Characterization of the promoter activity in Burkitt's lymphoma cell lines. Cytokine. 1995;7:1–7. doi: 10.1006/cyto.1995.1001. [DOI] [PubMed] [Google Scholar]
- 17.Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Moreira PR, de Sá AR, Xavier GM, et al. A functional interleukin-1β gene polymorphism is associated with chronic periodontitis in a sample of Brazilian individuals. J Periodontal Res. 2005;40:306–11. doi: 10.1111/j.1600-0765.2005.00801.x. [DOI] [PubMed] [Google Scholar]
- 19.Koch W, Kastrati A, Böttiger C, Mehilli J, von Beckerath N, Schömig A. Interleukin-10 and tumour necrosis factor gene polymorphisms and risk of coronary artery disease and myocardial infarction. Atherosclerosis. 2001;159:137–44. doi: 10.1016/s0021-9150(01)00467-1. [DOI] [PubMed] [Google Scholar]
- 20.Kornman KS, Crane A, Wang HY, et al. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol. 1997;24:72–7. doi: 10.1111/j.1600-051x.1997.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 21.Walker SJ, Van Dyke TE, Rich S, Kornman KS, di Giovine FS, Hart TC. Genetic polymorphisms of the IL-1α and Il-1β genes in African-American LJP patients and an African-American control population. J. Periodontol. 2000;71:723–8. doi: 10.1902/jop.2000.71.5.723. [DOI] [PubMed] [Google Scholar]
- 22.Yamasaki K, Tabeta K, Nakajima T, et al. Interleukin-10 gene promoter polymorphism in Japanese patients with adult and early-onset periodontitis. J Clin Periodontol. 2001;21:828–32. doi: 10.1034/j.1600-051x.2001.028009828.x. [DOI] [PubMed] [Google Scholar]
- 23.Gonzales JR, Michel J, Diete A, Herrmann JM, Bödeker RH, Meyle J. Analysis of genetic polymorphisms at the interleukin-10 loci aggressive and chronic periodontitis. J Clin Periodontol. 2002;29:816–22. doi: 10.1034/j.1600-051x.2002.290905.x. [DOI] [PubMed] [Google Scholar]
- 24.Kinane DF, Hodge P, Eskdale J, Ellis R, Gallagher G. Analysis of genetic polymorphisms at the interleukin-10 and tumour necrosis factor loci in early-onset periodontitis. J Periodontal Res. 1999;34:379–86. doi: 10.1111/j.1600-0765.1999.tb02270.x. [DOI] [PubMed] [Google Scholar]
- 25.Berglundh T, Donati M, Hahn-Zoric M, Hanson L, Padyukov L. Association of the-1087 IL 10 gene polymorphism with severe chronic periodontitis in Swedish Caucasians. J Clin Periodontol. 2003;30:249–54. doi: 10.1034/j.1600-051x.2003.10274.x. [DOI] [PubMed] [Google Scholar]
- 26.Settin A, Seif MA, Shahat M, El-Baz R, El-Kheir EA. Gene polymorphisms of TNF-α-308(G/A), IL-10-1082(G/A), IL6-174(G/C) and IL1Ra(VNTR) in Egyptian cases with adult and early onset periodontitis. Internet J Den Sci. 2006;4(1) Date of access: 2009 february 1. [Google Scholar]
- 27.Parra FC, Amado RC, Lambertucci JR, Rocha J, Antunes CM, Pena SD. Color and genomic ancestry in Brazilians. Proc Natl Acad Sci USA. 2003;100:177–82. doi: 10.1073/pnas.0126614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yilmaz V, Yentür SP, Saruhan-Direskeneli G. IL-12 and IL-10 polymorphisms and their effects on cytokine production. Cytokine. 2005;30:188–94. doi: 10.1016/j.cyto.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Cullinan MP, Westerman B, Hamlet SM, et al. Progression of periodontal disease and interleukin-10 gene polymorphism. J Periodontal Res. 2008;43(3):328–33. doi: 10.1111/j.1600-0765.2007.01034.x. [DOI] [PubMed] [Google Scholar]
- 30.Moraes MO, Santos AR, Schonkeren JJ, et al. Interleukin-10 promoter haplotypes are differently distributed in Brazilian versus the Dutch population. Immunogenetics. 2003;54:896–9. doi: 10.1007/s00251-003-0543-3. [DOI] [PubMed] [Google Scholar]
- 31.Lazarus R, Klimecki WT, Palmer LJ, et al. Single nucleotide polymorphisms in the interleukin-10 gene: differences in frequencies, linkage disequilibrium patterns and haplotypes in three United States Ethnic groups. Genomics. 2002;54:896–9. doi: 10.1006/geno.2002.6820. [DOI] [PubMed] [Google Scholar]
- 32.Gibson AW, Edberg JC, Wee J, Westendorp RGJ, Huizinga TWJ, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL10 promoter affect IL10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001;166:3915–22. doi: 10.4049/jimmunol.166.6.3915. [DOI] [PubMed] [Google Scholar]
- 33.Reichert S, Machulla HK, Klapproth J, et al. The interleukin-10 promoter haplotype ATA is a putative risk factor for aggressive periodontitis. J Periodontal Res. 2008;43(1):40–7. doi: 10.1111/j.1600-0765.2007.00992.x. [DOI] [PubMed] [Google Scholar]
- 34.Gallager G, Eskdale J, Bidwell JL. Cytokine genetics: polymorphisms, functional variations and disease associatio. In: Lotze M, editor. The cytokine handbook. USA: Elsevier Science Ltda; 2003. pp. 19–56. [Google Scholar]
- 35.Scarel-Caminaga RM, Trevilatto PC, Souza AP, Brito Jr RB, Camargo LE, Line SR. Interleukin 10 gene promoter polymorphisms are associated with chronic periodontitis. J Clin Periodontol. 2004;31:443–8. doi: 10.1111/j.1600-051X.2004.00500.x. [DOI] [PubMed] [Google Scholar]
- 36.Folwaczny M, Glas J, Torok HP, Mende M, Folwaczny C. Lack of association between the TNF α G –308 A promoter polymorphism and periodontal disease. J Clin Periodontol. 2004;31:449–53. doi: 10.1111/j.1600-051x.2004.00499.x. [DOI] [PubMed] [Google Scholar]
- 37.Maria de Freitas N, Imbronito AV, Neves AC, Nunes FD, Pustiglioni FE, Lotufo RF. Analysis of IL-1A (-889) and TNFA (-308) gene polymorphism in Brazilian patients with generalized aggressive periodontitis. Eur Cytokine Netw. 2007;18(3):142–7. doi: 10.1684/ecn.2007.0100. [DOI] [PubMed] [Google Scholar]
- 38.Craandijk J, Van Krugten MV, Verweij CL, Van Der Velden U, Loos BG. Tumor necrosis factor alpha gene polymorphisms in relation to periodontitis. J Clin Periodontol. 2002;29:28–34. doi: 10.1034/j.1600-051x.2002.290105.x. [DOI] [PubMed] [Google Scholar]
- 39.Fassmann A, Holla LI, Buckova D, Vasku A, Znojil V, Vanek J. Polymorphisms in the +252 (A/G) lymphotoxin-alpha and the –308 (A/G) tumor necrosis factor-alpha genes and susceptibility to chronic periodontitis in a Czech population. J Periodontal Res. 2003;38:394–9. doi: 10.1034/j.1600-0765.2003.00661.x. [DOI] [PubMed] [Google Scholar]
- 40.Moreira PR, Costa JE, Gomez RS, Gollob KJ, Dutra WO. The IL1A (-889) gene polymorphism is associated with chronic periodontal disease in a sample of Brazilian individuals. J Periodontal Res. 2007;42:23–30. doi: 10.1111/j.1600-0765.2006.00910.x. [DOI] [PubMed] [Google Scholar]
- 41.Moreira PR, Lima PMA, Sathler KOB, et al. Interleulin-6 expression and gene polymorphism are associated with severity of periodontal disease in a sample of Brazilian individuals. Clin Exp Immunol. 2007;148(1):119–26. doi: 10.1111/j.1365-2249.2007.03327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]