Abstract
Metaplastic breast carcinoma, a rare tumor composed of adenocarcinomatous and nonglandular growth patterns, is characterized by a propensity for distant metastases and resistance to standard anticancer therapies. We sought confirmation that this tumor is a basal-like breast cancer, expressing epidermal growth factor receptor (EGFR) and stem cell factor receptor (KIT). EGFR activating mutations and high copy number (associated with response to tyro-sine kinase inhibitor gefitinib) and KIT activating mutations (associated with imatinib sensitivity) were then investigated. Seventy-seven metaplastic cases were identified (1976-2006); 38 with tumor blocks available underwent pathologic confirmation before EGFR and KIT immunohistochemical analyses. A tissue microarray of malignant glandular and metaplastic elements was constructed and analyzed immunohistochemically for cytokeratin 5/6, estrogen receptor, progesterone receptor, and p63, and by fluorescence in situ hybridization for EGFR and HER-2/neu. DNA isolated from individual elements was assessed for EGFR and KIT activating mutations. All assessable cases were negative for estrogen receptor, progesterone receptor, and (except one) HER2. The majority were positive for cytokeratin 5/6 (58%), p63 (59%), and EGFR overexpression (66%); 24% were KIT positive. No EGFR or KIT activating mutations were present; 26% of the primary metaplastic breast carcinomas were fluorescence in situ hybridization-positive, displaying high EGFR copy number secondary to aneusomy (22%) and amplification (4%). We report here that metaplastic breast carcinoma is a basal-like breast cancer lacking EGFR and KIT activating mutations but exhibiting high EGFR copy number (primarily via aneusomy), suggesting that EGFR tyrosine kinase inhibitors should be evaluated in this molecular subset of breast carcinomas.
Introduction
Metaplastic breast carcinomas are a heterogeneous group of tumors in which the adenocarcinomatous element is admixed with one or more squamous, spindle, chondroid, or osseous neoplastic components (1, 2). Metaplastic breast cancer is rare, accounting for <5% of all breast malignancies. An earlier Mayo Clinic study indicated that although more frequently node-negative at presentation, metaplastic breast carcinoma is more aggressive than breast adenocarcinoma without metaplasia, having an increased risk of locally recurrent and metastatic disease (3). Furthermore, regimens conventionally employed for metastatic breast cancer appear to be less effective for metastatic metaplastic breast carcinoma in this series.
Comparison of gene expression profiles of breast carcinomas (4-6) has validated the traditional classification of these molecularly diverse tumors into two broad groups, those positive or those negative for estrogen receptor (ER) expression. ER-negative tumors have been subdivided into normal breast-like, basal epithelial-like, and HER2 (ErbB2) overexpressing subclasses (4). The basal epithelial-like subgroup of breast carcinomas is characteristically negative for ER, progesterone receptor (PR), and HER2-overexpression (that is, “triple negative”) but positive for EGFR (epidermal growth factor receptor 1, ErbB1, HER1), KIT (stem cell factor receptor; mast cell growth factor receptor), cytokeratin 5/6 (CK 5/6), and p63 (7, 8). Clinically, the basal-like breast tumor subtype is associated with a poorer prognosis in terms of relapse-free survival and overall survival (5, 6, 9-11). Early literature reports indicate that the vast majority of metaplastic breast carcinomas, the subject of this report, are also negative for ER, PR, and HER2-overexpression as well as positive for EGFR, CK 5/6, and p63 expression (12-14), suggesting that these tumors may exhibit characteristics associated with basal-like breast carcinomas (14).
Because metaplastic breast carcinomas are characteristically negative for ER and HER2 and because these tumors are often unresponsive to conventional chemotherapeutic regimens (3), treatment options are limited and new drug therapies are urgently needed. EGFR mutations in exons 18, 19, and 21 are associated with response to the tyrosine kinase inhibitor gefitinib in non-small cell lung cancer (NSCLC; refs. 15, 16). A recent report showed that although EGFR was overexpressed in 68% of metaplastic breast carcinomas, EGFR activating mutations in exons 18 to 21 were not present (17). Additionally, the above study reported various levels of EGFR amplification measured by chromogenic in situ hybridization (CISH) in 23% of metaplastic tumors. Because high EGFR copy number detected by fluorescence in situ hybridization (FISH) (either via gene amplification or high polysomy/aneusomy in which the increased number of EGFR copies is detected with a balanced increase in the number of chromosome 7 copies) is associated with gefitinib response in lung cancer (18, 19), FISH-positivity in metaplastic breast carcinoma may be a useful marker for identifying patients who may benefit from EGFR inhibitors but has never been analyzed. Analogously to EGFR, activating mutations in KIT (C-Kit, CD117) exons 9, 11, 13, and 17 are associated with response of gastrointestinal stromal tumors to the tyrosine kinase inhibitor imatinib (20), but their presence in metaplastic breast carcinoma is unknown. This study was conducted to examine the basal immunohistochemical profile, activating mutations in EGFR and KIT, and EGFR and HER-2/neu copy numbers by FISH in a panel of metaplastic breast carcinomas.
Materials and Methods
Patient Samples
The Mayo Clinic medical index was queried from 1976 to 2006 with the following terms: “metaplastic breast cancer,” “spindle cell cancer,” “squamous cell cancer,” “cancer with sarcomatoid features,” “chondroid metaplasia,” “bony or osseous metaplasia,” “breast cancer-chondroid metaplasia,” “breast cancer-sarcomatous metaplasia,” “breast cancer-spindle cell metaplasia,” or “breast cancer-squamous metaplasia.” A total of 77 patients were identified with one of these diagnoses. Of these, pathology slides were available for review in 49 patients. Before inclusion of a case in this study, an H&E slide from each associated block was reviewed by a pathologist to confirm the diagnosis of metaplastic breast carcinoma. Of these 49 cases, the diagnosis of metaplastic carcinoma was confirmed in 45 patients by the study breast pathologist (C.A.R.). Formalin-fixed, paraffin-embedded tumor blocks were available in 38 of 45. These 38 cases comprise the study cohort. The present study was reviewed and approved by the Mayo Clinic Institutional Review Board.
Immunohistochemical Analysis
Sections (5 μm) of the tumor blocks were analyzed by immunohistochemistry for KIT and EGFR. Immunohistochemical staining for EGFR was scored based on intensity from 0 to 3+ as per manufacturer’s guidelines, with 0 indicating absence of staining, and 1+, 2+, and 3+ representing weak, moderate, and strong staining intensity, respectively. EGFR overexpression was defined as a score of either 2+ or 3+. Figure 1 illustrates a representative case of metaplastic breast carcinoma for each level of EGFR immunohistochemical staining. KIT staining was scored as either negative or positive. Tissue microarray sections (5 μm) were stained for p63, CK 5/6, ER, and PR, with staining scored as negative or positive. If immunohistochemistry scores of any biomarker differed among a patient’s tumor sections, the greatest intensity score was reported (12 patients). All immunohistochemistry antibodies and methods are listed in Supplementary Table S1.8
Figure 1.
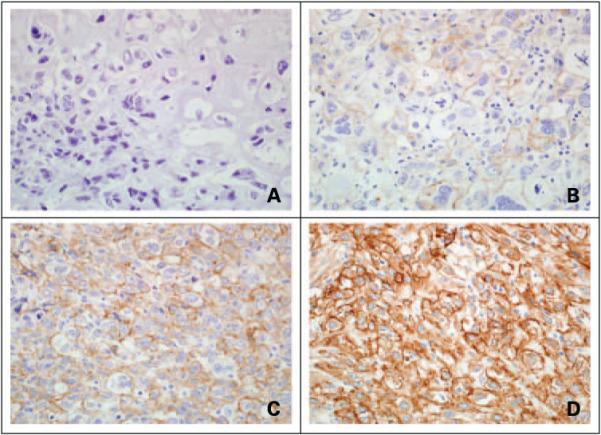
Immunohistochemical analysis of EGFR in metaplastic breast carcinomas. Individual specimens with staining intensity scores of (A)0, (B) 1+, (C) 2+, and (D) 3+, with EGFR overexpression being defined as an intensity of 2+ or 3+.
EGFR and KIT Mutational Analysis
Histologic review of H&E-stained sections from the 56 tumor blocks accessible for the 38 metaplastic breast carcinoma cases (1-3 blocks per patient) identified malignant glandular, squamous, spindle cell, chondroid, and osseous components. Each individual element was circled on an H&E slide, and the corresponding area of tissue was removed from three unstained (10 μm) sections of the associated tumor block. DNA was isolated from each of these individual elements with the QIAamp DNA Mini Kit (Qiagen) such that a total of 73 unique aliquots of DNA were collected for mutational analysis (that is, DNA samples from 13 glandular, 11 squamous, 38 spindle cell, 1 osseous, and 10 chondroid components).
EGFR and KIT exons of interest were amplified by use of PCR with a modification of the technique for EGFR mutational analysis of Gilbert et al. (21). PCR primers for the selected exons were designed to hybridize at locations that resulted in the production of amplicons containing predictive mutation sites as well as ≤181 bp of exon sequence (with the exception of the KIT exon 11 amplicon, which contained 206 bp of exon sequence). All primer sequences and annealing temperatures are listed in Supplementary Table S2.8 Amplifications were done with iQ Supermix (Bio-Rad) with 1.25 units (final) iTaq DNA Polymerase (Bio-Rad). All PCR amplifications were done in a Perkin-Elmer model 9700 thermal cycler.
Amplicons were sequenced on both strands in the Mayo DNA Sequencing Facility using the universal M13 forward and reverse sequences as primers (except for KIT exon 13, for which the forward PCR primer was employed instead of the M13 forward sequence). ABI BigDye Terminator sequencing chemistry was employed with an ABI 3730 DNA sequencer, and sequencing chromatograms were analyzed using Sequencher 4.5 (Gene Codes). NT_033968.5 and NM_005228.3 were the Genbank accession numbers for the EGFR reference sequences used in these studies, whereas NT_022853.14 and U63834 were the reference sequences employed for KIT.
Genomic DNA controls containing EGFR mutations encoding L858R in exon 21, delE746-A750 in exon 19, and delL747-P753insS in exon 19 were kind gifts of Drs. Daphne Bell and Daniel Haber (Massachusetts General Hospital). HMC-1 cells with KIT mutations encoding V560G in exon 11 and D816V in exon 17 were generously provided by Dr. Joseph Butterfield (Mayo Clinic Rochester).
Tissue Microarray Construction
A tissue microarray was constructed with a Beecher ATA-27 automated arrayer from the 73 individual malignant glandular and metaplastic elements identified previously in the tissue blocks of the 38 metaplastic breast carcinoma cases. Each of these individual elements was circled on an H&E slide, and triplicate 0.6-mm cores were removed from the corresponding area of tissue in the associated tumor block and placed into a single recipient paraffin block.
Gene Copy Number Analysis
Tissue microarray sections (5 μm) were analyzed by FISH for EGFR and HER-2/neu copy number. FISH for HER-2/neu (ERBB2) was done with the PathVysion HER2 DNA Probe Kit (Vysis; ref. 22). FISH for EGFR was done with the LSI EGFR/CEP 7 Probe (Vysis) as per manufacturer’s instructions. Thirty nuclei were scored per sample, and the number of HER-2/neu or EGFR (red) signals and chromosome 17 centromere or chromosome 7 centromere (green) signals, respectively, were recorded. A ratio of HER-2/neu:chromosome17 centromere or EGFR:chromosome 7 centromere of 0.8 to 1.30 was defined as normal, a ratio of <0.8 was interpreted as gene deletion, a ratio of 1.30 to 2.0 was defined as gene duplication, and a ratio ≥2.0 was interpreted as gene amplification. EGFR FISH-positive samples were those showing amplification or high aneusomy with ≥40% of cells having ≥4 copies of EGFR, where aneusomy was defined as a normal EGFR:chromosome 7 centromere ratio with >30% of cells having ≥3 chromosome 7 centromere signals (that is, a balanced gain in EGFR and chromosome 7 centromere copy numbers). If any element within a patient’s tumor was FISH-positive, the tumor was reported to be FISH-positive (3 patients).
Results
Clinical Characteristics of Metaplastic Breast Carcinoma Cases
Seventy-seven metaplastic cases were identified between 1976 and 2006 at Mayo Clinic. Of the 49 cases with pathology slides available, a diagnosis of metaplastic breast carcinoma was confirmed in 45 patients, 38 that had accessible tumor blocks. This study of 38 patients with metaplastic breast carcinoma includes 30 (cohort A) who underwent excision or reexcision of primary disease and 8 (cohort B) who underwent either excision of disease following neoadjuvant chemotherapy (2 patients) or excision of recurrent/metastatic disease (6 patients). The median age at surgery of cohort A was 61 years (range, 34-90); six of these patients had been previously diagnosed with another cancer. At presentation, cohort A tumors tended to be 2 to 5 cm in size (60%), to be Nottingham grade 3 (73%), to have a spindle cell component (83%), and to be node-negative (63%; Table 1). Cohort B were in various stages of their disease course and had received a variety of treatments before surgery for this instance of metaplastic breast cancer.
Table 1. Characteristics of 30 primary metaplastic breast carcinoma cases.
| Median age (range) | 61 y (34-90) |
| Year of surgery | |
| 1976-1979 | 10.0% |
| 1980-1989 | 6.7% |
| 1990-1999 | 33.3% |
| 2000-2005 | 50.0% |
| Prior history of cancer | 20.0% |
| Maximum tumor dimension (cm) | |
| <2 | 20% |
| 2-5 | 60% |
| >5 | 20% |
| No. positive nodes | |
| Not examined | 13.3% |
| 0 | 63.3% |
| 1-3 | 16.7% |
| 4-9 | 3.3% |
| 10+ | 3.3% |
| Nottingham grade | |
| 1 | 3.3% |
| 2 | 23.3% |
| 3 | 73.3% |
| Tumor histology | |
| Spindle cell | 43.3% |
| Chondroid | 3.3% |
| Mixture of glandular and metaplastic elements | |
| Glandular and squamous | 3.3% |
| Glandular and spindle cell | 13.3% |
| Glandular and chondroid | 6.7% |
| Glandular with squamous and spindle cell | 10.0% |
| Glandular with squamous and chondroid | 3.3% |
| Glandular with spindle cell and chondroid | 3.3% |
| Mixture of metaplastic elements | |
| Squamous and spindle cell | 6.7% |
| Spindle cell and chondroid | 3.3% |
| Spindle cell, chondroid, and osseous | 3.3% |
| Presence of metastatic disease | 3.3% |
Immunohistochemistry
Of 38 metaplastic breast carcinoma cases, all those assessable were negative by immunohistochemistry for ER and PR, and by FISH for HER-2/neu amplification (except for one primary). By immunohistochemistry, EGFR was overexpressed in 70% of cohort A, whereas 54% were CK 5/6 positive, 58% were p63 positive, and 23% were KIT positive. Of cohort B, ≥50% exhibited EGFR overexpression (4 of 8), CK 5/6 positivity (6 of 8), and p63 positivity (5 of 8). A summary of the immunohistochemical and FISH results for the 38 individual metaplastic breast carcinoma cases is shown in Table 2, and a subgroup analysis of biomarker expression for all 38 cases is provided in Table 3.
Table 2. Individual immunohistochemical and FISH results for metaplastic breast carcinoma cases (n = 38).
| Case no. | Tumor histology | EGFR* | EGFR FISH† | KIT* | CK 5/6‡ | p63‡ | ER‡ | PR‡ | HER2 FISH† |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Spindle cell | 1+ | - | - | - | - | - | - | - |
| 2 | Chondroid | 3+ | - | + | + | ND | ND | - | - |
| 3 | Glandular and spindle cell | 3+ | + | - | + | + | - | - | - |
| 4 | Spindle cell | 2+ | - | - | - | - | - | - | - |
| 5 | Spindle cell | 0 | - | - | + | - | - | - | - |
| 6 | Spindle cell, chondroid, and osseous | 1+ | - | - | - | - | - | - | - |
| 7 | Glandular with squamous and spindle cell | 3+ | - | - | + | + | - | - | - |
| 8 | Glandular and spindle cell | 3+ | + | - | - | + | - | - | - |
| 9 | Glandular with squamous and spindle cell | 2+ | ND§ | - | ND | ND | ND | - | - |
| 10 | Squamous | 1+ | - | - | + | + | - | - | - |
| 11 | Spindle cell | 1+ | - | - | + | - | - | - | - |
| 12 | Glandular with squamous and spindle cell | 3+ | - | + | + | + | - | - | - |
| 13 | Glandular and spindle cell | 2+ | - | - | + | + | - | - | - |
| 14 | Glandular and squamous | 3+ | - | + | + | + | - | - | - |
| 15 | Glandular with squamous and spindle cell | 3+ | - | - | + | + | - | - | - |
| 16 | Squamous and spindle cell | 1+ | + | - | + | + | - | - | - |
| 17 | Spindle cell | 2+ | - | - | - | + | - | - | - |
| 18 | Squamous and spindle cell | 3+ | - | - | + | + | - | - | - |
| 19 | Glandular and spindle cell | 1+ | - | + | - | - | - | - | - |
| 20 | Spindle cell | 1+ | - | - | - | - | - | - | - |
| 21 | Spindle cell | 3+ | - | - | - | - | - | - | - |
| 22 | Spindle cell | 3+ | - | - | - | + | - | - | - |
| 23 | Spindle cell and chondroid | 2+ | + | + | + | - | - | - | - |
| 24 | Glandular and chondroid | 1+ | - | + | - | - | - | - | - |
| 25 | Spindle cell | 0 | ND | - | ND | ND | ND | ND | ND |
| 26 | Spindle cell | 3+ | - | - | + | + | - | - | - |
| 27 | Glandular with squamous and spindle cell | 3+ | + | - | + | + | - | - | - |
| 28 | Glandular with spindle cell and chondroid | 1+ | - | + | + | + | - | - | - |
| 29 | Glandular and spindle cell | 3+ | ND | - | + | ND | ND | ND | - |
| 30 | Glandular and spindle cell | 2+ | - | + | + | + | - | - | + |
| 31 | Glandular and chondroid | 2+ | - | + | + | + | - | - | - |
| 32 | Spindle cell | 3+ | + | - | + | + | - | - | - |
| 33 | Glandular with squamous and chondroid | 2+ | + | - | + | - | - | - | - |
| 34 | Spindle cell | 3+ | + | - | - | + | - | - | - |
| 35 | Spindle cell | 3+ | - | - | - | + | - | - | - |
| 36 | Spindle cell | 3+ | - | - | - | - | - | - | - |
| 37 | Spindle cell | 1+ | - | - | - | - | - | - | - |
| 38 | Spindle cell | 1+ | - | - | - | - | - | - | - |
As determined by immunohistochemistry of sections from tumor blocks.
As determined by FISH of tissue microarray sections. “EGFR FISH,” determination of FISH-positivity (aneusomy with ≥40% of cells having ≥4 copies of EGFR or EGFR amplification); “HER2 FISH,” determination of HER-2/neu amplification.
As determined by immunohistochemistry of tissue microarray sections.
ND, not determined due to tissue dropout in the tissue microarray.
Table 3. Subgroup analysis of biomarker expression in metaplastic breast carcinomas (n = 38 cases).
| Tumor histology | EGFR overexpression* |
EGFR FISH-positive† |
KIT expression* |
CK 5/6 expression‡ |
p63 expression‡ |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Primary | Other | Primary | Other | Primary | Other | Primary | Other | Primary | Other | |
| Squamous | 0/1 | 0/1 | 0/1 | 1/1 | 1/1 | |||||
| Spindle cell | 8/13 | 1/3 | 2/12§ | 0/3 | 0/13 | 0/3 | 2/12§ | 2/3 | 5/12§ | 1/3 |
| Chondroid | 1/1 | 0/1 | 1/1 | 1/1 | § | |||||
| Mixture of glandular and metaplastic elements | ||||||||||
| Glandular and squamous | 1/1 | 0/1 | 1/1 | 1/1 | 1/1 | |||||
| Glandular and spindle cell | 4/4 | 1/2 | 1/3§ | 1/2 | 1/4 | 1/2 | 3/4 | 1/2 | 3/3§ | 1/2 |
| Glandular and chondroid | 1/2 | 0/2 | 2/2 | 1/2 | 1/2 | |||||
| Glandular with squamous and spindle cell | 3/3 | 2/2 | 1/2§ | 0/2 | 0/3 | 1/2 | 2/2§ | 2/2 | 2/2§ | 2/2 |
| Glandular with squamous and chondroid | 1/1 | 1/1 | 0/1 | 1/1 | 0/1 | |||||
| Glandular with spindle cell and chondroid | 0/1 | 0/1 | 1/1 | 1/1 | 1/1 | |||||
| Mixture of metaplastic elements | ||||||||||
| Squamous and spindle cell | 1/2 | 1/2 | 0/2 | 2/2 | 2/2 | |||||
| Spindle cell and chondroid | 1/1 | 1/1 | 1/1 | 1/1 | 0/1 | |||||
| Spindle cell, chondroid, and osseous | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | |||||
NOTE: Value represents (number positive) / (total number of patients per tumor subtype). Results were separated according to case type, either primary (cohort A) or other (cohort B). All assessable tumors were negative for ER, PR, and (except one) HER-2/neu amplification.
As determined by immunohistochemistry of sections from tumor blocks. “EGFR overexpression,” intensity of 2+ or 3+.
Aneusomy with ≥40% of cells having ≥4 copies of EGFR or (for one tumor) EGFR amplification, as determined by FISH analysis of tissue microarray sections.
As determined by immunohistochemistry of tissue microarray sections.
Results for some tumors not available due to tissue dropout in tissue microarray.
EGFR and KIT Mutational Analysis
EGFR exons 18, 19, and 21 and KIT exons 9, 11, 13, and 17 were assessed for mutations in 73 DNA samples isolated from malignant glandular and metaplastic components of 38 metaplastic breast carcinoma tumors. Sequence was obtained for all EGFR and KIT exons in all 73 DNA samples from the 38 cases, except for one sample (the spindle cell component of a tumor composed of spindle cell, chondroid, and osseous elements) in which no sequence for any of the EGFR or KIT exons could be obtained and one sample (the osseous component of the tumor composed of spindle cell, chondroid, and osseous elements) in which neither EGFR exon 21 nor KIT exon 11 could be sequenced. No EGFR or KIT activating mutations were found among these cases.
EGFR Copy Number Analysis
Of the 27 (of 30) primary metaplastic breast carcinoma cases assessable in the tissue microarray for EGFR copy number by FISH analysis, 7 (26%) displayed high EGFR copy number. One of the cohort B cases also showed high EGFR copy number. Of these eight FISH-positive metaplastic breast carcinomas, one (a primary) showed EGFR amplification and seven displayed aneusomy with ≥4 EGFR copies in ≥40% of cells—most frequently in the spindle cell component (75%). Figure 2 illustrates examples of the FISH-positive metaplastic breast tumors. Summaries of the EGFR FISH results for all of the metaplastic breast carcinomas by case and by cohort are provided in Tables 2 and 3, respectively, whereas Table 4 presents the characteristics of the individual FISH-positive metaplastic breast tumors.
Figure 2.
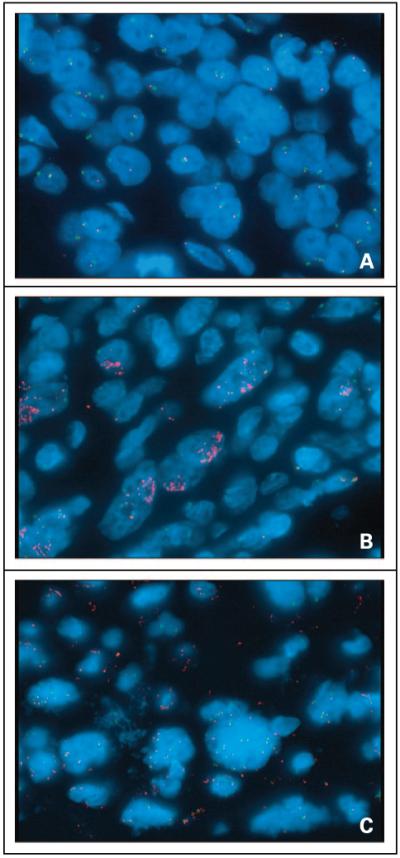
FISH analysis of EGFR copy number in metaplastic breast carcinomas. Red, EGFR signal; green, chromosome 7 signal. Individual specimens with (A) normal EGFR copy number, (B) amplification of EGFR, and (C) aneusomy with ≥40% of cells having ≥4 copies of EGFR.
Table 4. Characteristics of FISH-positive metaplastic breast carcinomas.
| Case no. | Tumor Histology | EGFR* | EGFR FISH† | KIT* | CK 5/6‡ | p63‡ | ER‡ | PR‡ | HER2 FISH† |
|---|---|---|---|---|---|---|---|---|---|
| 32 | Spindle cell | 3+ | Aneusomy | - | + | + | - | - | - |
| 34 | Spindle cell | 3+ | Amplification | - | - | + | - | - | - |
| 8 | Glandular and spindle cell | 3+ | Aneusomy | - | - | + | - | - | - |
| 3§ | Glandular and spindle cell | 3+ | Aneusomy | - | + | + | - | - | - |
| 27 | Glandular with squamous and spindle cell | 3+ | Aneusomy | - | + | + | - | - | - |
| 33 | Glandular with squamous and chondroid | 2+ | Aneusomy | - | + | - | - | - | - |
| 16 | Squamous and spindle cell | 1+ | Aneusomy | - | + | + | - | - | - |
| 23 | Spindle cell and chondroid | 2+ | Aneusomy | + | + | - | - | - | - |
NOTE: No FISH-positive tumors had EGFR (exons 18, 19, and 21) or KIT (exons 9, 11, 13, and 17) activating mutations.
As determined by immunohistochemistry of sections from tumor blocks. EGFR staining was scored based on intensity from 0 to 3+, with overexpression defined as intensity of 2+ or 3+.
As determined by FISH of tissue microarray sections. “EGFR FISH”, determination of FISH-positivity (aneusomy with ≥40% of cells having ≥4 copies of EGFR or EGFR amplification); “HER2 FISH,” determination of Her-2/neu amplification.
As determined by immunohistochemistry of tissue microarray sections.
Recurrent disease; all other cases were primaries.
Discussion
This molecular study of metaplastic breast carcinomas confirmed previous findings that metaplastic tumors exhibit many characteristics typical of basal-like breast carcinomas, that is, ER/PR/HER2-negativity, p63 and CK 5/6 positivity, EGFR overexpression, and (approximately one-quarter) KIT positivity (7, 14). Additionally, we confirm the findings of Reis-Filho et al. (17) that metaplastic carcinomas do not harbor activating mutations in EGFR. We are, however, the first to show the absence of activating mutations in KIT. Most importantly, our demonstration that the malignant histologic elements (predominantly spindle cell) displayed high EGFR copy number (primarily via aneusomy) suggests that EGFR inhibitors should be investigated as a potential therapeutic agent for this subtype of breast cancer.
The association of EGFR activating mutations with responsiveness to the tyrosine kinase inhibitor gefitinib observed in NSCLC (15, 16) has been the subject of numerous studies since the first publication in 2004. A recent review summarized mutational analyses of EGFR exons 18 to 21 in a total of 3,000 NSCLC cases (23); a retrospective comparison of mutational status with gefitinib response in 288 of these cases did not provide a perfect correlation although it did indicate that the majority of gefitinib-responsive NSCLC tumors harbored activating EGFR mutations. Subsequently, high EGFR gene copy number by FISH analysis has been associated with response to gefitinib in NSCLC (18) and bronchioloalveolar carcinoma subtypes (19). Specifically, EGFR FISH-positivity in tumors [defined as high balanced polysomy with ≥4 EGFR copies in ≥40% of cells or EGFR amplification (gene:chromosome ≥2 per cell, gene clusters, or ≥15 gene copies per cell in ≥10% of cells)] is associated with a higher gefitinib response rate in NSCLC (18). In bronchioloalveolar carcinoma subtypes of NSCLC, gefitinib responsiveness is also associated with EGFR FISH-positivity [high balanced polysomy with ≥4 EGFR copies in ≥40% of cells or EGFR amplification (gene:chromosome ≥2 per cell, gene clusters, or ≥15 gene copies per cell in z10% of cells; ref. 19)]. More recently, a prospective study of gefitinib in 42 NSCLC patients (24) reported objective responses in 48% of participants and confirmation of EGFR amplification and high polysomy as predictors of gefitinib responsiveness. Further, an association between gefitinib response and EGFR FISH-positivity in the absence of EGFR activating mutations was seen in a recent NSCLC study (25) in which 40% of patients lacking EGFR mutations exhibited FISH-positivity; notably, of the 21 nonmutants with high EGFR copy number, 24% responded to gefitinib treatment. Although the prognostic role of classical EGFR mutations and/or increased EGFR copy number requires further clarification, these factors are nonetheless considered predictive for tumor response to gefitinib (see ref. 26).
We report here that no EGFR activating mutations were found in these 38 metaplastic breast carcinoma cases. Our findings are comprehensive, because we sequenced DNA extracted from each of the malignant elements of metaplastic breast carcinoma (squamous, chondroid, adenocarcinomatous, osseous, and spindle). In addition, employing EGFR FISH criteria associated with gefitinib response in NSCLC (18, 19), EGFR FISH-positivity (amplification with gene:chromosome ≥2 per cell, or aneusomy with ≥4 EGFR copies in ≥40% of cells) was found in almost one-quarter of the assessable metaplastic breast carcinomas in this study (in 26% of the primary metaplastic breast carcinoma cases of cohort A). By comparison, one metaplastic breast carcinoma (spindle cell with focal squamous differentiation) included in a study of breast cancers by Bhargava et al. (27) showed high-level EGFR amplification by chromogenic in situ hybridization (“15 gene copies/nucleus”). Furthermore, this metaplastic tumor lacked activating EGFR mutations in the exons examined (19 and 21). Reis-Filho et al. (28) reported seven metaplastic breast carcinomas that showed by chromogenic in situ hybridization apparently low-level amplification (“>5 signals/nuclei”) to high-level amplification (“large gene signal clusters”): four spindle cell and three carcinomas with squamous elements. No indication was given, however, as to which of the seven showed the high-level EGFR amplification essential for the high EGFR copy number associated with gefitinib response in NSCLC. This same group did a follow-up study enlarging the sample size to 47 metaplastic carcinomas and showed (by chromogenic in situ hybridization) some level of EGFR amplification in 11 tumors but no EGFR activating mutations (17). It should be noted that neither the report by Bhargava et al. (27) nor the two studies by Reis-Filho et al. (17, 28) labeled the chromosome 7 centromere in addition to the EGFR gene during chromogenic in situ hybridization analyses. This is vitally important, as it allows the calculation of the ratio of number of EGFR copies to number of chromosome 7 copies per sample, as was done in the FISH analyses presented here. Thus, this report is the first description in metaplastic breast carcinoma samples of an increase in EGFR copy number due to a balanced increase in the number of chromosome 7 copies. Furthermore, in the present study, only one of the metaplastic carcinomas with high EGFR copy number displayed EGFR gene amplification—the majority of the FISH-positive tumors (seven) showed high aneusomy in which at least 40% of the cells contained at least 4 copies of the EGFR gene (Fig. 2). These findings suggest that EGFR amplification is rare in metaplastic breast carcinomas, and that aneusomy is the most likely mechanism for high EGFR copy number.
As has been seen for HER2-overexpression in breast tumors (22), a direct correlation between EGFR immunohistochemistry and EGFR FISH-positivity in the metaplastic breast carcinomas in this study was not consistently found. As indicated in Table 4, the FISH-positive metaplastic samples ranged from 1+ to 3+ in immunohistochemical staining intensity, where the 1+ tumor would be considered negative for EGFR-overexpression. Thus, immunohistochemical measurements of EGFR protein expression did not prove to be predictive of FISH results determining EGFR copy number in metaplastic breast carcinomas.
Finally, as also indicated in Table 4, seven of the eight metaplastic breast carcinomas with high EGFR copy number contained a spindle cell component. Although more than one element in these seven tumors may have had high EGFR copy number, the spindle cell component was (except in one case) always FISH-positive. These data suggest that the spindle cell element was important for EGFR FISH-positivity in these tumors.
Unlike the relatively infrequent classical EGFR mutations in NSCLC, activating KIT mutations occur in as many as 90% of gastrointestinal stromal tumors, predominantly in exons 9 and 11 (see ref. 29 for review). KIT activating mutations have not been readily found in other tumor types (30-32). In a breast cancer study, 3% of 1,654 tumors examined were KIT-positive, and mutational analysis of 10 of the strongly positive tumors was negative for KIT activating mutations in exons 2, 8, 9, 11, 13, and 17 (33). Although together these studies suggest that KIT mutations are uncommon in tumors other than gastrointestinal stromal tumors, none specifically report KIT mutational analysis of metaplastic breast cancer, a member of the basal-like subclass of breast carcinomas of which >30% are KIT-positive (7). For the first time, we report that despite the presence of KIT expression in 24% of metaplastic breast tumors, KIT activating mutations were not present.
In summary, we have shown that metaplastic breast carcinomas exhibit molecular characteristics most consistent with the basal subtype of breast cancer. Although activating mutations in EGFR and KIT were not found, the presence of high EGFR copy number by FISH warrants further study to determine the role of EGFR tyrosine kinase inhibitors in treatment of metaplastic breast carcinoma patients.
Acknowledgments
Grant support: Paul Calabresi Program in Clinical-Translational Research at Mayo Clinic grant CA 90628 (M.P. Goetz), Career Development Award from Mayo Cancer Center Breast Cancer SPORE grant CA 116201 (M.P. Goetz), and Mayo Comprehensive Cancer Center grant CA 15083 (M.M. Ames).
Footnotes
Note: J.A. Gilbert and M.P. Goetz contributed equally to this work.
Supplementary material for this article is available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
This work was presented in preliminary form at the 29th Annual San Antonio Breast Cancer Symposium in December 2006.
References
- 1.Rosen PP. Rosen’s breast pathology. Lippincott Williams & Wilkins; Philadelphia: 2001. Carcinoma with metaplasia; pp. 425–61. [Google Scholar]
- 2.Tavassoli FA. Classification of metaplastic carcinomas of the breast. Pathol Annu. 1992;27(Pt 2):89–119. [PubMed] [Google Scholar]
- 3.Rayson D, Adjei AA, Suman VJ, et al. Metaplastic breast cancer: prognosis and response to systemic therapy. Ann Oncol. 1999;10:413–9. doi: 10.1023/a:1008329910362. [DOI] [PubMed] [Google Scholar]
- 4.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sotiriou C, Neo S-Y, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100:10393–8. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–74. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 8.Livasy CA, Karaca G, Nanda R, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–71. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 9.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van de Rijn M, Perou CM, Tibshirani, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161:1991–6. doi: 10.1016/S0002-9440(10)64476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer sub-types, and survival in the Carolina breast cancer study. JAMA. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 12.Koker MM, Kleer CG. p63 Expression in breast cancer. A highly sensitive and specific marker of metaplastic carcinoma. Am J Surg Pathol. 2004;28:1506–12. doi: 10.1097/01.pas.0000138183.97366.fd. [DOI] [PubMed] [Google Scholar]
- 13.Leibl S, Moinfar F. Metaplastic breast carcinomas are negative for Her-2 but frequently express EGFR (Her-1): potential relevance to adjuvant treatment with EGFR tyrosine kinase inhibitors? J Clin Pathol. 2005;58:700–4. doi: 10.1136/jcp.2004.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reis-Filho JS, Milanezi F, Steele D, et al. Metaplastic breast carcinomas are basal-like tumours. Histopathology. 2006;49:10–21. doi: 10.1111/j.1365-2559.2006.02467.x. [DOI] [PubMed] [Google Scholar]
- 15.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 16.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–50. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 17.Reis-Filho JS, Pinheiro C, Lambros MBK, et al. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol. 2006;209:445–53. doi: 10.1002/path.2004. [DOI] [PubMed] [Google Scholar]
- 18.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–55. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a southwest oncology group study. J Clin Oncol. 2005;23:6838–45. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 20.Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–9. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert JA, Lloyd RV, Ames MM. Lack of mutations in EGFR in gastroenteropancreatic neuroendocrine tumors. N Engl J Med. 2005;353:209–10. doi: 10.1056/NEJM200507143530219. [DOI] [PubMed] [Google Scholar]
- 22.Perez EA, Roche PC, Jenkins RB, et al. HER2 testing in patients with breast cancer: poor correlation between weak positivity by immunohisto-chemistry and gene amplification by fluorescence in situ hybridization. Mayo Clin Proc. 2002;77:148–54. doi: 10.4065/77.2.148. [DOI] [PubMed] [Google Scholar]
- 23.Chan SK, Gullick WJ, Hill ME. Mutations of the epidermal growth factor receptor in non-small cell lung cancer—search and destroy. Eur J Cancer. 2006;42:17–23. doi: 10.1016/j.ejca.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 24.Cappuzzo F, Ligorio C, Janne PA, et al. Prospective study of gefitinib in epidermal growth factor receptor fluorescence in situ hybridization-positive/phospho-Akt-positive or never smoker patients with advanced non-small-cell lung cancer: the ONCOBELL trial. J Clin Oncol. 2007;25:2248–55. doi: 10.1200/JCO.2006.09.4300. [DOI] [PubMed] [Google Scholar]
- 25.Han S-W, Kim T-Y, Jeon YK, et al. Optimization of patient selection for gefitinib in non-small cell lung cancer by combined analysis of epidermal growth factor receptor mutation, K-ras mutation, and Akt phosphorylation. Clin Cancer Res. 2006;12:2538–44. doi: 10.1158/1078-0432.CCR-05-2845. [DOI] [PubMed] [Google Scholar]
- 26.Shepherd FA, Tsao M-S. Unraveling the mystery of prognostic and predictive factors in epidermal growth factor receptor therapy. J Clin Oncol. 2006;24:1219–20. doi: 10.1200/JCO.2005.04.4420. [DOI] [PubMed] [Google Scholar]
- 27.Bhargava R, Gerald WL, Li AR, et al. EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol. 2005;18:1027–33. doi: 10.1038/modpathol.3800438. [DOI] [PubMed] [Google Scholar]
- 28.Reis-Filho JS, Milanezi F, Carvalho S, et al. Metaplastic breast carcinomas exhibit EGFR, but not HER2, gene amplification and over-expression: immunohistochemical and chromogenic in situ hybridization analysis. Breast Cancer Res. 2005;7:R1028–35. doi: 10.1186/bcr1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Zwan SM, DeMatteo RP. Gastrointestinal stromal tumor: 5 years later. Cancer. 2005;104:1781–8. doi: 10.1002/cncr.21419. [DOI] [PubMed] [Google Scholar]
- 30.Went PT, Dirnhofer S, Bundi M, et al. Prevalence of KIT expression in human tumors. J Clin Oncol. 2004;22:4514–22. doi: 10.1200/JCO.2004.10.125. [DOI] [PubMed] [Google Scholar]
- 31.Burger H, den Bakker MA, Kros JM, et al. Activating mutations in c-KIT and PDGFRα are exclusively found in gastrointestinal stromal tumors and not in other tumors overexpressing these imatinib mesylate target genes. Cancer Biol Ther. 2005;4:1270–4. doi: 10.4161/cbt.4.11.2253. [DOI] [PubMed] [Google Scholar]
- 32.Sihto H, Sarlomo-Rikala M, Tynninen O, et al. KIT and platelet-derived growth factor receptor α tyrosine kinase gene mutations and KIT amplifications in human solid tumors. J Clin Oncol. 2005;23:49–57. doi: 10.1200/JCO.2005.02.093. [DOI] [PubMed] [Google Scholar]
- 33.Simon R, Panussis S, Maurer R, et al. KIT (CD117)-positive breast cancers are infrequent and lack KIT gene mutations. Clin Cancer Res. 2004;10:178–83. doi: 10.1158/1078-0432.ccr-0597-3. [DOI] [PubMed] [Google Scholar]


