Abstract
Structure-based design, synthesis and X-ray structure of protein-ligand complexes of memapsin 2 are described. The inhibitors are designed specifically to interact with S2 and S3-active site residues to provide selectivity over memapsin 1 and cathepsin D. Inhibitor 6 has exhibited exceedingly potent inhibitory activity against memapsin 2 and selectivity over memapsin 1 (>3800-fold) and cathepsin D (>2500-fold). A protein-ligand crystal structure revealed cooperative interactions in the S2 and S3 active sites of memapsin 2. These interactions may serve as an important guide to design selectivity over memapsin 1 and cathepsin D.
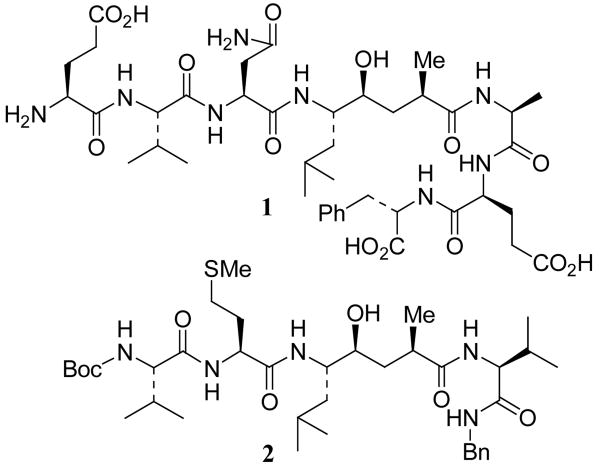
The proteolytic enzyme memapsin 2 (β-secretase, BACE-1) has emerged as a leading target for therapeutic intervention of Alzheimer's disease (AD).1 It is one of two proteases that cleave the β-amyloid precursor protein (APP) to generate the 40/42 residue amyloid-β peptide (Aβ). The excess level of Aβ leads to formation of amyloid plaques and neurofibrillary tangles in the brain.2, 3 The neurotoxicity of Aβ eventually leads to the death of neurons, inflammation of the brain, dementia and AD.4 Based upon initial kinetics and substrate specificity data,5 we designed a number of potent inhibitors incorporating a nonhydrolyzable Leu-Ala hydroxyethylene dipeptide isostere.6 One such inhibitor is OM99-2 (1, Figure 1) which has a Ki value of 1.6 nM for human memapsin 2.6a An X-ray crystal structure of 1-bound memapsin 2 was determined at 1.9 Å resolution.7 The structure provided molecular insight into the ligand-binding site interactions of the memapsin 2 active site.
Figure 1.
Structure of Inhibitors 1 and 2
Subsequently, our preliminary structure-activity relationship studies led to the design of potent peptidomimetic inhibitor 2 with a Ki value of 2.5 nM against memapsin 2.6b However, it displayed no selectivity over memapsin 1 (BACE-2) or cathepsin D. From a therapeutic point of view, the selectivity of memapsin 2 inhibitors over other human aspartic proteases is expected to be important, especially memapsin 1 and cathepsin D.
Memapsin 1, which has specificity similarity with memapsin 2,8 has independent physiological functions. Cathepsin D, which is abundant in all cells, plays an important role in cellular protein catabolism.9 Its inhibition would likely consume inhibitor drugs as well as lead to probable toxicity.
The X-ray structure of 1-bound memapsin 2 revealed a number of critical ligand-binding site interactions in the S2 and S3-subsites.7 Based upon examination of this X-ray structure and a model of memapsin 1, it appears that the residues in the S2 and S3-subsites may be suitable for building selectivity over memapsin 1 and cathepsin D. Herein we report our structure-based design and synthesis of novel memapsin 2 inhibitors that incorporate methylsulfonyl alanine as the P2-ligand and pyrazole and oxazole-derived heterocycles as the P3-ligands. The corresponding inhibitors have exhibited enhanced potency against memapsin 2 and excellent selectivity over memapsin 1 and cathepsin D. Furthermore, the protein-ligand X-ray structure of the pyrazole-bearing inhibitor provided important molecular insight into the specific cooperative ligand-binding site interactions for selectivity design.
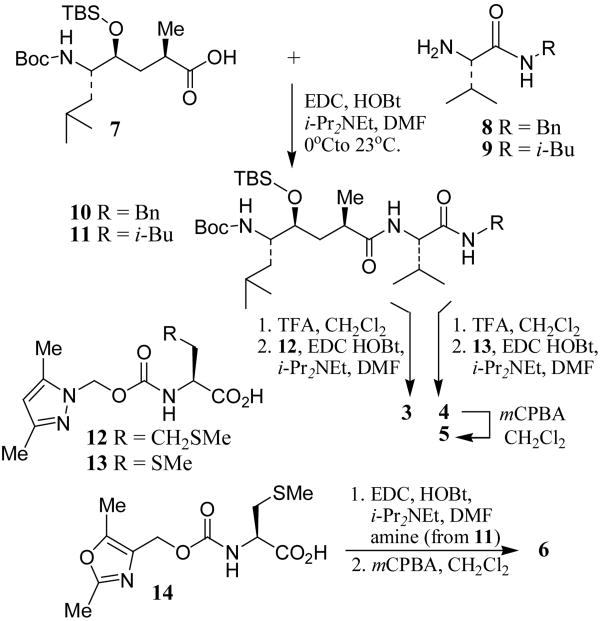
The synthesis of inhibitors 3-6 is outlined in Scheme 1. Coupling of previously described6 Leu-Ala dipeptide isostere 7 with valine derivatives 8 and 9 using EDC and HOBt in the presence of i-Pr2NEt provided derivatives 10 and 11 (71-95%). Urethanes 12 and 13 were prepared by treatment of 2,5-dimethylpyrazolylmethanol with triphosgene in CH2Cl2 followed by addition of methionine and methylcysteine methyl esters to provide the corresponding urethanes.10 Saponification of resulting methyl esters with aqueous lithium hydroxide provided 12 and 13 (36-44%). Removal of Boc and TBS groups by exposure of 10 and 11 to trifluoroacetic acid and coupling of the resulting amines with the corresponding acids using EDC and HOBt afforded inhibitors 3 and 4 (40-64%). Oxidation of sulfide 4 with mCPBA in a mixture (6:1) of CH2Cl2 and MeOH furnished sulfone 5 (86%). Acid 14 was prepared by alkoxycarbonylation10 of 2,5-dimethyl-4-oxazolemethanol11 and methylcysteine methyl ester followed by saponification of the resulting ester (see supporting information for details). It was converted to inhibitor 6 by analogous procedures described above.
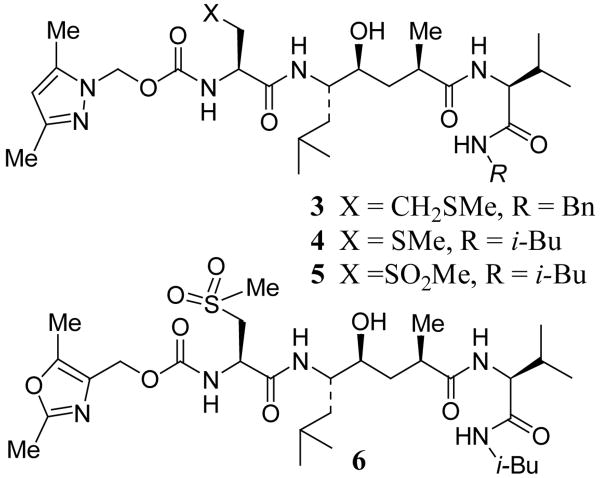
Scheme 1.
Synthesis of Inhibitors 3-6
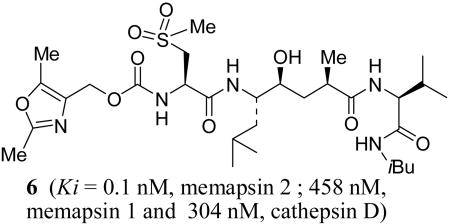
Potencies of various inhibitors were determined against recombinant memapsin 2, memapsin 1 and human cathepsin D. The results are shown in Table 1. As shown, inhibitor 2 with P3-Boc-Val is more potent for memapsin 1 than memapsin 2. Incorporation of pyrazolylmethyl urethane in place of P3-Boc-Val provided inhibitor 3 having a >5-fold reduction in potency for memapsin 2 compared to inhibitor 2. Inhibitor 3 also showed significantly reduced activity against M1 with a Ki value of 811 nM (58-fold selectivity over M1), but showed little selectivity for M2 over CD. Inhibitor 4 with a P3′-isobutylamide and P2-methylcysteine has shown a 3-fold enhancement of M2 potency. It has also shown >36-fold selectivity over M1 and a modest 3-fold selectivity over CD. Based upon inhibitor models, the P2-methionine in 3 or P2-methylcysteine side chain in 4 does not appear to be forming a hydrogen bond with Arg-235 of memapsin 2. However, the corresponding P2-sulfone of 4 appears to be in close proximity to Arg-235 of memapsin 2. Indeed, oxidation of the P2-methyl cysteine provided inhibitor 5 (MW 658) with very impressive potency (M2 Ki 0.3 nM) and selectivity for memapsin 2. It displayed 1186-fold selectivity over M1 and 436-fold selectivity over CD.
Table 1.
Ki values and selectivities for Inhibitors 2-6a
| Compd | M2 (nM) | M1 (nM) | CD (nM) | Selectivity Ratios | IC50 (μM) | |
|---|---|---|---|---|---|---|
| M1/M2 | CD/M2 | |||||
| 2 | 2.5 | 1.2 | nd | -- | -- | 6.5 |
| 3 | 14 | 811 | 25 | 58 | 1 | nd |
| 4 | 4.4 | 161 | 15 | 36 | 3 | nd |
| 5 | 0.3 | 356 | 131 | 1186 | 436 | 1.4 |
| 6 | 0.12 | 458 | 304 | >3800 | >2500 | 1.7 |
Data represent the mean value of 3-5 determinations; memapsin 2 (M2), memapsin 1 (M1), cathepsin D (CD); nd, not determined
To gain further molecular insight, the 5-bound memapsin 2 X-ray structure was determined at 1.8 Å resolution (Figure 3).12 As it appears from the structure, one of the P3-pyrazole nitrogens is within hydrogen bonding distance to Thr-232 with one of the dimethyl groups effectively filling in the shallow hydrophobic pocket in the S3′-subsite and the other occupying the hydrophobic S3-subsite. In addition, the P2-sulfone functionality is within hydrogen bonding distance to Arg-235 as well as with a tight-bound water molecule in the S2-subsite. These interactions of the P2 and P3 ligands are presumably responsible for the observed enhanced selectivity for inhibitor 5 compared to inhibitor 4.
Figure 3.
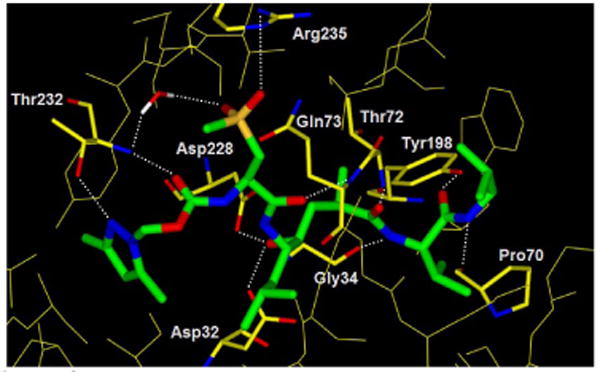
Inhibitor-5-bound X-ray structure of memapsin 2
Based upon this molecular insight into the selectivity, we designed the oxazolylmethyl P3-ligand for inhibitor 6 (MW 659). This provided by far the most potent (M2 Ki 0.12 nM) and selective inhibitor (>3800-fold over M1 and >2500-fold over CD). We have also determined the cellular inhibition of memapsin 2 by inhibitors 5 and 6 in Chinese hamster ovary cells. They have shown an average cellular IC50 value of 1.4 μM and 1.7 μM respectively compared to an IC50 value of 45 μM for 1.13
In conclusion, our structure-based design led to the development of very potent and highly selective memapsin 2 inhibitors. Furthermore, our X-ray structural analysis of protein-inhibitor complexes has uncovered potentially important molecular interactions useful in the design of selectivity against other aspartyl proteases. Additional studies to further elucidate the role of these and other interactions important for selectivity are in progress.
Supplementary Material
Figure 2.
Structure of Inhibitors 3-6
Acknowledgments
Financial support by the National Institutes of Health (AG 18933) is gratefully acknowledged.
Footnotes
Supporting Information Available: Experimental procedures and spectral data for 3-6, 10-14 and X-ray information and complete reference 3a. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Vassar R, Citron M. Neuron. 2000;27:419. doi: 10.1016/s0896-6273(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 2.(a) Roggo A. Curr Top Med Chem. 2002;2 doi: 10.2174/1568026024607490. [DOI] [PubMed] [Google Scholar]; (b) Olson RE, Thompson LA. Ann Rep Med Chem. 2000;35:31. [Google Scholar]
- 3.(a) Vassar R, et al. Science. 1999;286 [Google Scholar]; (b) Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Proc Natl Acad Sci USA. 2000;97 doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sinha S, Lieberburg I. Proc Natl Acad Sci USA. 1999;96:11049. doi: 10.1073/pnas.96.20.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Selkoe DJ. Nature. 1999;399A doi: 10.1038/399a023. [DOI] [PubMed] [Google Scholar]; (b) Selkoe D. Physiol Rev. 2001;81:741. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 5.Turner R, Koelsch G, Hong L, Castenheira P, Ghosh AK, Tang J. Biochemistry. 2001;40:10001. doi: 10.1021/bi015546s. [DOI] [PubMed] [Google Scholar]
- 6.(a) Ghosh AK, Shin D, Downs D, Koelsch G, Lin X, Ermolieff J, Tang J. J Am Chem Soc. 2000;122 doi: 10.1021/ja000300g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ghosh AK, Bilcer G, Harwood C, Kawahama R, Shin D, Downs D, Hussain KA, Hong L, Loy JA, Nguyen C, Koelsch G, Ermolieff J, Tang J. J Med Chem. 2001;44:2865. doi: 10.1021/jm0101803. [DOI] [PubMed] [Google Scholar]
- 7.Hong L, Koelsch G, Lin X, Wu S, Terzyan S, Ghosh AK, Zhang XC, Tang J. Science. 2000;290:150. doi: 10.1126/science.290.5489.150. [DOI] [PubMed] [Google Scholar]
- 8.Turner RT, Loy JA, Nguyen C, Devasamudram T, Ghosh AK, Koelsch G, Tang J. Biochemistry. 2002;41:8742. doi: 10.1021/bi025926t. [DOI] [PubMed] [Google Scholar]
- 9.Diment S, Leech MS, Stahl PD. J Biol Chem. 1988;263:6901. [PubMed] [Google Scholar]
- 10.Ghosh AK, Duong TT, McKee SP, Thompson WJ. Tetrahedron Lett. 1992;33:2781. doi: 10.1016/S0040-4039(00)78856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kukla MJ, Fortunato JM. J Org Chem. 1984;49:5003. [Google Scholar]
- 12.The protein-ligand X-ray structure of 5–bound memapsin 2 has been deposited in PDB (PDB ID: 2G94). For a stereoview and crystallographic information, please see supporting information
- 13.Chang WP, Koelsch G, Wong S, Downs D, Da H, Weerasena V, Gordon B, Devasamudram T, Bilcer G, Ghosh AK, Tang J. J Neurochem. 2004;89:1409. doi: 10.1111/j.1471-4159.2004.02452.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.