Abstract
Gene expression quantitative trait locus (eQTL) mapping has become a powerful tool in systems biology. While many authors have made important discoveries using this approach, one persistent challenge in eQTL studies is the selection of loci and genes that should receive further biological investigation. In this study, we compared eQTL generated from gene expression profiling in the livers of two panels of mouse strains, 41 BXD recombinant inbred and 36 mouse diversity panel (MDP) strains. Cis-eQTL, loci in which the transcript and its maximum QTL are co-located, have been shown to be more reproducible than trans-eQTL, which are not co-located with the transcript. We observed that 9.9% of cis-eQTL and 2.0% of trans-eQTL replicated between the two panels. Notably, a significant eQTL hotspot on distal chromosome 12 observed in the BXD panel was reproduced in the MDP. Furthermore, the shorter linkage disequilibrium in the MDP strains allowed us to considerably narrow the locus and limit the number of candidate genes to a cluster of Serpin genes, which code for extracellular proteases. We conclude that this strategy has some utility in increasing confidence and resolution in eQTL mapping studies; however, due to the high false positive rate in the MDP, eQTL mapping in inbred strains is best carried out in combination with an eQTL linkage study.
Introduction
Gene expression quantitative trait locus (eQTL) mapping is a statistical technique that correlates quantitative measurements of mRNA expression with genetic polymorphisms segregating in a population to locate genomic intervals that are likely to regulate the expression of each transcript (Farrall 2004; Gilad et al. 2008). When transcript expression is measured using microarrays, the result consists of tens of thousands of genome-wide eQTL profiles, one for each transcript. The goal of such an analysis is to identify clusters of co-regulated genes, to discover candidate genes that may regulate the expression of these clusters, to elucidate normal tissue-specific physiology and seek candidate genes underlying disease-related phenotypes. eQTL mapping has been successfully applied in yeast (Brem et al. 2002), arabidopsis (West et al. 2007), maize (Shi et al. 2007), mice (Bystrykh et al. 2005; Chesler et al. 2005; Schadt et al. 2003) and humans (Monks et al. 2004).
Two common features of eQTL studies are the existence of cis-eQTL, genes which are regulated by loci co-located within 5 Mb of the transcript, and trans-eQTL, genes which are regulated by distant loci (>5 Mb or on different chromosomes) (Kliebenstein 2008; Peirce et al. 2006). In general, cis-eQTL tend to produce stronger statistical associations than trans-eQTL (Doss et al. 2005), and this is regarded as evidence of greater biological plausibility for the existence of true functional cis-eQTL. Trans-eQTL can occur individually at a single genomic locus or can occur collectively as part of eQTL trans-bands. The latter are thought to be genomic loci that control the expression of a larger number of genes than expected by chance. Several reports have suggested that most eQTL trans-bands are likely to be spurious (Breitling et al. 2008; de Koning et al. 2005; Kliebenstein 2008) and may be attributed to the correlation structure among transcript expression. Briefly, if one transcript is spuriously associated with a locus, then all transcripts that are highly correlated with the first transcript will also exhibit spurious association. The difficulty is that clusters of transcripts truly associated with a causative locus will also show this same pattern, making the detection of true positives quite difficult. To date, one eQTL trans-band has been biologically validated using small interfering RNA (siRNA) knockdown of the candidate gene to demonstrate a change in the predicted function of the genes in the trans-band (Wu et al. 2008).
While linkage studies in F2 or recombinant inbred (RI) lines are thought to produce more robust eQTL results, the limited number of recombination events in such crosses produces large QTL intervals and makes the selection of candidate genes difficult. One approach that has been used to narrow individual QTL intervals is in silico haplotype mapping in laboratory inbred strains (Burgess-Herbert et al. 2008; Dipetrillo et al. 2005). Another approach relies upon the combination of data from multiple studies to both narrow the QTL interval and select QTL that are reproducible. For a single phenotype, several methods have been described that can be used to combine QTL data from different crosses (Li et al. 2005; Malmanger et al. 2006; Peirce et al. 2007; Walling et al. 2000). However, due to the high cost of replicating a large eQTL study and the computational challenge of combining data for thousands of transcripts, these methods are difficult to apply to most eQTL studies.
The reproducibility of eQTL in two panels of closely related mice, BXD recombinant inbreds (RI) (Taylor et al. 1999) and an F2 cross between the same parental strains, has been reported to be high (Peirce et al. 2006). However, it is not clear whether eQTL will replicate in more diverse populations within the same species. Recent work has shown that genome wide association (GWA) mapping in panels of inbred strains suffers from a high false-positive rate (Manenti et al. 2009) but suggests a combined approach using GWA and classical linkage mapping in a genetic cross. Here, we apply this technique to investigate the reproducibility of mouse liver eQTL between a linkage study in BXD RI lines (Gatti et al. 2007) and a GWA study in inbred strains of the Mouse Diversity Panel (MDP) (Paigen et al. 2000). We observed that 9.9% of cis-eQTL and 2.0% of trans-eQTL replicate between the two data sets and we use the finer haplotype structure of the MDP to narrow the eQTL intervals. We also find that an eQTL trans-band on distal chromosome 12 is reproducible, and conclude that this approach should be added to the array of tools used to select candidate eQTL for biological validation.
Methods
BXD Strains
The details of breeding, housing, RNA isolation and gene expression measurements in these mice are described in (Gatti et al. 2007). Briefly, 38 strains of male BXD RI mice, C57BL/6J and DBA/2J parentals, and B6D2F1 were used to perform genome-wide eQTL mapping for 20,868 transcripts using Agilent G4121A microarrays (Santa Clara, CA).
BXD QTL Mapping
eQTL mapping in the BXD panel was carried out using FastMap (Gatti et al. 2009) configured to perform single-marker mapping and 1,000 permutations per transcript to produce per-transcript significance thresholds. A subset of 2,486 transcripts (out of 20,868 on the array) were selected by retaining all transcripts with known genomic locations (in Mouse Genome Build 36) and a maximum eQTL peak with a p-value ≤ 0.05. This subset was used in the analysis of eQTL replication in the MDP eQTL data. Cis-eQTL were defined as those for which the maximum QTL and the transcript were co-located within 5 Mb. The QTL interval was taken as the peak-width at 1 log-of-the-odds score (1-LOD) below the eQTL peak maximum. Subsets of eQTL were selected at per-transcript p-values of 0.001, 0.01 and 0.05.
Inbred strains of the MDP
Male mice (aged 7-9 weeks) were obtained from The Jackson Laboratory and housed in polycarbonate cages on Sani-Chips irradiated hardwood bedding (P.J. Murphy Forest Products Corp., Montville, NJ). Animals were fed NTP-2000 wafer feed (Zeigler Brothers, Inc., Gardners, PA) and water ad libitum, and maintained on a 12 h light-dark cycle. Mice utilized in this study comprise 31 inbred strains that are priority strains for the Mouse Phenome Project (Paigen et al. 2000): 129S1/SvImJ, A/J, AKR/J, BALB/cByJ, BTBR T+ tf/J, BUB/BnJ, CAST/EiJ, C3H/HeJ, C57BL/10J, C57BL/6J, C57BLKS/J, C57BR/CdJ, C57L/J, CBA/J, CZECHII/EiJ, DBA/2J, FVB/NJ, JF1/Ms, KK/HlJ, LP/J, MA/MyJ, MSM/Ms, NOD/ShiLtJ (formerly NOD/LtJ), NON/LtJ, NZO/H1LtJ, NZW/LacJ, P/J, PERA/EiJ, PL/J, PWD/PhJ, RIIIS/J, SEA/GnJ, SJL/J, SM/J, SWR/J, and WSB/EiJ. Care of mice followed institutional guidelines under a protocol approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill.
RNA isolation
To minimize variability in transcript expression that might arise due to circadian rhythms or lobular variation, animals were sacrificed between 9 AM and 11AM and the left liver lobe was selected for analysis of gene expression. RNA was extracted from 30 mg of liver tissue using the Qiagen RNeasy kit (Qiagen, Valencia, CA). RNA concentrations were measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and quality was verified using the Agilent Bio-Analyzer (Agilent Technologies, Palo Alto, CA).
Microarray hybridizations
In this study, all RNA samples were hybridized to arrays individually; no samples were pooled. RNA amplifications and labeling were performed using Low RNA Input Linear Amplification kits (Agilent Technologies). For hybridization, 750 ng of total RNA from each mouse liver was amplified and labeled with Cy5 fluorescent dye. In parallel, 750 ng of a common reference RNA (Icoria Inc., RTP, NC) was labeled with Cy3 fluorescent dye in order to standardize analysis of global gene expression between mouse strains(Bammler et al. 2005). Labeled cRNA was then processed and hybridized to Agilent Mouse Toxicology Arrays (catalog# 4121A; 20,868 transcripts) according the manufacturer's protocol. Following hybridization, arrays were washed using a custom protocol developed by Icoria, Inc. Briefly, array gaskets are removed 1 (6× SSPE, 0.005% N-Lauroylsarcosine). Arrays were washed with Wash Solution 1 and incubated for one minute with gentle agitation on a magnetic stir plate. A second incubation was performed in Wash Solution 2 (0.06× SSPE, 0.005% N-Lauroylsarcosine).
Microarray Data Analysis
Raw microarray intensity values were obtained from Agilent Feature Extraction software (v8.5) and archived in the UNC Microarray Database (http://genome.unc.edu). The log2 ratio of Cy5/Cy3 intensity was normalized using LOWESS smoothing to eliminate intensity bias of features. Intensity ratios were transformed to eliminate hybridization batch effects using the Batch Normalization feature in Partek Genomics Suite (Partek Inc., St. Louis, MO). The strain means were obtained be averaging all arrays for each stain (1 or 2 per strain) and were used in the subsequent eQTL analysis. The raw microarray data is available from the Gene Expression Omnibus (GSE14563) as well as from WebQTL (Wang et al. 2003).
MDP QTL Mapping
High density single nucleotide polymorphism (SNP) data was used to perform eQTL mapping in the MDP (McClurg et al. 2007). Association mapping was carried out using FastMap (Gatti et al. 2009) as detailed above. Population structure was identified using a PCA plot of the SNP data and two major strata were identified; C57BL/6J, C57BL/10J, C57BLKS/J, C57BR/cdJ & C57L/J were in one stratum and the remaining strains were in the other. We subtracted the mean gene expression value of each stratum from the strains in each respective stratum before mapping. Significant eQTL were selected at a p ≤ 0.001, 0.01 and 0.05 levels. After mapping, transcripts with significant eQTL on more than 5 chromosomes were considered to have to high a rate of false positives and were discarded.
Determination of eQTL Reproducibility
The eQTL intervals for three sets of transcripts, selected at increasing degrees of stringency (p ≤ 0.001, 0.01, 0.05) in the BXD panel were intersected with eQTL intervals for those same transcripts in the MDP. The BXD eQTL were intersected with three sets of eQTL from the MDP selected at increasing levels of stringency (p ≤ 0.001, 0.01, 0.05). An eQTL was considered to be replicated when 1) the eQTL was significant in both data sets at the current significance levels and 2) the minimum p-value on the chromosome where the eQTL occurred in the MDP intersected the BXD eQTL interval.
Determination of Null Probability of Replication
In order to assess the probability of an eQTL replicating between the BXD panel and the MDP, we selected one transcript at random from the significant BXD eQTL and one transcript at random from the significant MDP eQTL and looked for an eQTL within 5.0 Mb in both panels. This process was repeated 1,000,000 times. We found that eQTL intersected only 0.097% of the time and concluded that the chance of eQTL replication occurring by chance was quite low.
Determination of Null Probability of eQTL trans-bands
Following the procedure outlined in (Breitling et al. 2008), we permuted the strains in the SNP data while holding the strain order in the gene expression data constant and performed eQTL mapping 100 times using FastMap. We then counted the number of permutations in which an eQTL trans-band of size n occurred at least once.
Identical By Descent (IBD) Regions
The software available at http://compgen.unc.edu/DisplayIntervals/DisplayIntervals.html was used to determine which regions of the genome are IBD (Zhang et al. 2009). This tool uses the SNP data produced by (Szatkiewicz et al. 2008) and calls a region IBD if there are 100 or more consecutive, non-polymorphic SNPs between the two strains.
SNPs in Probe Sequences
The genomic locations of the probes on the Agilent G4121A microarray were obtained from Agilent (Santa Clara, CA). High density mouse SNP data containing 7.87 × 106 SNPs was obtained from (Szatkiewicz et al. 2008). Probe sequences containing SNPs were found and intersected with the reproducible cis-eQTL. For each cis-eQTL, we performed a one-sided Student's T-test at the SNP of highest association between the expression of strains with the C57BL/6J allele and those with the other allele to determine if the eQTL was “C57BL/6J allele high” or not. Fisher's exact test was used to test the null hypothesis that a SNP in the probe sequence was equally likely to occur in cis-eQTL that are C57BL/6J-high versus DBA/2J-high.
Results and Discussion
eQTL Mapping in the Mouse Diversity Panel
eQTL studies are expensive and time consuming to carry out. In the mouse, while experimental cost remains considerable for data collection, no genotyping is generally required as many large panels of inbred mice have been genotyped and this data is publicly available (Roberts et al. 2007b). Examples include RI lines such as the BXD (Peirce et al. 2004), BXH and LXS (Shifman et al. 2006) strains. While these strains are useful, the large sizes of their recombination block structures are not conducive to identifying quantitative trait genes (QTG). For example, the BXD strains have a mean distance of 324, 493 base pairs (bp) between informative markers (www.genenetwork.org/mouseCross.html#BXD). In turn, higher density genotype data containing 156,525 SNPs in 71 inbred strains has a mean intermarker distance of 16,611 bp between informative markers (Roberts et al. 2007a). While this SNP resolution in panels of inbred strains is almost 20 times greater, population stratification, haplotype block structure and local linkage disequilibrium may also confound the ability to find the QTG.
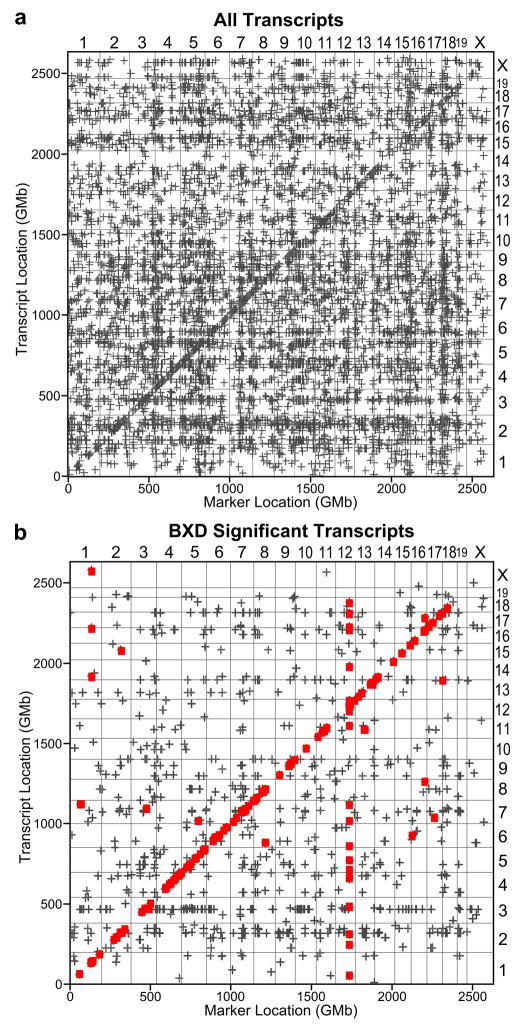
eQTL mapping in the MDP has been proposed as a way to both increase the resolution of RI-based eQTL mapping and limit the need for multiple crosses and additional breeding (McClurg et al. 2007). Here, we performed eQTL mapping on liver gene expression data obtained from 36 naïve inbred mouse strains in order to test the hypothesis that these strains can be successfully used for genome-wide eQTL mapping. As observed in a previous eQTL study in mouse hypothalamus (McClurg et al. 2007), we found that noise, lack of statistical power and population stratification present a formidable challenge to data interpretation whereby the false positive rate is likely to be unacceptably high. For example, at a genome-wide significance level of 0.05, there were 2,453,897 significant loci for 4,061 transcripts and the mean number of significant loci per transcript was 604. In fact, one transcript had over 26,000 significant loci. These loci were not clustered in a few locations, but were distributed throughout the genome (Figure 1a).
Figure 1.
a) Transcriptome map of all 20,868 transcripts on the microarray using 31 M. m. domesticus derived strains at a per-transcript p =< 0.05 significance level. b) Transcriptome map of significant eQTL in the BXD strains (grey crosses) at p =<0.05 with replicated eQTL using in the laboratory inbreds overlaid (red squares) at p =<0.05.
In agreement with recent criticism of the pitfalls of eQTL mapping in an MDP (Breitling et al. 2008), we reason that eQTL mapping in a panel of inbred strains may not be the best independent way to discover eQTL. First, the MDP do not form a segregating population in which each allele can be assigned to a specific progenitor, which means that phenotypic associations demonstrate that an allele is identical-by-state rather than identical-by-descent. Second, the MDP has a complex breeding history and performing association mapping in any subset may be similar to selecting the same number of outbred mice for association mapping; the resulting power is likely to be low. Third, population stratification between M. m. domesticus and non-M. m. domesticus strains is difficult to overcome in a computationally feasible manner when performing mapping on 20,868 transcripts. Fourth, while there are 156,525 SNPs in the data set, there are only 59,255 unique strain distribution patterns (SDP), which creates a high probability that a given transcript will map to more than one locus. Often, the SDPs cluster in the same region of the genome, but there are many cases where they are distributed throughout the genome. Thus, while eQTL mapping in some subset of the MDP may be appealing, new mouse resources such as the Collaborative Cross (Churchill et al. 2004; Threadgill et al. 2002) are likely to provide much greater power and resolution for genome-wide systems genetics.
Because of the limitations of the MDP, the effect of population structure on the eQTL mapping results needed to be removed. Previously, it has been suggested that removing distantly related strains will reduce the presence of false positives introduced by population stratification (Wu et al. 2008). We removed the 5 non-M. m. domesticus derived strains (CAST/EiJ, CZECHII/EiJ, JF1/Ms, MSM/Ms, PWD/PhJ) from the data set and performed eQTL mapping in the remaining 31 M. m. domesticus inbred strains of the MDP for all 20,868 transcripts on the array. At an α=0.05 significance threshold there were 12,749 significant loci for 1,582 transcripts (mean of 8 loci per transcript) with a median eQTL interval of 24,832 bp. The resulting transcriptome map (Figure 1a) is difficult to interpret due to a large number of likely false positives. It has been shown for individual phenotypes that data from a linkage mapping study can be used in conjunction with an association study to produce more robust results (Manenti et al. 2009); thus, we reasoned that an independent mouse liver eQTL data obtained in BXD RI panel might be used to replicate eQTL and narrow the width of the candidate loci.
Using the BXD eQTL data to inform MDP Mapping
FastMap (Gatti et al. 2009) was used to perform eQTL mapping for each transcript using data from a BXD liver eQTL study (Gatti et al. 2007). In order to reduce the number of statistical tests performed in the MDP, we first selected 2,486 transcripts in the BXD panel that met eQTL significance thresholds of p ≤ 0.05. We performed eQTL mapping using FastMap in the MDP using these 2,486 transcripts and retained all peaks with p-values ≤ 0.05. We then selected three significance levels in the BXD panel (p ≤ 0.001, 0.01, 0.05) and the MDP (p ≤ 0.001, 0.01, 0.05) and compared the number of replicated eQTL at these thresholds (Table 1). At all MDP p-value thresholds, the number of replicated eQTL decreases with increasing stringency of the BXD threshold and cis-eQTL are more reproducible than trans-eQTL.
Table 1.
Total number of eQTL that replicate between the two data sets at varying significance levels in the BXD and MDP. Percent values are the number of eQTL that replicate divided by the number of significant eQTL in the BXD panel.
| BXD p-value | Sig. BXD transcripts | Inbred p-value | ||
|---|---|---|---|---|
| 0.001 (%) | 0.01 (%) | 0.05 (%) | ||
| 0.001 | 854 | 29 (3.4) | 69 (8.1) | 104 (12.2) |
| 0.01 | 1,338 | 29 (2.2) | 72 (5.4) | 119 (8.9) |
| 0.05 | 2,486 | 31 (1.2) | 77 (3.1) | 128 (5.1) |
| Sig. Inbred transcripts | 238 | 1,190 | 2,981 | |
Next, we selected a BXD threshold of p ≤ 0.05 and an MDP threshold of p ≤ 0.05 and compared eQTL peaks between the two data sets. We removed any transcripts from the MDP that showed significant loci on more than 5 chromosomes. At these thresholds, there were 2,981 significant loci in the MDP data for 369 transcripts with a mean number of significant loci per transcript of 8 (median = 3). Consistent with previous studies, we found cis-eQTL to be more reproducible than trans-eQTL, with 9.9% of cis-eQTL and 2.0% of trans-eQTL replicating between data sets (Table 2). We assessed the null probability of eQTL replication between the two panels of mice by selecting two transcripts at random from the BXD and MDP data sets and searching for a co-located eQTL in both panels within a 5 Mb window. Spurious overlap occurred only 971 times in 106 trials (0.097%), suggesting that the observed reproducibility in eQTL is not due to chance. In contrast, a study comparing eQTL in a BXD RI panel with those in a C57BL/5J × DBA/2J F2 cross found that 67% of the cis-eQTL and 23% of trans-eQTL replicated (Peirce et al. 2006). This is not surprising since it would be expected that there would be greater reproducibility between two panels derived from similar parental strains than between two panels with different breeding histories and different distributions of polymorphisms.
Table 2.
The number of replicated cis and trans eQTL between the BXD data set and the MDP at an inbred threshold of p ≤ 0.05 and several BXD p-values thresholds. Percent values are the number of replicated cis or trans eQTL divided by the number of cis or trans eQTL respectively in the BXD panel.
| BXD p-value | BXD eQTL | Replicated eQTL | |||
|---|---|---|---|---|---|
| Cis | Trans | Total | Cis (% of BXD cis) | Trans (% of BXD trans) | |
| 0.001 | 639 | 215 | 854 | 77 (12.1) | 27 (12.6) |
| 0.01 | 827 | 511 | 1,338 | 91 (11.0) | 28 (5.5) |
| 0.05 | 988 | 1,498 | 2,486 | 98 (9.9) | 30 (2.0) |
We next plotted the MDP eQTL location against the transcript location for each transcript that was significant in the BXD strains (Figure 1b, grey crosses), and overlaid eQTL that replicated between the two data sets on this plot (Figure 1b, red squares). Previous eQTL studies have found that the two most common features of transcriptome maps are 1) a band of cis-eQTL along the diagonal and 2) vertical trans-eQTL bands that represent eQTL hotspots (Kliebenstein 2008). Both of these features appear in the transcriptome map that results from the replicated eQTL between the BXD and MDP. Previously, we showed that there is a strong eQTL hotspot in the BXD panel on distal Chr 12 that involves over 100 transcripts (Gatti et al. 2007). In the male BXD strains there are 130 transcripts that have a maximum eQTL on Chr12 between 103 and 108 Mb at a p-value of 0.05 (104 transcripts at p ≤ 0.01). There is also an eQTL trans-band on Chr 7 in the BXD male data set. While the trans-band on Chr 7 does not replicate, we found that the eQTL trans-band on Chr 12 replicates in the independent MDP with 19 of the transcripts having a maximum eQTL on Chr12 between 104 and 105 Mb at a p-value ≤ 0.05 In order to assess the probability of seeing an eQTL trans-band of size 19 by chance, we applied a permutation technique that involves re-running the eQTL analysis repeatedly, reordering the strain names in the SNP data each time, and counting the number of eQTL that occur at each SNP (Breitling et al. 2008). We found that an eQTL trans-band of size 19 never occurred and that the largest eQTL trans-band (17 transcripts) occurred only once in 100 permutations (Sup. Fig. 1). From this, we conclude that the BXD eQTL trans-band found to replicate in the MDP is unlikely to have occurred by chance.
It has been demonstrated previously that cis-eQTL may be spuriously caused by polymorphisms that occur in transcript probe sequences (Alberts et al. 2007; Peirce et al. 2006). If this were the case, it would not be surprising to see these cis-eQTL replicate in the two panels of mice. Using a high density SNP data set (see Methods), we searched for polymorphisms within the probe sequences of the 91 cis-eQTL transcripts and found 28 with at least one SNP in the probe. We categorized the cis-eQTL as “C57BL/6J allele high” if a Student's T-test between the expression of strains with the C57BL/6J allele and those with the other allele produced a p-value < 0.5. We then performed Fisher's exact test to test the null hypothesis that a SNP in the probe sequence is equally likely to occur in a C57BL/6J-allele-high cis-eQTL as in a cis-eQTL that is high when the non-C57BL/6J allele is present. The result was not significant (p = 0.65), leading us to conclude that SNPs within the probe sequences are not responsible for the reproducibility of cis-eQTL between the two panels, a result concordant with other studies that have searched for cis-eQTL bias on the Agilent platform (Doss et al. 2005).
Narrowing eQTL using the MDP
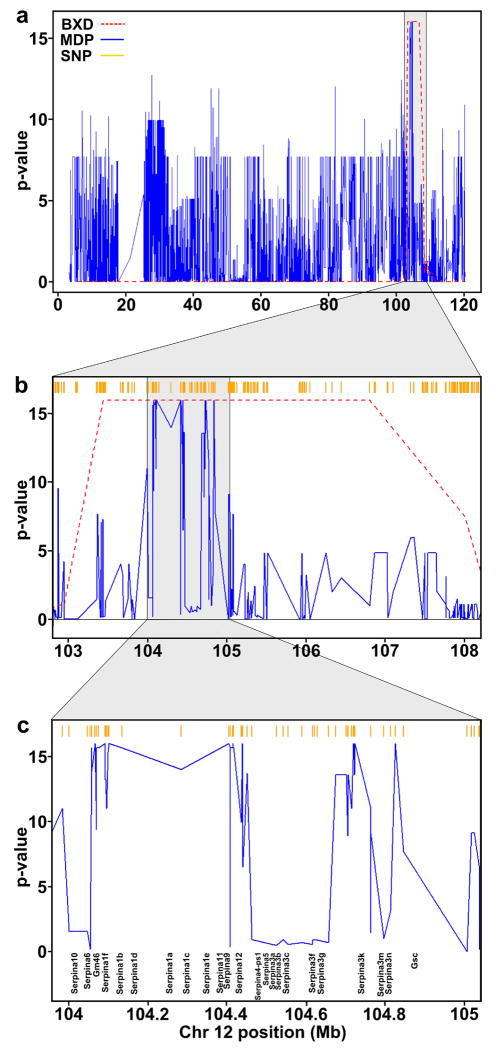
As mentioned above, while the BXD strains are an excellent resource for QTL mapping, the recombination block structure among the strains remains large. For example, the eQTL hotspot interval on distal Chr 12 is approximately 5 Mb wide in the BXD panel (Figure 2a, b), while being only ∼1 Mb in the MDP (Figure 2c). Similarly, among all significant eQTL in the BXD panel, the mean width of the QTL interval was 16.6 Mb (median = 12 Mb), while that of the replicated eQTL in the MDP was 0.33 Mb (median = 0.32 Mb). This reduction in size is likely due to the finer haplotype block structure resulting from the random mating during the development of these strains (Roberts et al. 2007b). The improved resolution provided by the MDP reduced the number of candidate QTGs that may be responsible for the Chr 12 eQTL hotspot from 60 to 11 genes, all of which are part of the serine protease inhibitor (serpin) family of genes.
Figure 2.
Reduction in eQTL hotspot width using laboratory inbred strains. Panel (a) shows the eQTL profile on Chr 12 for the BXD strains (red) and the laboratory inbred strains (black) using Fisher's combined method to aggregate the p-values of all transcripts that have a significant QTL at this locus in each panel. SNP density in the 156K data is shown in orange. Panels (b) & (c) show successively zoomed in regions of the hotspot and demonstrate the narrower eQTL interval produced by the inbred strains.
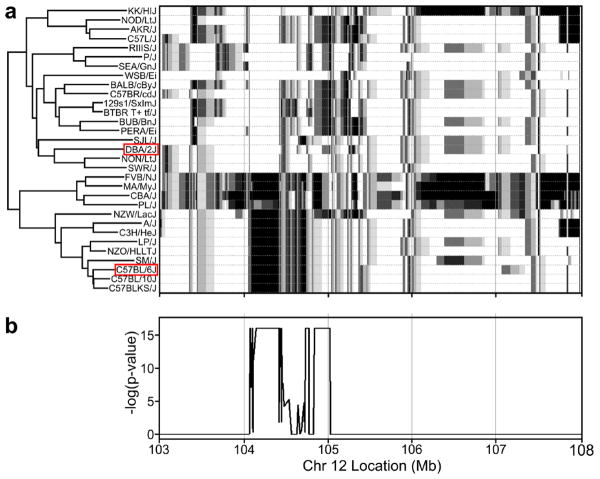
The serpin genes at this locus consist of a cluster of Serpina1 genes, which are the mouse orthologs of human α1-antitrypsin, and Serpina6, 9, 10, 11 and 12. A detailed analysis of the Serpina1 locus showed that there are seven paralogs of these genes, denoted as Serpina1a-e and Serpina-DOM6 & DOM7. DOM6 and DOM7 are two new isoforms of Serpina1 that were first identified by (Barbour et al. 2002). The quantity and membership of Serpina1 genes varies across the MDP. Some strains, including the C57BL/6J reference strain, contain Serpina1a through e. Other strains, including DBA/2J, contain Seripina1a,b and DOM6 or DOM7 (Barbour et al. 2002). To further elucidate possible candidate QTGs we performed a haplotype analysis of the SNPs between 103 & 108 Mb on Chr 12, which is the region of significance in the BXD strains, and found that strains in the MDP cluster (Figure 3a) in a manner similar to that detailed in (Barbour et al. 2002). We also plotted the aggregate significance score, represented by Fisher's combined statistic, of the 19 transcripts that replicate between mouse panels at the Chr 12 locus. The peak falls clearly between 104 and 104.5 Mb. C57BL/6J clusters with the minor allele whereas DBA/2J clusters with the major allele. Fisher's combined method was used to aggregate the p-values of the 19 transcripts in the MDP that have a significant eQTL at this locus and this was plotted on the same scale (Figure 3b). While it is possible that any gene that is in strong linkage disequilibrium with the Serpina1 genes are candidate eQTL regulators, we believe that this division and clustering of the inbred strains is consistent with the hypothesis that Serpina1 ortholog variation regulates the expression of the Chr12 hotspot.
Figure 3.
Haplotype analysis of Chr 12 eQTL hotspot. a) Grey shading represents the 8 possible haplotypes in a 3 SNP window for the 31 strains used in this study, clustered by haplotype pattern. b) –log(p-value) of Fisher's combined statistic for the 19 transcripts that replicate the Chr 12 eQTL hotspot between the BXD and inbred strains.
One advantage of using the commonly available MDP for eQTL replication is that the data need be collected only once for each organ. We have produced and made public gene expression data in the livers of 36 MDP (see Methods). As other investigators produce data for additional tissues in these strains, eQTL mapping can be performed to search for replicated eQTL across different tissues. Another advantage is that the QTL intervals in the MDP are narrower than in RI and F2 crosses, which reduces the number of candidate genes that the investigator must pursue.
At the same time, there are many potential pitfalls in eQTL mapping using the MDP (Breitling et al. 2008; Chesler et al. 2001). One drawback to the approach of selection of eQTL using independent strain panels is that a failure to replicate an eQTL is uninformative. For example, it has been estimated that 57% of the C57BL/6J and DBA/2J genomes are IBD (Doss et al. 2005). This means that many genes which are transcriptional regulators will not contain polymorphisms in crosses of these two strains and thus will not produce a differential effect in transcriptional regulation. Among the 128 reproducible eQTL, 4 (3.1%) occur in a region that was called IBD between C57BL/6J and DBA/2J. In contrast, among the 241 eQTL that are significant in the MDP but not in the BXD panel, 127 (52.7%) were in regions that are IBD between C57BL/6J and DBA/2J.
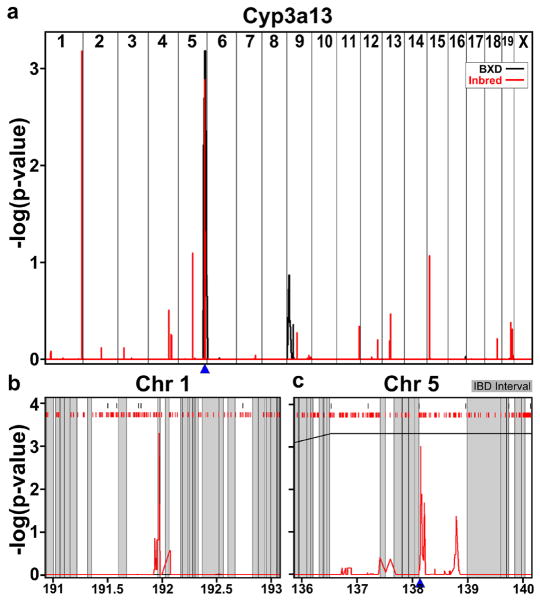
This reasoning can be illustrated with the following example. Cytochrome P450, family 3, subfamily a, polypeptide 13 (Cyp3a13) has one reproducible cis-eQTL on distal Chr 5 (Figure 4a, c) that falls in a region that is not IBD. In addition to the Chr 5 eQTL, the MDP also has a trans-eQTL on distal Chr 1 (Figure 4b) that is located in a region that is IBD in the BXD panel and therefore could not be detected due to the lack of polymorphisms. This locus contains one gene, prospero-related homeobox 1 (Prox1), which is a transcription factor involved in liver morphogenesis (Papoutsi et al. 2007) and tumor suppression (Shimoda et al. 2006). However, we were unable to find any connection between Prox1 and Cyp3a13 in the literature and therefore cannot indicate whether this is a true or false positive.
Figure 4.
a) Cyp3a13 QTL plot with the BXD p-values in black and the laboratory inbred p-values in red. Cyp3a13 location is shown by the blue triangle. b) Cyp3a13 QTL plot on Chr 1 with IBD intervals between C57BL/6J and DBA/2J shaded in grey, BXD markers along the top in black, laboratory inbred markers in red. c) Cyp3a13 QTL plot on Chr5.
Failure to replicate may also be due to variations in allele frequency across the genome in the MDP (Payseur et al. 2007). The average minor allele frequency (MAF) in the BXD panel is 0.48 whereas it is 0.26 in the MDP. A MAF closer to 0.5 will allow for greater power to detect expression differences between alleles and is one of the reasons that the BXD panel is a better mapping population. To see if this effect was partially responsible for eQTL that did not replicate, we calculated the MAF in the MDP at the locus that was most significant in the BXD panel for eQTL that replicated and for those that did not. Of the eQTL that replicated, 43% had a MAF between 0.4 and 0.6 in the MDP whereas only 24% had a MAF in this range among eQTL that did not replicate. This suggests that the variation in power due to MAF was partially responsible for eQTL that did not replicate between the panels.
Furthermore, an eQTL may not be replicable between the two data sets because there are complex interactions between genes that regulate gene expression. Due to the limited power provided by sampling between 30 and 40 strains and the fitting of a single locus model, it is unlikely that we can detect more than two loci with strong effect sizes for any transcript. It is also possible that an eQTL will fail to replicate because the expression of a transcript is spuriously associated with a genotype (Peng et al. 2007; Perez-Enciso et al. 2007). Such a false association may even occur for an eQTL hotspot since the expression of all transcripts that map to that locus is highly correlated. This means that if one of the transcripts is spuriously associated with a locus, then all other highly correlated transcripts are also associated that locus. The BXD male strains have another eQTL hotspot on proximal Chr 7 that fails to reproduce in the MDP and this hotspot may represent a false positive eQTL hotspot. However, the reproduction of the Chr 12 hotspot in two separate panels of mice increased the likelihood that it is an important regulatory locus in the mouse liver.
Conclusion
The selection of gene expression QTL that warrant further biological investigation is an arduous task. Here, we used MDP mice to determine what eQTL replicate with a previous linkage study in order to guide the selection of eQTL for further biological confirmation. Consistent with other reports, we found that cis-eQTL replicate with greater frequency than trans-eQTL. Importantly, we confirmed that a major liver-specific eQTL hotspot on distal Chr 12 is replicated in both data sets and used MDP mapping results to considerably narrow this interval of interest. When eQTL are reproducible, they offer investigators a set of candidates with which to pursue further analyses such as RNA interference knockdowns.
Supplementary Material
Acknowledgments
We thank several anonymous reviewers for excellent suggestions and criticism which strengthened the final manuscript. Financial support for these studies was provided, in part, by the United States Environmental Protection Agency grants RD833825 & F08D20579. However, the research described in this article has not been subjected to the Agency's peer review and policy review and therefore does not necessarily reflect the views of the Agency and no official endorsement should be inferred. DMG was also supported by the UNC Environmental Sciences & Engineering Interdisciplinary Fellowship.
References
- 1.Alberts R, Terpstra P, Li Y, Breitling R, Nap JP, et al. Sequence polymorphisms cause many false cis eQTL. PLoS ONE. 2007;2:e622. doi: 10.1371/journal.pone.0000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, et al. Standardizing global gene expression analysis between laboratories and across platforms. Nat Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- 3.Barbour KW, Wei F, Brannan C, Flotte TR, Baumann H, et al. The murine alpha(1)-proteinase inhibitor gene family: polymorphism, chromosomal location, and structure. Genomics. 2002;80:515–522. [PubMed] [Google Scholar]
- 4.Breitling R, Li Y, Tesson BM, Fu J, Wu C, et al. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 2008;4:e1000232. doi: 10.1371/journal.pgen.1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 6.Burgess-Herbert SL, Cox A, Tsaih SW, Paigen B. Practical applications of the bioinformatics toolbox for narrowing quantitative trait Loci. Genetics. 2008;180:2227–2235. doi: 10.1534/genetics.108.090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using “genetical genomics”. Nat Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- 8.Chesler EJ, Lu L, Shou S, Qu Y, Gu J, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 9.Chesler EJ, Rodriguez-Saz SL, Mogil JS. In Silico Mapping of Mouse Quantitative Trait Loci. Science. 2001;294:2423–2423. doi: 10.1126/science.294.5551.2423a. [DOI] [PubMed] [Google Scholar]
- 10.Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 11.de Koning DJ, Haley CS. Genetical genomics in humans and model organisms. Trends Genet. 2005;21:377–381. doi: 10.1016/j.tig.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Dipetrillo K, Wang X, Stylianou IM, Paigen B. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 2005;21:683–692. doi: 10.1016/j.tig.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrall M. Quantitative genetic variation: a post-modern view. Hum Mol Genet. 2004;13:R1–R7. doi: 10.1093/hmg/ddh084. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 15.Gatti D, Maki A, Chesler EJ, Kirova R, Kosyk O, et al. Genome-level analysis of genetic regulation of liver gene expression networks. Hepatology. 2007;46:548–557. doi: 10.1002/hep.21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatti DM, Shabalin AA, Lam TC, Wright FA, Rusyn I, et al. FastMap: fast eQTL mapping in homozygous populations. Bioinformatics. 2009;25:482–489. doi: 10.1093/bioinformatics/btn648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kliebenstein D. Quantitative Genomics: Analyzing Intraspecific Variation Using Global Gene Expression Polymorphisms or eQTL. Annu Rev Plant Biol. 2008 doi: 10.1146/annurev.arplant.043008.092114. [DOI] [PubMed] [Google Scholar]
- 19.Li R, Lyons MA, Wittenburg H, Paigen B, Churchill GA. Combining data from multiple inbred line crosses improves the power and resolution of quantitative trait loci mapping. Genetics. 2005;169:1699–1709. doi: 10.1534/genetics.104.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malmanger B, Lawler M, Coulombe S, Murray R, Cooper S, et al. Further studies on using multiple-cross mapping (MCM) to map quantitative trait loci. Mamm Genome. 2006;17:1193–1204. doi: 10.1007/s00335-006-0070-2. [DOI] [PubMed] [Google Scholar]
- 21.Manenti G, Galvan A, Pettinicchio A, Trincucci G, Spada E, et al. Mouse genome-wide association mapping needs linkage analysis to avoid false-positive Loci. PLoS Genet. 2009;5:e1000331. doi: 10.1371/journal.pgen.1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClurg P, Janes J, Wu C, Delano DL, Walker JR, et al. Genomewide association analysis in diverse inbred mice: power and population structure. Genetics. 2007;176:675–683. doi: 10.1534/genetics.106.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, et al. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paigen K, Eppig JT. A mouse phenome project. Mamm Genome. 2000;11:715–717. doi: 10.1007/s003350010152. [DOI] [PubMed] [Google Scholar]
- 25.Papoutsi M, Dudas J, Becker J, Tripodi M, Opitz L, et al. Gene regulation by homeobox transcription factor Prox1 in murine hepatoblasts. Cell Tissue Res. 2007;330:209–220. doi: 10.1007/s00441-007-0477-4. [DOI] [PubMed] [Google Scholar]
- 26.Payseur BA, Place M. Prospects for association mapping in classical inbred mouse strains. Genetics. 2007;175:1999–2008. doi: 10.1534/genetics.106.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peirce JL, Broman KW, Lu L, Williams RW. A simple method for combining genetic mapping data from multiple crosses and experimental designs. PLoS ONE. 2007;2:e1036. doi: 10.1371/journal.pone.0001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peirce JL, Li H, Wang J, Manly KF, Hitzemann RJ, et al. How replicable are mRNA expression QTL? Mamm Genome. 2006;17:643–656. doi: 10.1007/s00335-005-0187-8. [DOI] [PubMed] [Google Scholar]
- 29.Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng J, Wang P, Tang H. Controlling for false positive findings of trans-hubs in expression quantitative trait loci mapping. BMC Proc. 2007;1 1:S157. doi: 10.1186/1753-6561-1-s1-s157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Enciso M, Quevedo JR, Bahamonde A. Genetical genomics: use all data. BMC Genomics. 2007;8:69. doi: 10.1186/1471-2164-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts A, McMillan L, Wang W, Parker J, Rusyn I, et al. Inferring missing genotypes in large SNP panels using fast nearest-neighbor searches over sliding windows. Bioinformatics. 2007a;23:i401–i407. doi: 10.1093/bioinformatics/btm220. [DOI] [PubMed] [Google Scholar]
- 33.Roberts A, Pardo-Manuel d V, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome. 2007b;18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 35.Shi C, Uzarowska A, Ouzunova M, Landbeck M, Wenzel G, et al. Identification of candidate genes associated with cell wall digestibility and eQTL (expression quantitative trait loci) analysis in a Flint × Flint maize recombinant inbred line population. BMC Genomics. 2007;8:22. doi: 10.1186/1471-2164-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shifman S, Bell JT, Copley RR, Taylor MS, Williams RW, et al. A High-Resolution Single Nucleotide Polymorphism Genetic Map of the Mouse Genome. PLoS Biol. 2006;4:e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimoda M, Takahashi M, Yoshimoto T, Kono T, Ikai I, et al. A homeobox protein, prox1, is involved in the differentiation, proliferation, and prognosis in hepatocellular carcinoma. Clin Cancer Res. 2006;12:6005–6011. doi: 10.1158/1078-0432.CCR-06-0712. [DOI] [PubMed] [Google Scholar]
- 38.Szatkiewicz JP, Beane GL, Ding Y, Hutchins L, Pardo-Manuel d V, et al. An imputed genotype resource for the laboratory mouse. Mamm Genome. 2008;19:199–208. doi: 10.1007/s00335-008-9098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor BA, Wnek C, Kotlus BS, Roemer N, MacTaggart T, et al. Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm Genome. 1999;10:335–348. doi: 10.1007/s003359900998. [DOI] [PubMed] [Google Scholar]
- 40.Threadgill DW, Hunter KW, Williams RW. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm Genome. 2002;13:175–178. doi: 10.1007/s00335-001-4001-Y. [DOI] [PubMed] [Google Scholar]
- 41.Walling GA, Visscher PM, Andersson L, Rothschild MF, Wang L, et al. Combined analyses of data from quantitative trait loci mapping studies. Chromosome 4 effects on porcine growth and fatness. Genetics. 2000;155:1369–1378. doi: 10.1093/genetics/155.3.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
- 43.West MA, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, et al. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics. 2007;175:1441–1450. doi: 10.1534/genetics.106.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu C, Delano DL, Mitro N, Su SV, Janes J, et al. Gene set enrichment in eQTL data identifies novel annotations and pathway regulators. PLoS Genet. 2008;4:e1000070. doi: 10.1371/journal.pgen.1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q, McMillan L, Pardo-Manuel de Villena F, Threadgill DW, Wang W. Inferring genome-wide mosaic structure. Proceedings of the 14th Pacific Symposium on Biocomputing (PSB) 2009;14:150–161. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.