Abstract
It is thought that bacteria excrete redox-active pigments as antibiotics to inhibit competitors. In Pseudomonas aeruginosa, the endogenous antibiotic pyocyanin activates SoxR, a transcription factor conserved in Proteo- and Actinobacteria. In Escherichia coli, SoxR regulates the superoxide stress response. Bioinformatic analysis coupled with gene expression studies in P. aeruginosa and Streptomyces coelicolor revealed that the majority of SoxR regulons in bacteria lack the genes required for stress responses, despite the fact that many of these organisms still produce redox-active small molecules, which indicates that redox-active pigments play a role independent of oxidative stress. These compounds had profound effects on the structural organization of colony biofilms in both P. aeruginosa and S. coelicolor, which shows that “secondary metabolites” play important conserved roles in gene expression and development.
The opportunistic pathogen Pseudomonas aeruginosa releases phenazines, redox-active antibiotics (1, 2). Historically, attention has focused on their toxicity in bacteria and eukaryotes, which arises from the production of superoxide (3, 4). More recently, however, it has been recognized that these compounds have diverse physiological functions, particularly under oxygen-limited conditions (2, 5-7). We found that the blue phenazine pyocyanin is an intercellular signal that triggers a specific response in P. aeruginosa, with only 22 genes up-regulated, including the complete SoxR regulon (8). The transcription factor SoxR is well-characterized in the enteric bacteria Escherichia coli and Salmonella enterica serovar Typhimurium as a stress-response regulator. In these bacteria, SoxR activates the transcription factor SoxS, which controls genes involved in the removal of superoxide and nitric oxide and protection from organic solvents and antibiotics. That SoxR-regulated genes were triggered by pyocyanin was, therefore, initially not surprising, as this would be consistent with the conventional view of phenazines as toxic compounds (9-11).
However, recent studies of the SoxR regulons in pseudomonads indicate an alternative role to the E. coli SoxR-SoxS paradigm. First, superoxide is not the sole activator of SoxR, as P. aeruginosa pyocyanin also induces the expression of its regulon under anoxic conditions (8). Second, SoxRs from Pseudomonas putida (12) and P. aeruginosa (8, 13, 14) do not control any of the genes typically involved in superoxide resistance and detoxification, rather, SoxR from P. aeruginosa up-regulates expression of two transporters and a putative monooxygenase (fig. S1A). Third, P. aeruginosa soxR mutants show no decrease in resistance to superoxide, unlike E. coli soxR mutants (14). These observations led us to hypothesize that redox-active signaling molecules, such as phenazines, might control other aspects of microbial behavior.
In this study, we investigated the distribution of the E. coli—type oxidative stress response by performing a BLAST search for SoxR and SoxS in the bacterial domain (15). SoxR was found in sequences from 176 strains in the phyla Proteobacteria and Actinobacteria (Fig. 1A), 123 of which come from completed genomes. The occurrence of SoxS was restricted to the family Enterobacteriaceae. To identify alternative SoxR targets in non-enterics, we searched all available complete bacterial genomes (616) for the presence of soxRboxes (i.e., SoxR-binding sites in the promoter regions of target genes) using a position weight matrix (PWM) derived from the soxRbox sequences of 12 diverse SoxR-containing bacteria (fig. S1B). This PWM permits statistically robust predictions of SoxR binding to a soxRbox. Of the 123 soxR-containing genomes, 121 contain soxRboxes. SoxRboxes were also found in 27 genomes (19 were Firmicutes) that do not contain a soxR homolog. The results of our analysis (table S1 and http://soxRbox.mit.edu) were consistent with gene expression studies made in the Gram-negative bacteria E. coli, S. enterica (10), P. aeruginosa (8, 13, 14), and Agrobacterium tumefaciens (16), which validates our search algorithm.
Fig. 1.
(A) Distribution of SoxR and SoxS among phyla of the domain Bacteria. A BLAST search for E. coli SoxR and SoxS was performed, and SoxS was found only in enterics. SoxR homologs were identified in 176 α-, β-, δ-, and γ-Proteobacteria and Actinobacteria. All of these homologs contain the SoxR-specific cysteine motif CI[G/Q]CGC[L/M][S/L]XXXC required for binding of the [2Fe-2S] cluster (31). The number of hits within respective phyla are indicated, followed by the total number of genomes surveyed. Members of these phyla (in black) are noted for their ability to produce and excrete redox-active small molecules, such as phenazines (18) and actinorhodin (20). Representative structures are shown. The tree was constructed using the ARB neighbor joining method from 16S ribosomal RNAs of 604 bacterial species. The bar represents 0.1 base substitutions per nucleotide. (B) Gene categories regulated by SoxR. Only in enterics are soxRboxes located upstream of soxS, which confirms the uniqueness of this network. In all other soxR-containing Proteo- and Actinobacteria, soxRboxes are mainly found upstream of five gene types as indicated; 100% correspond to 16 α-Proetobacteria, 18 β-Proteobacteria, 27 enteric, 38 non-enteric γ-Proteobacteria, or 22 Actinobacteria. “Dehydr.” stands for putative dehydrogenases; “oxygen.” for putative mono- or dioxygenases; “L-PSP” putative L-PSP endoribonucleases, a family of ribonucleases that share homology with the rat liver perchloric acid-soluble protein, L-PSP; and “methyl./acetylase” putative methyl- or acetyltransferases. Additional annotation information can be found at http://soxRbox.mit.edu.
The organization found in E. coli (fig. S1), with one soxRbox upstream of soxS and no other soxRboxes in the genome, occurred only in enterics (27 genomes) (Fig. 1B). Two enterics contained an additional soxRbox upstream of putative dioxygenases. The remaining organisms contained one or more soxRboxes upstream of genes other than soxS. These SoxR target genes fell into five main categories, including transporters, oxygenases, dehydrogenases, putative acetyl- or methyltransferases, and L-PSP endoribonucleases (L-PSP is defined in the legend to Fig. 1B), all of which are potentially involved in the transformation or transport of small molecules, such as antibiotics (17). The occurrence of soxR upstream of soxS in enterics thus appears to be an evolutionary exception confirmed by the unique branching of the enteric orthologs on a SoxR phylogenetic tree (fig. S2).
Given that many of the bacteria that contain soxRboxes are producers of redox-active antibiotics (18) (Fig. 1A), it seems reasonable that SoxR may have evolved to regulate their transport and/or turnover. We chose to work with the Gram-positive actinomycete Streptomyces coelicolor A3(2) to test whether the SoxR regulon is up-regulated in response to endogenous small molecules, because members of this phylum are widely recognized as important sources of antibiotics (19). S. coelicolor A3(2) produces the blue pigment actinorhodin and the red undecylprodigiosin (fig. S3) (20). On the basis of our analysis, we predicted a SoxR regulon comprising two genes for S. coelicolor A3(2), encoding putative redox enzymes (Fig. 2A). Expression of these genes in the wild type (strain M145) was compared with that in a mutant that does not produce the two pigments (strain M512) (21). Both predicted SoxR-regulated genes were significantly up-regulated in the wild type relative to the pigment-null mutant (∼250- to 6000-fold), as determined by quantitative reverse transcription polymerase chain reaction (RT-PCR) (Fig. 2B), which confirmed that pigment production stimulated gene expression via SoxR. Hence, the primary function of SoxR in S. coelicolor, as in P. aeruginosa, is not to activate a response to superoxide but to mediate a response to endogenous pigments.
Fig. 2.
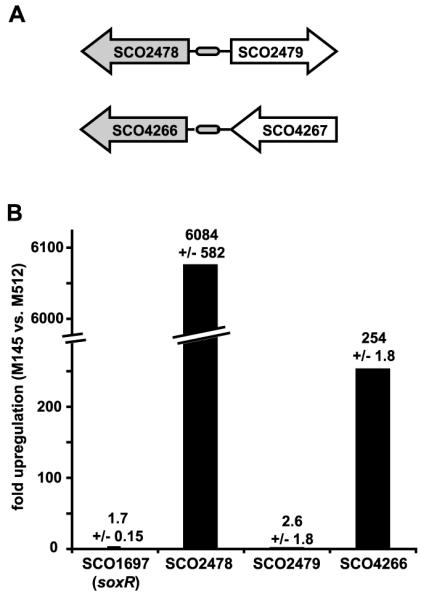
The putative S. coelicolor A3(2) SoxR regulon is specifically up-regulated by pigments. (A) Genes predicted to be regulated by SoxR are shown in gray. (B) RNA extracted from plate-grown S. coelicolor A3(2) M145 and the pigment-null mutant M512 was used to generate cDNA for quantitative RT-PCR (15). Signals were standardized to SCO4548 (32). The experiment was done in triplicate, and data reported represent the mean ±SD. SoxR itself (SCO1697) was also tested for changes in gene expression.
Recently, we showed that phenazines are terminal signals in P. aeruginosa’s quorumsensing cascade (8). The importance of quorum sensing for the coordination of many bacterial communities is well established (22). Moreover, a phenazine-dependent effect on biofilm formation has been reported in P. aureofaciens (23). Together with our bioinformatic SoxR results, these observations led us to hypothesize that redox-active pigments might act as signals to modulate the structural organization of cellular communities.
To investigate the effect of extracellular pigments on community development, we began by focusing on P. aeruginosa PA14. We spotted 10-μl aliquots of late exponential-phase cultures onto agar plates and incubated them at room temperature for 8 days. Under these conditions, wild-type cells initially formed smooth colonies (Fig. 3). After 4 days of incubation, the colonies began to wrinkle and reached a maximum area of ∼2.5 cm2 (Figs. 3 and 4A). However, the phenazine-null mutant formed severely wrinkled colonies within 2 days of incubation (Fig. 3), which subsequently flattened and spread to ∼3.5 cm2 (Fig. 4A). In contrast, a mutant that over-produced pyocyanin (DKN370) remained smooth and compact (Figs. 3 and 4A). These results demonstrated a role for phenazines in controlling bacterial colony size and structure.
Fig. 3.
Phenazine production modulates colony morphology in P. aeruginosa PA14. P. aeruginosa cultures were spotted onto agar plates containing Congo Red and Coomassie Blue, and incubated at 20°C for 6 days. The phenazine null strain (Δphz) started to wrinkle on day 2, the wild type (wt) wrinkled on day 3, and the soxR and mexGHI-opmD deletion strains wrinkled on day 5, whereas a pyocyanin overproducer (DKN370) remained smooth and white after 6 days.
Fig. 4.
(A) Surface coverage of 35 colonies per strain monitored over 8 days (±SD). (B) Concentration of pyocyanin release from three colonies into 10 ml agar supplemented with Congo Red and Coomassie Blue. After 5 days of growth at room temperature, the cells were scraped off, pyocyanin was extracted from the agar using chloroform, and extracts were analyzed by high-performance liquid chromatography. The data reported represent the mean ±SD. (C) Spore suspensions of S. coelicolor A3(2) M145 and the pigment mutant M512 were spotted and incubated for 5 days on R5- medium at room temperature. The pigment mutant exhibits a wrinkled morphology, whereas the wild type takes on a smoother phenotype. Scale bar is 0.5 cm.
Phenazines are diffusible molecules that may influence phenotype over distance. Indeed, we found that adding pyocyanin to the growth medium (fig. S4A) or spotting the phenazine overproducer next to the phenazine deletion mutant (fig. S4B) resulted in the formation of smooth compact colonies. We tested the role of SoxR in mediating the effect of phenazines on colony morphology by making a SoxR-deletion mutant. However, the mutant behaved similarly to the pyocyanin overproducer, and colonies remained smooth for 4 days (Fig. 3). As for the overproducer, ΔsoxR released more pyocyanin into the agar than the wild type (Fig. 4B). There thus appears to be a direct correlation between pyocyanin release and colony smoothness.
To further analyze the ΔsoxR phenotype, we tested P. aeruginosa mutants disrupted in the SoxR target genes PA14_35160 (encoding a putative monooxygenase), mexGHI-opmD [encoding a resistance-nodulation—cell division (RND) efflux pump], and PA14_16310 [encoding a major facilitator superfamily (MFS) transporter]. Deletions of PA14_35160 and PA14_16310 did not affect colony morphology; however, the loss of mexGHI-opmD produced a phenotype that looked like the ΔsoxR mutant, i.e., wrinkling was slow (Fig. 3), and was accompanied by a slightly elevated pyocyanin release (Fig. 4B). By contrast, the release of the yellow phenazine-1-carboxylate (PCA) and an unidentified red phenazine (possibly 5-methyl-PCA), decreased by 10 and 60%, respectively, in the mexGHI-opmD mutant relative to the wild type, which indicates that mexGHI-opmD is a general phenazine transporter. Antibiotic biosynthetic genes are often found adjacent to their cognate transporter (24), so it is interesting to note that the mexGHI-opmD operon is clustered with the phenazine biosynthetic genes phzM, phzA1-G1, and phzS (fig. S5A).
PhzA1-G1 synthesizes the yellow phenazine PCA, and PhzM methylates PCA to yield the red phenazine 5-methyl-PCA, which is then hydroxylated by PhzS to form pyocyanin. Transposon insertion mutants in mexI and opmD of P. aeruginosa PAO1 are known to accumulate an unidentified toxic compound that causes an elongated lag phase in planktonic cultures (25). We found a similar phenotype in P. aeruginosa PA14 (fig. S5B), which is probably caused by an intracellular accumulation of phenazines. Our experiments showed that SoxR target genes do not directly influence colony development; instead, SoxR regulates the efflux of phenazines via the RND transporter MexGHI-OpmD. Although yellow PCA and red phenazine are retained in the mexGHI-opmD mutant, the release of pyocyanin indicates an alternative efflux mechanism favoring pyocyanin. Compensatory changes in expression of RND efflux pumps are well known to occur in P. aeruginosa (26).
To determine whether the phenotypic effects of pigment production observed for P. aeruginosa were unique to this organism or more generalizable, we performed analogous experiments with S. coelicolor A3(2). As for P. aeruginosa, a pigment-defective mutant of S. coelicolor adopted a more wrinkled morphology than the respective wild type (Fig. 4C). The mechanisms whereby pigments control colony morphology are not understood, but are likely to be complex. For P. aeruginosa PA14, we know that pyocyanin affects the expression of at least 35 genes other than those in the SoxR regulon (8)and has profound effects on the cell’s physiology, including the redox state of the intracellular nicotinamide adenine dinucleotide [NAD(H)] pool (27). Any number of these effects may contribute, both directly and indirectly, to the ultimate architectures observed. One component that is likely involved is extracellular polysaccharide (EPS). Congo Red, a constituent of the agar used in the experiments shown in Fig. 3, is known to bind the gluose-rich exopolysaccharide PEL (28). Because the phenazine-null mutant is bright red, whereas the pyocyanin overproducer is pale, we infer there is an inverse relationship between phenazine and PEL production (Fig. 3). How phenazines affect the pel genes and how such changes in EPS composition contribute to colony morphogenesis remain to be determined.
Pigments excreted by bacteria have long been assumed to be “secondary” metabolites or even waste products, owing to the sporadic strain- and condition-dependent nature of their production (29). Many of these redox-active compounds are known to have antibiotic activities toward competing cells (1, 20), but until recently, their potential to directly participate in the physiology of the producing organism has been largely neglected (7). We now know that small molecules initially characterized as antibiotics allow intercellular communication within bacterial populations (30), and this work implies a conserved function for redox-active pigment antibiotics of the Gram-negative bacterium P. aeruginosa and the Gram-positive bacterium S. coelicolor A3(2). These pigments influence transcriptional regulation and modulate the physical characteristics of communities of their producers at later stages in their development. Rather than being “secondary,” diverse redox-active antibiotics may share similar functions of primary importance throughout the bacterial domain.
Supplementary Material
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/321/5893/1203/DC1 Materials and Methods Figures S1 to S5 Table S1 References
References and Notes
- 1.Mavrodi DV, Blankenfeldt W, Thomashow LS. Annu. Rev. Phytopathol. 2006;44:417. doi: 10.1146/annurev.phyto.44.013106.145710. [DOI] [PubMed] [Google Scholar]
- 2.Price-Whelan A, Dietrich LE, Newman DK. Nat. Chem. Biol. 2006;2:71. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 3.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Cell. 1999;96:47. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 4.Mazzola M, Cook RJ, Thomashow LS, Weller DM, Pierson LS., 3rd Appl. Environ. Microbiol. 1992;58:2616. doi: 10.1128/aem.58.8.2616-2624.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez ME, Kappler A, Newman DK. Appl. Environ. Microbiol. 2004;70:921. doi: 10.1128/AEM.70.2.921-928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Newman DK. Environ. Sci. Technol. 2008;42:2380. doi: 10.1021/es702290a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez ME, Newman DK. Cell. Mol. Life Sci. 2001;58:1562. doi: 10.1007/PL00000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. Mol. Microbiol. 2006;61:1308. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 9.Liochev SI, Benov L, Touati D, Fridovich I. J. Biol. Chem. 1999;274:9479. doi: 10.1074/jbc.274.14.9479. [DOI] [PubMed] [Google Scholar]
- 10.Pomposiello PJ, Demple B. J. Bacteriol. 2000;182:23. doi: 10.1128/jb.182.1.23-29.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsaneva IR, Weiss B. J. Bacteriol. 1990;172:4197. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park W, Pena-Llopis S, Lee Y, Demple B. Biochem. Biophys. Res. Commun. 2006;341:51. doi: 10.1016/j.bbrc.2005.12.142. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi K, Tagawa S. J. Biochem. 2004;136:607. doi: 10.1093/jb/mvh168. [DOI] [PubMed] [Google Scholar]
- 14.Palma M, et al. Infect. Immun. 2005;73:2958. doi: 10.1128/IAI.73.5.2958-2966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Materials and methods are available as supporting material on Science Online.
- 16.Eiamphungporn W, Charoenlap N, Vattanaviboon P, Mongkolsuk S. J. Bacteriol. 2006;188:8669. doi: 10.1128/JB.00856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cundliffe E. Annu. Rev. Microbiol. 1989;43:207. doi: 10.1146/annurev.mi.43.100189.001231. [DOI] [PubMed] [Google Scholar]
- 18.Turner JM, Messenger AJ. Adv. Microb. Physiol. 1986;27:211. doi: 10.1016/s0065-2911(08)60306-9. [DOI] [PubMed] [Google Scholar]
- 19.Hopwood DA. Streptomyces in Nature and Medicine: The Antibiotic Makers. Oxford Univ. Press; Oxford: 2007. [Google Scholar]
- 20.Chater KF. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:761. doi: 10.1098/rstb.2005.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Floriano B, Bibb M. Mol. Microbiol. 1996;21:385. doi: 10.1046/j.1365-2958.1996.6491364.x. [DOI] [PubMed] [Google Scholar]
- 22.Davies DG, et al. Science. 1998;280:295. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 23.Maddula VS, Zhang Z, Pierson EA, Pierson LS., 3rd Microb. Ecol. 2006;52:289. doi: 10.1007/s00248-006-9064-6. [DOI] [PubMed] [Google Scholar]
- 24.Tahlan K, et al. Mol. Microbiol. 2007;63:951. doi: 10.1111/j.1365-2958.2006.05559.x. [DOI] [PubMed] [Google Scholar]
- 25.Aendekerk S, et al. Microbiology. 2005;151:1113. doi: 10.1099/mic.0.27631-0. [DOI] [PubMed] [Google Scholar]
- 26.Li XZ, Barre N, Poole K. J. Antimicrob. Chemother. 2000;46:885. doi: 10.1093/jac/46.6.885. [DOI] [PubMed] [Google Scholar]
- 27.Price-Whelan A, Dietrich LE, Newman DK. J. Bacteriol. 2007;189:6372. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman L, Kolter R. Mol. Microbiol. 2004;51:675. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 29.Williams RP. Bacteriol. Rev. 1956;20:282. doi: 10.1128/br.20.4.282-284.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yim G, Wang HH, Davies J. Philos. Trans. R. Soc. London B Biol. Sci. 2007;362:1195. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Single-letter abbreviations for the amino acid residues are as follows:A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; Y, Tyr; and X, any amino acid.
- 32.Mehra S, et al. J. Ind. Microbiol. Biotechnol. 2006;33:159. doi: 10.1007/s10295-005-0034-7. [DOI] [PubMed] [Google Scholar]
- 33.We thank P. Straight (Harvard University); M. Bibb and A. Hesketh (John Innes Centre, UK) for providing S. coelicolor A3(2) strains M145 and M512; and N. C. Caiazza, C. T. Brown, and B. Wold for helpful discussions. This work was supported by an EMBO Long Term Fellowship (L.E.P.D.) and grants from the Packard Foundation and Howard Hughes Medical Institute (D.K.N.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





