Abstract
We used digital long serial analysis of gene expression to discover gene expression differences between node-negative and node-positive colorectal tumors and developed a multigene classifier able to discriminate between these two tumor types. We prepared and sequenced long serial analysis of gene expression libraries from one node-negative and one node-positive colorectal tumor, sequenced to a depth of 26,060 unique tags, and identified 262 tags significantly differentially expressed between these two tumors (P < 2 x 10-6). We confirmed the tag-to-gene assignments and differential expression of 31 genes by quantitative real-time polymerase chain reaction, 12 of which were elevated in the node-positive tumor. We analyzed the expression levels of these 12 upregulated genes in a validation panel of 23 additional tumors and developed an optimized seven-gene logistic regression classifier. The classifier discriminated between node-negative and node-positive tumors with 86% sensitivity and 80% specificity. Receiver operating characteristic analysis of the classifier revealed an area under the curve of 0.86. Experimental manipulation of the function of one classification gene, Fibronectin, caused profound effects on invasion and migration of colorectal cancer cells in vitro. These results suggest that the development of node-positive colorectal cancer occurs in part through elevated epithelial FN1 expression and suggest novel strategies for the diagnosis and treatment of advanced disease.
Introduction
Colorectal cancer is the third leading cause of cancer deaths in the United States. It is estimated that 149,000 individuals will have their conditions diagnosed with colorectal cancer during 2008 and that 50,000 deaths will result from this disease. Diagnosis before spread to the regional lymph nodes (node-negative disease) is associated with a 90% 5-year survival rate, whereas diagnosis after lymphatic spread (node-positive disease) decreases survival to a 67% 5-year rate [1]. The diagnosis of node-positive disease is accomplished by histologic examination of regional lymph nodes available within the surgically excised tissue. The probability of disease-free survival of patients diagnosed with node-negative disease increases with the number of negative nodes observed [2]. The mechanism by which node sampling is related to outcome is not clear, but one possibility is that a portion of node-negative individuals is understaged because of insufficient sampling depth. Current guidelines recommend that colorectal cancer patients have a minimum of 12 lymph nodes examined; however, only 37% of patients receive the proper level of lymph node evaluation [3]. Despite careful attention to node status at the time of surgery, a significant number of node-negative patients experience a recurrence of their disease within 5 years and may have been understaged. A molecular test that identifies node-positive colorectal cancer would therefore be of substantial clinical value.
To identify gene expression markers of lymph node involvement, expression profiling of colorectal cancers has been performed and genes associated with node status were reported [4–6]. In addition, gene expression profiling has been used to identify markers associated with node status in other epithelial neoplasms such as pancreatic cancer [7], oral squamous cell carcinoma [8], and invasive breast cancer [9]; however, the mechanisms by which these marker genes influence pathology remain to be elucidated.
We applied high-throughput pyrosequencing to long serial analysis of gene expression (SAGE) to obtain deep expression profiles of lymph node-negative and lymph node-positive human colorectal cancers. From this, we observed 262 tags that were significantly differentially expressed and confirmed the tag-to-gene assignments of 30 genes with altered expression between these two tumor types. In particular, we found that node-negative tumor epithelial cells express low levels of FN1 messenger mRNA (mRNA) and protein, whereas node-positive tumor epithelial cells express high levels of FN1 mRNA and protein. We subsequently demonstrated that forced overexpression of FN1 dramatically increased the migratory and invasive properties of SW480 colorectal cancer cells. These results indicate that elevated epithelial FN1 expression in primary colorectal cancers may be a useful molecular marker of positive lymph node status and suggest that fibronectin-integrin antagonists may be useful additions to standard chemotherapy treatments for node-positive colorectal cancer patients.
Materials and Methods
Tumor Specimens and Laser Capture Microdissection
Twenty-five primary colorectal cancers were collected at the time of surgery by the South Carolina Biorepository System and immediately frozen at -80°C. Tissue samples were embedded in frozen-section medium (RA Scientific, Kalamazoo, MI), cut into 25 x 20-µm sections, and fixed onto silane-prep glass slides (Sigma, St Louis, MO) by sequential dehydration in baths of 75%, 95%, and 100% ethanol followed by xylene. Slides were air-dried and desiccated before laser capture microdissection. Guide slides were cut and stained with hematoxylin and eosin to aid in the identification of epithelial cells during laser capture dissection of unstained sections. Tumor epithelial cells were captured onto CapSure Macro LCM Caps (Molecular Devices, Sunnyvale, CA) using an ArcturusPixCellIIe laser capture microdissector. Caps containing excised cells were immediately placed onto 0.5-ml Eppendorf tubes containing 200 µl of lysis/binding buffer (Invitrogen, Carlsbad, CA) up to five aliquots of lysis buffer were sequentially used to lyse five caps each to create a total lysate of approximately 1 ml.
Genomic DNA Purification, Quantification, and Mismatch Repair Proficiency Testing
Genomic DNA was purified from microdissected epithelial cell lysates using QIAamp DNA Micro Kit (Qiagen, Valencia, CA). DNA was quantitated using real-time polymerase chain reaction (PCR) primers directed against long interspersed nuclear element sequences (LINEF-AAAGCCGCTCAACTACATGG, LINER-CTCTATTTCCTTCAGTTCTGCTC; Integrated DNA Technologies, Coralville, IA). PCR was performed using 6.25 µl of iTaqSupermix (Bio-Rad, Hercules, CA), 1.25 µl of 2 µM LINEF, 1.25 µl of 2 µM LINER, 2.5 µl of PCR water (Invitrogen), and 1.25 µl of DNA template. Thermal cycling was performed using a MyIQ Thermal Cycler (Bio-Rad) and the following protocol: 1 cycle at 95°C for 1 minute, 60 cycles at 94°C for 10 seconds and at 59°C for 30 seconds, and 1 cycle at 70°C for 5 minutes.
Mismatch repair-proficient and -deficient tumors were identified by assessing the stability of the BAT26 microsatellite locus in each tumor. BAT26 microsatellite PCR products were prepared using 6.25 µl of iTaqSupermix (Bio-Rad), 1.25 µl of 2 µM BAT26F (TGACTACTTTTGACTTCAGCC), 1.25 µl of 2 µM BAT26R (AACCATTCAACATTTTTAACCC), 2.5 µl of PCR water (Invitrogen), and 1.25 µl of DNA template. Thermal cycling was performed MyIQ Thermal Cycler (Bio-Rad) using the following protocol: 1 cycle at 95°C for 1 minute, 60 cycles at 94°C for 10 seconds and at 59°C for 45 seconds, and 1 cycle at 68°C for 5 minutes. PCR products were analyzed by electrophoresis on a 3% agarose gel and photographed using an Alpha Imager and Quantity One software (Alpha Innotech, San Leandro, CA). Only microsatellite stable tumors were used for gene expression studies.
Messenger RNA Purification and Complementary DNA Synthesis
Messenger RNA was isolated from microdissected epithelial cells using mRNA direct beads from the I-SAGE Long Kit (Invitrogen) according to the manufacturer's instructions. Complementary DNA (cDNA) was prepared by reverse transcription in a 90-µl reaction volume containing 18 µl of 5x first-strand buffer, 1 µl of RNAse OUT, 54.5 µl of diethylpyrocarbonate water, 9 µl of 0.1 M DTT, 4.5 µl of 10 mM deoxyribonucleotide triphosphate mix, and 3 µl of Superscript II Reverse Transcriptase (Invitrogen) incubated at a temperature of 42°C for 1 hour. Quantity and quality of the resulting cDNA were determined for each sample by quantitative real-time PCR against the 5′ and 3′ portions of the EEF1A1 transcript. Quantitative realtime PCR was performed using 12.5 µl of iTaqSupermix (Bio-Rad), 2.5 µl of 2 µM forward primer, 2.5 µl of 2 µM reverse primer, and 6.5 µl of nuclease-free water per reaction. Thermal cycling was performed using a MyIQ Thermal Cycler (Bio-Rad) and the following protocol: 1 cycle at 95°C for 1 minute and 50 cycles at 94°C for 10 seconds and at 60°C for 30 seconds. A standard curve was created from 5 µg of normal colon cDNA (Clontech, Palo Alto, CA) diluted serially at 1:4 for a total of five standards. Quantity was determined by the concentration of EEF1A1 as reported from the standard curve using MyIQ software version 1.0 (Bio-Rad). The reactions were performed in triplicate, and the concentrations were averaged. mRNA quality was determined by comparing the ratios of the quantities of 5′ and 3′ segments of the control EEF1A1 transcript, and samples of sufficient quality (5′EEF1A1/3′EEF1A1) greater than 0.5 were used for second-strand cDNA synthesis, RNA amplification, and SAGE library construction.
Second-strand cDNA synthesis was performed by adding 465 µl of diethylpyrocarbonate water, 150 µl of 5x second-strand buffer, 15 µl of deoxyribonucleotide triphosphate mix, 5 µl of Escherichia coli DNA ligase, 20 µl of E. coli DNA polymerase, and 5 µl of E. coli RNase H (Invitrogen) to the first-strand cDNA synthesis reactions. The reaction mixture was incubated at 16°C for 2 hours. The solid-phase double-stranded cDNA was washed and digested in a reaction mixture containing 172 µl of 3 mM Tris-HCl, 0.3 mM EDTA, pH 8.0; 2 µl of 100x BSA; 20 µl of 10x buffer 4; and 6 µl of NlaIII enzyme (Invitrogen).
T7 RNA Transcription/RNA Amplification
To amplify the 3′ ends of mRNA for SAGE, we prepared a linker containing the T7 RNA polymerase promoter sequence and ligated this linker to NlaIII-digested double-stranded cDNA. Forty nanograms of T7 LongSAGE adapter (T7 FWD: 5′-CAGAGAATGCATAATACGTACTCACTATAGGGATCCACAAGAACTACTACATG-3′; and T7 REV: 5′PO4TAGTAGTTCTTGTGGATCCCTATAGTGAGTCGTATTATGCATTCTCTG-3′) was ligated to bead-bound NlaIII-digested double-stranded cDNA in a reaction containing 14.5 µl of 3 Mm Tris-HCl, 0.3 mM EDTA, pH 8.0; 1 µl of T7 adapter; 2 µl of 10x ligase buffer; and 2.5 µl of T4 DNA ligase (Invitrogen). Excess adapter was removed by washing the bead-bound double-stranded cDNA three times with wash buffer D (Invitrogen). RNA was amplified using the T7 MEGAscript kit (Ambion, Austin, TX) in a reaction containing 2 µl of adenosine 5′-triphosphate solution, 2 µl of cytidine 5′-triphosphate solution, 2 µl of guanosine 5′-triphosphate solution, 2 µl of uridine 5′-triphosphate solution, 2 µl of 10x reaction buffer, 2 µl of enzyme mix, and 8 µl of nuclease-free water. The reaction mixture was incubated at 37°C overnight. Amplified RNA generated by the in vitro transcription reaction was eluted from the beads using 40 µl of nuclease-free water at 70°C for 5 minutes.
LongSAGE
One-tenth of the total transcribed RNA containing supernatant was reverse-transcribed and quantified by real-time PCR. An aliquot equivalent of 20 µg of total RNA was used from the remaining supernatant as an input for the I-SAGE Long Kit. LongSAGE libraries were created using the manufacturer's instructions with the following alterations:
Primers used for DiTAG amplification were modified to contain a sequencing site for recognition by 454 Life Sciences (Branford, CT). The primer sequences used were 454 LongSAGEA2 5′-GCC TCC CTC GCG CCA TCA GTT GGA TTT GCT GGT GCA GTA-3′ and 454 LongSAGEB1 5′-GCC TTG CCA GCC CGC TCA GCG AAT TCA AGC TTC TAA CGATG-3′. The resulting DiTAGs were directly sequenced on a GS20 without concatamerization or cloning into bacteria.
Real-time PCR
Quantitative real-time PCR was used to quantify FN1, PITX2, FLJ22104, RPL39, EIF1AX, AP3S1, NDUFA8, and EEF1A1 expression using the following primers:
FN1 FWD 5′-TGG CCAGTCCTACAACCAGT-3′;
FN1 REV 5′-CGGGAATCTTCTCTGTCAGC-3′;
PITX2 FWD 5′-ATGGAGACCAACTGCCGCAA-3′;
PITX2 REV 5′-TCACACGGGCCGGTCCACTGC-3′;
FLJ22104 FWD 5′-GCAGCTGTCATGGAAGTTCA-3′;
FLJ22104 REV 5′-CATCAAGGACTTTTCGGTTCA-3′;
RPL39 FWD 5′-CTCGCCATGTCTTCTCACAA-3′;
RPL39 REV 5′-CCAGCTTGGTTCTTCTCCAA-3′;
EIF1AX FWD 5′-GCAGTGTACTGGAGAGGGGA-3′;
EIF1AX REV 5′-TGAAGCTGAGACAAGCAGGA-3′;
AP3S1 FWD 5′-TGATGCACAAAATAAGCTGGA-3′;
AP3S1 REV 5′-TTGGGATCTCAGGAAGATTCA-3′;
NDUFA8 FWD 5′-GTGTGTGCTGGACAAACTGG-3′;
NDUFA8 REV 5′-GGGATTCTCCGGTAAAGGTC-3′;
EEF1A1 FWD 5′-CAATGCTTCCACCAACTCGT-3′;
EEF1A1 REV 5′-TCTTGACATTGAAGCCCA-3′.
PCR was performed using 12.5 µl of iTaqSupermix (Bio-Rad), 2.5 µl of 2 µM forward primer, 2.5 µl of 2 µM reverse primer, and 6.5 µl of nuclease-free water per reaction. Thermal cycling was performed using a MyIQ Thermal Cycler (Bio-Rad) and the following protocol: 1 cycle at 95°C for 1 minute and 50 cycles at 94°C for 10 seconds and at 60°C for 30 seconds. Gene expression levels were standardized to cDNA concentration by computing the difference between the cycle threshold (Ct) value of each classifier gene and the cycle threshold (Ct) value for a control gene (EEF1A1).
Logistic Regression Classifier
Denote the standardized expression levels of FN1, FLJ22104, RPL39, PITX2, EIF1AX, AP3S1, and NDUFA8 by X1 to X7 as shown.
Denote the gene expression level vector by X = (X1, …, X7). Let Y be the node status variable:Y = 1 means node-positive and Y = 0 node-negative. The logistic regression (LR) model specifies the probability that a colorectal tumor is node-positive as a function of gene expression levels given by the following:
Given the gene expression level vector and the node status of n = 23 tumors, denoted by (yi, xi), i = 1, 2, …, 23, the maximum likelihood estimates of the LR coefficients β0, β1, …, β7, denoted by b0, b1, …, b7, can be obtained through iterative procedures that are implemented in the R statistical package [10], the package that we used [11].
Given a new tumor with unobserved node status Y* and an observed gene expression level vector X* =x*, we estimate the probability that the tumor is node-positive through
The LR classifier is of the form
| (1) |
where c is between 0 and 1, a properly chosen cutoff or threshold. This means that if the estimated probability of node-positive status is at least c, we classify the tumor as node-positive.
If we denote a gene expression vector
then the estimated probability that the tumor is node-positive is given by
The LR classifier becomes
| (2) |
The cutoff or threshold d is related to the original cutoff c through
Usually, the cutoff c would be chosen to be 0.5, but the determination of the appropriate cutoff for the LR classifier is tied in to the desired performance of the classifier. We assessed the performance of the classifier with regards to its false-positive rate (FPR) and its false-negative rate (FNR) by constructing and evaluating a receiver operating characteristic (ROC) curve.
For a given tumor with node status Y0 and gene expression level vector X0, the conditional FPR associated with the LR classifier with cutoff c is defined as the probability that it is classified to be node-positive given that it is actually node-negative, whereas the conditional FNR is defined as the probability that it is classified to be node-negative given that it is actually node-positive. Taking averages of these quantities with respect to the distribution of (Y0, X0), we obtain the FPR and FNR, respectively, each of which is a function of the cutoff value c. The ROC curve is defined as the graph of FPR(c) versus 1 - FNR(c), where c ranges from 0 to 1. Or equivalently, it is the graph of the false-positive fraction versus the true-positive fraction at different cutoff values of c. The area under this curve is called the AUROC and serves as a measure of the quality of the class of classifiers.
Observe that from the definitions of the FPR and the FNR, the appropriate way to estimate FPR and FNR is to evaluate independent test data that are different from the training data used to generate the classifier. However, our sample data set is small and we did not have an independent test data set from which to measure the FPR and FNR functions. We therefore used the training data (Yi, Xi), i = 1, 2, …, 23, to estimate FPR(c) by FP̂R(c), which is the observed proportion of false-positives, whereas estimate FNR(c) by FP̂R(c), which is the observed proportion of false-negatives. The empirical ROC curve is then given by the graph of FP̂R(c) versus 1 - FP̂R(c) (or the observed false-positive fraction versus the observed true-positive fraction), and the empirical AUROC is given by the area under the empirical ROC curve.
We determined the appropriate value of the cutoff c (denoted as c*) from the empirical ROC by imposing an upper limit on the value of the FPR and to subsequently minimize the FNR. The approach adopted here is from clinical considerations that lead us to expect up to 20% false-positives (because of misdiagnosed node-negative disease), hence the need to control the FPR at the outset through the specification of the FPR threshold.
On determination of c* through the previously mentioned approach, the final LR classifier becomes δc*(x*) as given in (1) with c replaced with c*. Or, in the alternative form in (2), we use in this case the cutoff value of d* = log(c* / (1 -c*)) .b0 for d.
Fibronectin Immunofluorescence Microscopy
Frozen tumor specimens were cut into 10-µm sections and fixed onto silane-prep slides (Sigma). Sections were fixed in 95% ethanol, washed with PBS, and blocked with normal goat serum (Biogenex, San Ramon, CA) for 1 hour at room temperature. Samples were probed with either a 1:200 dilution of rabbit antifibronectin primary or a 1:500 dilution of mouse anti-BerEP4 primary antibody for 1 hour at room temperature. Samples were washed with PBS and then incubated with a 1:500 dilution of the appropriate AlexaFluor 488-conjugated secondary (Invitrogen). Samples were washed in PBS and coverslipped using 4′,6-diamidino-2-phenylindole, dihydrochloride-containing mounting medium.
Fibronectin Western Blot Analysis
Cell lysates from SW480 colorectal cancer cells and FN1-transfected subclones were lysed directly in Laemmli sample buffer. Cell numbers were quantitated by purifying genomic DNA and performing LINE PCR as described. Equivalent cell numbers were electrophoresed on 8% SDS-polyacrylamide gel and transferred electrophoretically to a polyvinylidine fluoride membrane at 80 mA overnight. Blots were probed with polyclonal rabbit anti-FN1 primary antibody overnight at 4°C (1:1000 dilution in 5% milk; part no. F3468; Sigma) or monoclonal mouse antitubulin antibody (1:20,000 dilution in 5% milk; part no. T9026; Sigma) for 1 hour at room temperature. Blots were washed and then probed with HRP-conjugated goat antirabbit immunoglobulin G secondary antibody (1:10,000 diluted in 5% milk; part no. PI-1000; Vector Laboratories, Burlingame, CA) for 2.5 hours at room temperature or HRP-conjugated goat antimouse secondary antibody (1:20,000 diluted in 5% milk) for 1 hour at room temperature. Bands were imaged by incubating the blots in ECL Western Blotting Detecting Kit (RPN21061; GE Healthcare Bio-Sciences, Piscataway, NJ) and exposing to x-ray film (Kodak Biomax XAR Film part no. 05-728-41; Thermo Fisher Scientific, Waltham, MA) for 5 minutes.
Cell Culture Methods
SW480 cells were a kind gift from Dr. Marj Pena. The cells were maintained in McCoy's 5A medium (Invitrogen) supplemented with 1x penicillin/streptomycin (Gibco, Carlsbad, CA) and 10% fetal bovine serum (Invitrogen) in a humidified incubator with 5% CO2.
Generation of FN-Expressing Clones
A full-length cDNA encoding adult Fibronectin (FN1) was cloned into the pIRES-Neo3 vector (Clontech, Mountain View, CA) and transfected into SW480 cells using Lipofectamine (Invitrogen). Single clones were selected by limiting dilution and growth using 1000 µg/ml G418. Clones were tested for FN1 mRNA expression by real-time PCR.
Migration and Invasion Assays
Invasion assays were performed using BD BioCoatM atrigel Invasion Chambers (BD Biosciences, San Jose, CA) according to the manufacturer's instructions. Either 50,000 cells (migration) or 100,000 cells (invasion) were seeded into the top of empty transwell migration chambers (migration) or chambers containing Matrigel inserts (invasion) and placed into culture wells containing complete medium with 10% fetal bovine serum as a chemoattractant. Assays were performed in the absence or presence of 4.5 µM of an inhibitory cyclic RGDfV peptide [12,13]. Cells were allowed to migrate or invade for 3 days, after which the filters were removed, stained with crystal violet, and visualized by microscopy. Cell counts were performed in triplicate and averaged.
Results
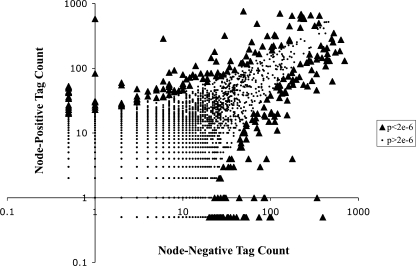
To identify genes that are differentially expressed during the progression of colorectal cancer, we modified the LongSAGE tags for amplicon sequencing on a 454 GS20 genome sequencer (454 Life Sciences). We laser capture microdissected tumor epithelial cells, purified and amplified mRNA, and prepared LongSAGE libraries from one node-negative and one node-positive colorectal tumor. Specific tumors were chosen based on the quantity and quality of mRNA recovered. LongSAGE tags were modified for 454 sequencing as described. One sequencing run on the 454 GS20 platform produced 351,943 sequencing reads from which 327,294 total tags were extracted, 26,060 of which were unique and expressed two or more times (Table 1A). One hundred fifty tags were differentially expressed by at least 25-fold (Table 1B). To control against contaminant inflation of type I error from multiple testing, we used a Bonferroni correction to arrive at a cutoff value of 0.05/26,060 = 1.9 x 10-6 for the P values for each pairwise comparison [14]. Under this conservative approach, 262 tags were defined to be statistically significantly differentially expressed between node-negative and node-positive colorectal tumors (Figure 1).
Table 1.
Summary of LongSAGE Data.
| (A) | ||
| Library | Total Reads | Total Tags |
| 18964 node- | 162,576 | 170,372 |
| 29271 node+ | 189,367 | 195,452 |
| Combined | 351,943 | 365,824 |
| (B) | ||
| Fold Differential Expression | No. of Tags Elevated in Node-Positive | No. of Tags Elevated in Node-Negative |
| 50 | 12 | 22 |
| 25 | 6 | 74 |
| 10 | 644 | 505 |
| 5 | 3876 | 2659 |
Tumor no. 18964 (node-negative) and tumor no. 29271 (node-positive) were sequenced to a depth of 365,824 total tags. To minimize sequencing errors, only tags that were observed two or more times were counted.
Figure 1.
Distribution of SAGE tags. Tag counts of zero were converted to 0.5 to plot on log scale and to compute expression ratios. Significantly differentially expressed tags (binomial P < 2 x 10-6) are shown as triangles.
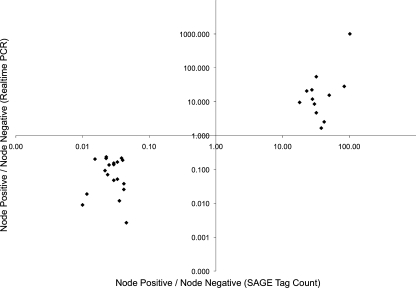
To validate the tag-to-gene assignments and eliminate potential bias introduced during RNA amplification, we analyzed the best gene matches for the top 75 most differentially expressed tags (binomial P < 2 x 10-6, ratio > 20) in unamplified cDNA from the profiled tumors using quantitative real-time PCR. From this analysis, we confirmed the tag-to-gene assignments and differential expression of 30 genes. The expression ratios observed by real-time PCR were in good agreement with the expression ratios observed by SAGE (Figure 2 andTable 2).
Figure 2.
Confirmed differentially expressed genes. Expression levels were determined by quantitative real-time PCR, and the ratio of node-positive to node-negative gene expression levels is plotted versus the ratio of node-positive to node-negative tag counts.
Table 2.
Differentially Expressed LongSAGE Tags and Corresponding Tag-to-Gene Matches.
| SAGE Tag | Gene Symbol | SAGE N(+)/N(-) | Real-time PCR N(+)/N(-) |
| TGTACCTCAGCTTTTTC | ORM2 | 102.00 | 1002.93 |
| ATTTTTACTAATGTATT | UBD | 84.00 | 28.25 |
| AAAACATTATGACTTTT | AP3S1 | 50.00 | 15.51 |
| AATTAACTCCGTTAAAA | ALDH1B1 | 42.00 | 2.53 |
| CGGTTTGCATCGACTGA | NDUFA8 | 38.00 | 1.66 |
| CCACTGCACTCCAGCAG | FLJ22104 | 32.00 | 54.57 |
| TGTCAGAATTTCATTCC | CTPS2 | 32.00 | 4.69 |
| GCGAGCAGCGGAGTCAA | RPL39 | 30.00 | 8.51 |
| ACAGCTAATTAGTACTA | EIF1AX | 28.00 | 11.88 |
| AGAATCACTTGAACCCA | HRH1 | 27.50 | 22.32 |
| ATCTTGTTACTGTGATA | FN1 | 23.00 | 20.66 |
| GGAGTAAAATATACTGC | PITX2 | 18.00 | 9.51 |
| CGTGCGAGACACGTGTG | C1orf30 | 0.05 | 0.00 |
| GTAGCGCCTCCTAACAG | CST7 | 0.04 | 0.03 |
| TTGATGGGCGACTTCAA | DNASE1 | 0.04 | 0.04 |
| GGTACCCATTTGATAAG | DUSP6 | 0.04 | 0.19 |
| ACAAGATATTTCTACCT | CASP4 | 0.04 | 0.22 |
| GACCAGTGGCTGGTCTC | GPA33 | 0.04 | 0.01 |
| GGTATTAACCACAGATT | DEFA6 | 0.03 | 0.05 |
| AACAGCAAGGAGTGTTT | APCDD1 | 0.03 | 0.16 |
| GCCAAGGAGTTCCAGGA | GPR35 | 0.03 | 0.05 |
| AACAAAGATATATTTTC | KIAAI199 | 0.03 | 0.14 |
| CTGCTATGGTCACTGAG | NRN1 | 0.03 | 0.15 |
| TTCCTGGAAACCTACGG | GOLTIA | 0.03 | 0.14 |
| AACCACTGCTACTCCCG | ID3 | 0.02 | 0.07 |
| GCCTGTTTGGGAGTGCG | UGTIA6 | 0.02 | 0.23 |
| AGCTCTTGGAGGCACCA | NDRG2 | 0.02 | 0.21 |
| TAGAAGATCTATGGAAA | NKD1 | 0.02 | 0.09 |
| TGAGAGGAGATGGACCC | NDUFB5 | 0.02 | 0.20 |
| CGTTCCTGCGGACGATC | ID1 | 0.01 | 0.02 |
| TACAAAATCGATTGGCT | IGF2 | 0.01 | 0.01 |
Expression levels were determined by quantitative real-time PCR, and the ratio of node-positive to node-negative tumors was calculated. Tag expression levels of zero were converted to 0.5 to compute expression ratios.
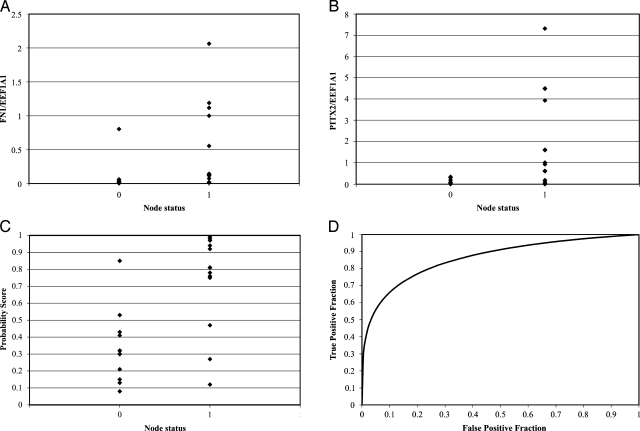
We then analyzed the expression of the 12 genes that were increased in node-positive colorectal cancer in a panel of 23 additional colorectal tumors (Table W1). Using an LR approach, we developed a seven-gene classifier, as described in the Materials and Methods section, capable of discriminating between node-negative and node-positive tumors (Figures 3, A–D, W1, A–E). In this set of tumors, the probability that an individual colorectal cancer was node-positive could be estimated using a function of the seven-gene expression values relative to the control gene, EEF1A1. For each tumor, we compute a composite gene expression value, V, according to the normalized cycle threshold (Ct) values observed for the seven best genes through the formula
Figure 3.
FN1 (A) and PITX2 (B) mRNA expressions in the epithelial cells of 23 colon cancer specimens. Probability of node-positive diagnosis based on LR classifier (C) and ROC analysis (D). Cutoff values associated with 20% FPRs for the multigene classifier produced optimal separation between node-negative and node-positive tumors.
The estimated probability of metastatic disease is then given by
As described in the Materials and Methods section, based on this value of P, a classifier could be developed, which is of the form where a tumor is classified as node-positive whenever the estimated probability P is greater than or equal to the cutoff value c. We evaluated the sensitivity and specificity of this class of LR classifiers at different cutoff values c and formed the empirical ROC curve. Because the recurrence rate from node-negative colorectal cancer can approach 20% [15], we set this value as an upper limit for the number of truly node-positive tumors that were inappropriately given a node-negative diagnosis. We therefore imposed the constraint that the estimated FPR (node-negative colon cancers receiving a positive gene expression score) should be no more than 20%, which corresponds to 80% specificity. This led to an optimal cutoff value of c = 0.43, and the resulting LR classifier has a sensitivity of 86%. ROC analysis of the class of LR classifiers revealed an area under the curve of 0.86 (Figure 3D). The selected LR classifier with cutoff of c = 0.43 correctly identified 21 of 25 tumors. The classifier performs significantly better than a random classifier (binomial P = .00046). Interestingly, one node-negative tumor (15095) scored very high on the LR classifier scale (with an estimated probability of being node-positive of 0.85), suggesting that this individual was misdiagnosed, harbors occult node-positive disease, and is at high risk for recurrence.
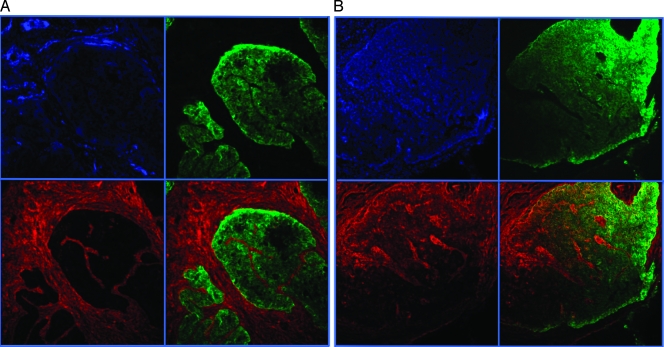
Because of its role in normal cell adhesion and its association with melanoma metastasis [16], we focused our attention on the Fibronectin gene (FN1), which was upregulated more than 20-fold in the node-positive LongSAGE library. To determine if the fibronectin protein is also upregulated in node-positive tumors, we performed immunofluorescence microscopy on node-positive and node-negative colorectal tumors. Samples were triple labeled with anti-FN1, anti-BerEP4 epithelial marker, and a DAPI nuclear stain. As previously reported [17], we observed dramatic expression of fibronectin protein in the stromal fibroblasts of normal colon and both node-negative and node-positive colon cancers. In addition, we observed a dramatic increase in epithelial fibronectin in node-positive lesions (Figure 4).
Figure 4.
(A) Colorectal tumors 30449 (node-negative) and (B) 29203 (node-positive). Nuclei are shown in blue, epithelial cells are shown in green, and fibronectin is shown in red. The lower right quadrant is an overlay of Fibronectin and BerEP4 staining.
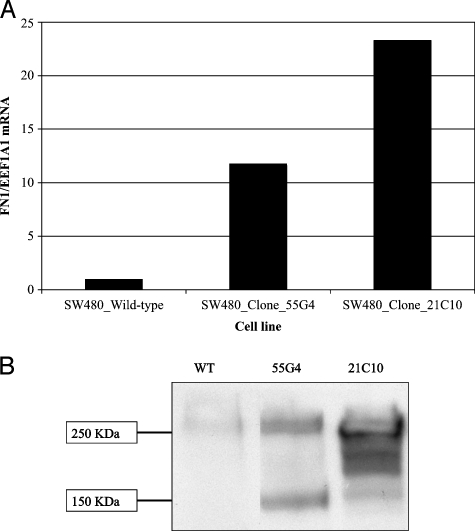
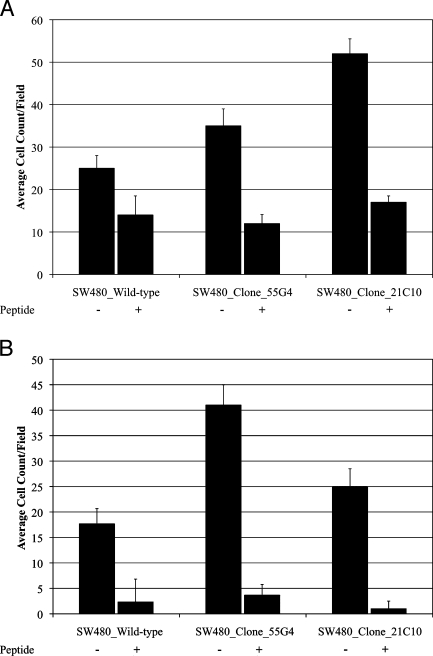
We sought to determine whether inappropriate fibronectin expression by tumor epithelial cells would cause in vitro effects consistent with lymph node metastasis such as enhancing cell migration or improving cell survival. To test this hypothesis, we cloned a full-length cDNA encoding the adult isoform of the FN1 transcript into pIRES-Neo3, generated stable subclones of SW480 colorectal cancer cells that overexpress the FN1 mRNA and protein relative to wild-type SW480 cells (Figure 5, A and B). We performed Boyden chamber migration and Matrigel invasion assays in the absence and presence of a cyclic RGD-containing peptide. The FN1-overexpressing clones demonstrated a marked increased ability to migrate through empty Boyden chambers and invade through Matrigel barriers in vitro (Figure 6, A and B). This property was completely abolished by the inclusion of 4.5 µM cyclic RGDfV peptides into the culture medium. These results demonstrate that increased fibronectin expression by tumor epithelial cells can cause enhanced migration and invasion and suggest that targeting a specific subpopulation of colorectal cancer patients with RGD peptide-based therapeutics could be more successful than treating unselected patients.
Figure 5.
FN1 mRNA (A) and protein (B) expressions in SW480 colorectal cancer cells transfected with pIRES-Neo3-FN1 or an empty vector control.
Figure 6.
Migration (A) and invasion (B) of wild-type and fibronectin-expressing clones of SW480 colorectal cancer cells in the absence (-) and presence (+) of inhibitory peptide RGDfV.
Discussion
It is estimated that 149,000 cases of colorectal cancer will be diagnosed in the United States in 2008 and that 60,000 of these tumors will be detected before regional lymph node involvement [1]. The 5-year survival rate for node-negative colorectal cancer is at best 90%. This means that approximately 6000 people will experience relapse from early-stage disease. It is possible that at least some of these individuals were understaged because of occult node-positive cancer and were therefore undertreated. Adjuvant chemotherapy can decrease the chances of recurrence for node-negative patients [18]; however, effective methods of identifying the node-negative patients who are at highest risk are needed. Applying our seven-gene classifier to node-negative patients may represent a successful strategy for identifying these patients. Because gene expression alterations are caused by gene mutations, it will be important to evaluate the performance of this classifier in the context of somatic mutations to RAS, RAF, and CSMD1 [19].
The FN1 gene is not expressed by normal colonic epithelial cells but is overexpressed by the myofibroblasts and epithelial cells of some colorectal adenocarcinomas [20].FN1 expression is downregulated as colon epithelial cells differentiate, suggesting that reexpression in adenocarcinoma is indicative of epithelial-to-mesenchymal transition [21].FN1 expression is associated with poor outcome in other cancers including melanoma [16], ovarian and breast cancer [22,23], and lymphoma [24]. It has also been shown that tumor cell fibronectin production is necessary for tumor cell migration in vitro [25]. It is possible that the progression of colorectal cancer from node-negative to node-positive disease may be facilitated in part by FN1 deregulation and subsequent enhanced tumor cell migration.
The mechanism by which FN1 is increased during colorectal cancer progression may involve stepwise broadening of WNT signaling. The WNT pathway is activated by mutations in adenomatous polyposis coli or β-catenin and is an initiating event in colorectal tumor formation leading to enhanced β-catenin/transcription factor 4 transcription [26]. However, these events alone are not sufficient to activate epithelial FN1 expression. Rather, FN1 expression develops later in a subset of intestinal tumors, which suggests that secondary genetic events are necessary to cause an epithelial-to-mesenchymal transition [27]. WNT signaling is able to induce FN1 expression in fibroblasts through the action of β-catenin/lymphoid enhancer-binding factor 1 (LEF1) complexes. Normal epithelial cells and early colorectal tumors lack LEF1 and, therefore, do not express FN1 [28]. SW480 colon cancer cells contain activating mutations in β-catenin, express transcription factor 4, and, therefore, show high levels of activity of reporter constructs containing TCF binding sites [29]. SW480 cells are unable to transactivate FN1 unless exogenous LEF1 is added [28]. Interestingly, the LEF1 gene is itself transactivated by β-catenin/LEF1/PITX2 complexes [30,31]. We found PITX2 to be increased in expression only in node-positive colorectal tumors, which is consistent with the idea that FN1 and other LEF1-dependent WNT targets could be increased during disease progression by elevated PITX2.
Gene expression profiling has resulted in the discovery of markers of early and late colorectal cancer progression [32–35] as well as good and poor clinical outcome [36]. Genes that contribute to the underlying mechanism represent attractive targets for diagnostic and therapeutic purposes. We believe our results indicate that FN1 up-regulation is an important step in the transition from lymph node-negative to lymph node-positive colorectal cancer. Tumor epithelial cells that have acquired elevated levels of FN1 migrate more efficiently and may escape anoikis by propagating survival signals through integrin receptors [37]. This is but one possible mechanism by which tumor cells metastasize to distant organs. Other possible mechanisms, such as inactivating mutations in anoikis pathway genes, may explain lymph node metastases in tumors that do not upregulate FN1 [38]. Because node-negative colon cancers that display elevated FN1 and PITX2 may be at high risk for recurrence, future studies must focus on determining the rates of relapse among large numbers of these types of patients. Finally, these results suggest that antagonists of fibronectin-integrin interaction, particularly those that do not activate downstream survival signals, may be effective agents for the treatment of colorectal cancers with increased expression of fibronectin. Cyclic RGD peptides that target fibronectin-integrin interaction have been tested in preclinical models of nonmetastatic, invasive colon cancer and have shown some positive effects [39]. It is possible that preselecting colon cancer patients for elevated FN1 expression will result in significant performance gains for cyclic RGD peptides and related compounds.
Supplementary Material
| Predictor | Description |
| X1 | FN1-EEF1A1 |
| X2 | FLJ22104-EEF1A1 |
| X3 | RPL39-EEF1A1 |
| X4 | PIT X2-EEF1A1 |
| X5 | EIF1A X-EEF1A1 |
| X6 | AP3S1-EEF1A1 |
| X7 | NDUFA8-EEF1A1 |
Acknowledgments
The authors thank Valerie Kennedy, histology technician (1966–2008).
Abbreviations
- SAGE
serial analysis of gene expression
Footnotes
This work was supported by the National Institutes of Health Centers of Biomedical Research Excellence grant P20 RR17698 and National Institutes of Health grant 1R21CA127683.
This article refers to supplementary materials, which are designated byTable W1 andFigure W1 and are available online atwww.neoplasia.com.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Govindarajan A, Baxter NN. Lymph node evaluation in early-stage colon cancer. Clin Colorectal Cancer. 2008;7(4):240–246. doi: 10.3816/CCC.2008.n.031. [DOI] [PubMed] [Google Scholar]
- 3.Baxter NN, Virnig DJ, Rothenberger DA, Morris AM, Jessurun J, Virnig BA. Lymph node evaluation in colorectal cancer patients: a population-based study. J Natl Cancer Inst. 2005;97(3):219–225. doi: 10.1093/jnci/dji020. [DOI] [PubMed] [Google Scholar]
- 4.Kwon HC, Kim SH, Roh MS, Kim JS, Lee HS, Choi HJ, Jeong JS, Kim HJ, Hwang TH. Gene expression profiling in lymph node-positive and lymph node-negative colorectal cancer. Dis Colon Rectum. 2004;47(2):141–152. doi: 10.1007/s10350-003-0032-7. [DOI] [PubMed] [Google Scholar]
- 5.Croner RS, Peters A, Brueckl WM, Matzel KE, Klein-Hitpass L, Brabletz T, Papadopoulos T, Hohenberger W, Reingruber B, Lausen B. Microarray versus conventional prediction of lymph node metastasis in colorectal carcinoma. Cancer. 2005;104(2):395–404. doi: 10.1002/cncr.21170. [DOI] [PubMed] [Google Scholar]
- 6.Grade M, Hormann P, Becker S, Hummon AB, Wangsa D, Varma S, Simon R, Liersch T, Becker H, Difilippantonio MJ, et al. Gene expression profiling reveals a massive, aneuploidy-dependent transcriptional deregulation and distinct differences between lymph node-negative and lymph node-positive colon carcinomas. Cancer Res. 2007;67(1):41–56. doi: 10.1158/0008-5472.CAN-06-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HN, Choi DW, Lee KT, Lee JK, Heo JS, Choi SH, Paik SW, Rhee JC, Lowe AW. Gene expression profiling in lymph node-positive and lymph node-negative pancreatic cancer. Pancreas. 2007;34(3):325–334. doi: 10.1097/MPA.0b013e3180317b01. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen ST, Hasegawa S, Tsuda H, Tomioka H, Ushijima M, Noda M, Omura K, Miki Y. Identification of a predictive gene expression signature of cervical lymph node metastasis in oral squamous cell carcinoma. Cancer Sci. 2007;98(5):740–746. doi: 10.1111/j.1349-7006.2007.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abba MC, Sun H, Hawkins KA, Drake JA, Hu Y, Nunez MI, Gaddis S, Shi T, Horvath S, Sahin A, et al. Breast cancer molecular signatures as determined by SAGE: correlation with lymph node status. Mol Cancer Res. 2007;5(9):881–890. doi: 10.1158/1541-7786.MCR-07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 11.McCullagh P, Nelder J. Generalized Linear Models. London, UK: Chapman & Hall; 1983. [Google Scholar]
- 12.Dechantsreiter MA, Planker E, Matha B, Lohof E, Holzemann G, Jonczyk A, Goodman SL, Kessler H. N-methylated cyclic RGD peptides as highly active and selective α(V)β(3) integrin antagonists. J Med Chem. 1999;42(16):3033–3040. doi: 10.1021/jm970832g. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann S, He S, Jin M, Ehren M, Wiedemann P, Ryan SJ, Hinton DR. A selective cyclic integrin antagonist blocks the integrin receptors αvβ3 and αvβ5 and inhibits retinal pigment epithelium cell attachment, migration and invasion. BMC Ophthalmol. 2005;5:16. doi: 10.1186/1471-2415-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonferroni CE. Studi in Onore del Professore Salvatore Ortu Carboni. Rome, Italy: 1935. Il calcolo delle assicurazioni su gruppi di teste; pp. 13–60. [Google Scholar]
- 15.Sarli L, Bader G, Iusco D, Salvemini C, Mauro DD, Mazzeo A, Regina G, Roncoroni L. Number of lymph nodes examined and prognosis of TNM stage II colorectal cancer. Eur J Cancer. 2005;41(2):272–279. doi: 10.1016/j.ejca.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406(6795):532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 17.Pujuguet P, Hammann A, Moutet M, Samuel JL, Martin F, Martin M. Expression of fibronectin ED-A+ and ED-B+ isoforms by human and experimental colorectal cancer. Contribution of cancer cells and tumor-associated myofibroblasts. Am J Pathol. 1996;148(2):579–592. [PMC free article] [PubMed] [Google Scholar]
- 18.Quasar Collaborative, author. Gray GR, Barnwell J, McConkey C, Hills RK, Williams NS, Kerr DJ. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370(9604):2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 19.Farrell C, Crimm H, Meeh P, Croshaw R, Barber T, Vandersteenhoven J, Butler W, Buckhaults P. Somatic mutations to CSMD1 in colorectal adenocarcinomas. Cancer Biol Ther. 2008;7:609–613. doi: 10.4161/cbt.7.4.5623. [DOI] [PubMed] [Google Scholar]
- 20.Hanamura N, Yoshida T, Matsumoto E, Kawarada Y, Sakakura T. Expression of fibronectin and tenascin-C mRNA by myofibroblasts, vascular cells and epithelial cells in human colon adenomas and carcinomas. Int J Cancer. 1997;73(1):10–15. doi: 10.1002/(sici)1097-0215(19970926)73:1<10::aid-ijc2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Vachon PH, Simoneau A, Herring-Gillam FE, Beaulieu JF. Cellular fibronectin expression is down-regulated at the mRNA level in differentiating human intestinal epithelial cells. Exp Cell Res. 1995;216(1):30–34. doi: 10.1006/excr.1995.1004. [DOI] [PubMed] [Google Scholar]
- 22.Helleman J, Jansen MP, Span PN, van Staveren IL, Massuger LF, Meijer-van Gelder ME, Sweep FC, Ewing PC, van der Burg ME, Stoter G, et al. Molecular profiling of platinum resistant ovarian cancer. Int J Cancer. 2006;118(8):1963–1971. doi: 10.1002/ijc.21599. [DOI] [PubMed] [Google Scholar]
- 23.Helleman J, Jansen MP, Ruigrok-Ritstier K, van Staveren IL, Look MP, Meijervan Gelder ME, Sieuwerts AM, Klijn JG, Sleijfer S, Foekens JA, et al. Association of an extracellular matrix gene cluster with breast cancer prognosis and endocrine therapy response. Clin Cancer Res. 2008;14(17):5555–5564. doi: 10.1158/1078-0432.CCR-08-0555. [DOI] [PubMed] [Google Scholar]
- 24.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350(18):1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 25.Nabeshima K, Inoue T, Shimao Y, Kataoka H, Koono M. Cohort migration of carcinoma cells: differentiated colorectal carcinoma cells move as coherent cell clusters or sheets. Histol Histopathol. 1999;14(4):1183–1197. doi: 10.14670/HH-14.1183. [DOI] [PubMed] [Google Scholar]
- 26.Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17(1):45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Halberg RB, Burch RP, Dove WF. Intestinal adenomagenesis involves core molecular signatures of the epithelial-mesenchymal transition. J Mol Histol. 2008;39(3):283–294. doi: 10.1007/s10735-008-9164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gradl D, Kuhl M, Wedlich D. The Wnt/Wg signal transducer β-catenin controls fibronectin expression. Mol Cell Biol. 1999;19(8):5576–5587. doi: 10.1128/mcb.19.8.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275(5307):1787–1790. doi: 10.1126/science.275.5307.1787. [see comment] [DOI] [PubMed] [Google Scholar]
- 30.Amen M, Liu X, Vadlamudi U, Elizondo G, Diamond E, Engelhardt JF, Amendt BA. PITX2 and β-catenin interactions regulate Lef-1 isoform expression. Mol Cell Biol. 2007;27(21):7560–7573. doi: 10.1128/MCB.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vadlamudi U, Espinoza HM, Ganga M, Martin DM, Liu X, Engelhardt JF, Amendt BA. PITX2, β-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J Cell Sci. 2005;118(Pt 6):1129–1137. doi: 10.1242/jcs.01706. [DOI] [PubMed] [Google Scholar]
- 32.Buckhaults P. Gene expression determinants of clinical outcome. Curr Opin Oncol. 2006;18(1):57–61. doi: 10.1097/01.cco.0000198022.76476.b2. [DOI] [PubMed] [Google Scholar]
- 33.Buckhaults P, Rago C, St Croix B, Romans KE, Saha S, Zhang L, Vogelstein B, Kinzler KW. Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res. 2001;61(19):6996–7001. [PubMed] [Google Scholar]
- 34.Buckhaults P, Zhang Z, Chen YC, Wang TL, St Croix B, Saha S, Bardelli A, Morin PJ, Polyak K, Hruban RH, et al. Identifying tumor origin using a gene expression-based classification map. Cancer Res. 2003;63(14):4144–4149. [PubMed] [Google Scholar]
- 35.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61(7):3124–3130. [PubMed] [Google Scholar]
- 36.Eschrich S, Yang I, Bloom G, Kwong KY, Boulware D, Cantor A, Coppola D, Kruhoffer M, Aaltonen L, Orntoft TF, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol. 2005;23(15):3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 37.Ruoslahti E. Fibronectin and its integrin receptors in cancer. Adv Cancer Res. 1999;76:1–20. doi: 10.1016/s0065-230x(08)60772-1. [DOI] [PubMed] [Google Scholar]
- 38.Jan Y, Matter M, Pai JT, Chen YL, Pilch J, Komatsu M, Ong E, Fukuda M, Ruoslahti E. A mitochondrial protein, Bit1, mediates apoptosis regulated by integrins and Groucho/TLE corepressors. Cell. 2004;116(5):751–762. doi: 10.1016/s0092-8674(04)00204-1. [DOI] [PubMed] [Google Scholar]
- 39.Haier J, Goldmann U, Hotz B, Runkel N, Keilholz U. Inhibition of tumor progression and neoangiogenesis using cyclic RGD-peptides in a chemically induced colon carcinoma in rats. Clin Exp Metastasis. 2002;19(8):665–672. doi: 10.1023/a:1021316531912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.