Abstract
Leishmania parasites primarily infect cells of macrophage lineage and can cause leishmaniasis in the skin, mucosal, and visceral organs, depending on both host- and parasite-derived factors. The protein disulfide isomerases (PDIs) are thiol-disulfide oxidoreductases that catalyze the formation, reduction, and isomerization of disulfide bonds of proteins in cells. Although four Leishmania PDI genes are functionally inferred from homology in the genome sequences, only two of them have been expressed as active proteins to date. The functional relationship among various PDI enzymes remains largely unclear. In this study, we expressed and partially characterized all four L. amazonensis PDIs encoding 52-, 47-, 40-, and 15-kDa proteins. Homology analysis showed that the sequence identity between L. amazonensis (New World) PDIs and their counterpart PDI sequences from L. major (Old World) ranged from 76% to 99%. Kinetic characterization indicated that while the 15-, 40-, and 47- kDa PDI proteins displayed both insulin isomerase and reductase activities, the 52-kDa protein had only isomerase activity with no detectable reductase activity. All four PDI proteins were recognized by sera from L. amazonensis-infected mice and were sensitive to inhibition by standard PDI inhibitors. This study describes the enzymatic activities of recombinant L. amazonensis PDIs and suggests a role for these proteins in parasite development.
Keywords: Leishmania, Protein disulfide isomerase (PDI)
Introduction
Leishmaniasis is caused by infection with Leishmania spp., a diverse group of intracellular protozoan parasites transmitted by sand flies. The infection can lead to various clinical manifestations, including ulcerative skin lesions, destructive mucosal inflammation, and disseminated visceral infection (Handman 2001). It is estimated that 2 million individuals develop symptomatic disease (cutaneous leishmaniasis, 1–1.5 million; visceral leishmaniasis, 0.5 million) each year and that the incidence of infection is substantial when subclinical infections are included. Worldwide, leishmaniasis is associated with about 2.4 million disability-adjusted life years and around 70,000 deaths per year (Desjeux 2004). Visceral leishmaniasis (occasionally other forms of leishmaniasis) is emerging as an important opportunistic infection among people with HIV-1 infection (Alvar et al. 1997). Most species of the subgenus Leishmania (Leishmania) cause cutaneous leishmaniasis (CL) in the Old World (e.g., L. major, L. tropica, and L. aethiopica) and New World (e.g., L. mexicana and L. amazonensis), respectively. Some Old World species (e.g., L. arabica, L. gerbilli, and L. turanica) do not cause diseases in humans. Species in the L. donovani complex, such as L. donovani and L. infantum (syn. L. chagasi) can cause visceral leishmaniasis (Kuhls et al. 2005; Mauricio et al. 2006). Parasite factors and host immune status are inextricably linked to disease pathogenesis.
The life cycle of Leishmania parasites involves two hosts. The promastigote stage primarily survives extracellularly in the midgut of the sand fly. When it infects a vertebrate host, the parasite alternates as an obligatory intracellular amastigote, replicating within the phagolysosomal compartment of macrophages. To initially establish an infection, promastigotes enter macrophages silently to evade a triggering host response (Engwerda et al. 2004; Sacks and Sher 2002); a progressive intracellular (amastigote) infection depends on the maintenance of macrophages in an inert, deactivated state (Belkaid et al. 2000; McMahon-Pratt and Alexander 2004).
PDI belongs to a member of the thioredoxin superfamily which can catalyze the physiological oxidation, reduction, and isomerization of disulfide bonds of proteins in prokaryotic and eukaryotic cells. Therefore, PDIs are involved in many aspects of cell function and development. For example, PDI on the sperm surface can induce gamete fusion (Ellerman et al. 2006) and enhance protein secretion from the endoplasmic reticulum (ER) (Papp et al. 2006). Recently, several studies have suggested the involvement of PDI activities in a number of disorders and infectious diseases. In neurodegenerative diseases, PDI proteins act as chaperones ameliorating the accumulation of misfolded proteins triggered by oxidative or nitrosative stress, or of mutated gene products (Uehara et al. 2006). This fusionlike function has also been found in relation to HIV infection in CD4+ T cells as a coreceptor (Barbouche et al. 2005). In prokaryotes, such as Neisseria meningitidis, two membrane PDI proteins have been found to be essential for bacterial growth and biogenesis of functional type IV pili (Tinsley et al. 2004). In parasites such as Neospora caninum and Toxoplasma gondii, PDI was reported as playing a role in the parasite-host cell interaction (Liao et al. 2006; Robinson and Roy 2006). Moreover, it has recently been shown that native PDI on the cell surface is required for the effective attachment of Chlamydia, an obligate intracellular bacterial pathogen of eukaryotic cells (Conant and Stephens 2007). Likewise, a 52-kDa PDI of N. caninum techyzoites is involved in the adhesion of parasites to host cells (Naguleswaran et al. 2005). The detection of anti-PDI IgA in tears of bovines infected with N. caninum suggests that PDI-specific antibodies may constitute part of the mucosal antibody repertoire, which is possibly involved in defense against protozoan parasites (Liao et al. 2006). In addition, PDI involvement has also been reported in parasite development. Some PDIs seen in protozoans (e.g., Caenorhabditis elegans) (Eschenlauer and Page 2003) were found to be related to the formation of the cuticle.
Recently, two groups have reported a 52-kDa PDI in L. major (Ben Achour et al. 2002) and an atypical PDI with only one catalyzing site “CGHC” in L. donovani (Padilla et al. 2003); however, there is no systemic description of the PDI family in Leishmania parasites, nor a detailed comparison in terms of enzymatic activities and its relevance in infection. In this report, we characterized the PDI family within several Leishmania species known to cause diverse clinical forms and examined in detail both the enzymatic activities of L. amazonensis PDI proteins, using two classical assays, and the potential functions of PDI proteins in parasite growth and infection.
Materials and methods
Parasites and mouse serum
L. amazonensis (MHOM/BR/77/LTB0016), L. major (MHOM/IL/80/Friedlin), L. infantum (MCAN/ES/89/IPZ229/1/89), and L. braziliensis (MHOM/BR/1975/M2903) were used in this study, and their promastigotes were routinely maintained in our laboratory. Promastigotes were cultured in Schneider's Drosophila growth medium (Life Technologies, Rockville, MD, USA), pH 7.0, supplemented with 20% FBS, 2 mM l-glutamine, and 50 µg/ml gentamicin. To maintain the infectivity of the parasites, BALB/c mice were infected subcutaneously in the hind foot with 2 × l06 stationary-phase promastigotes, and lesion-derived parasites were recovered at 6–10 weeks post infection. Mice were purchased from Harlan Sprague-Dawley (Indianapolis, ID, USA), maintained under specific pathogen-free conditions, and used at 6–7 weeks of age under protocols approved by the Animal Care and Use Committee of the University of Texas Medical Branch, Galveston, TX, USA. Mouse serum samples were harvested from L. amazonensis-infected BALB/c mice at 8 weeks post infection.
Oligonucleotides
Four pairs of primers designed for the amplification of PDIs in L. amazonensis were inferred from homology analysis of putative PDI genes in L. major, L. infantum, and L. braziliensis. All oligonucleotides were synthesized by Sigma (St. Louis, MO, USA). Primers for cloning and expression LaPDI52 (52-kDa PDI of L. amazonensis were: 5′GCGGAATTCGGAGGTGCAGGTG3′, 5′CGCAAGCTTCAAATCTTCCTC3′ (the EcoRI and HindIII sites were italicized); for LaPDI47 were: 5′GCGCCATGGACCCGTATGGACGGTCGTCGG3′, 5′GCGCGGCCGCCGCAGCTCCAACGCGC3′ (the NcoI and NotI sites were italicized); for LaPDI40 were: 5′GCGGAATTCGGAAGACCCGGG3′, 5′GCGAAGCTTCTTCATGTGCTTCTGGAT3′ (the EcoRI and HindIII sites were italicized); for LaPDI15 were: 5′GCGGAATTCGGAGATTGTCGAGCTCAACC3′, 5′CGCCTCGAGCTGCTTGTTGGCCGC3′ (the EcoRI and Xhol sites were italicized).
PCR amplification of PDI genes
Total RNA was isolated from 2 × l07 L. amazonensis promastigotes using an RNeasy Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocols. First-strand cDNA synthesis was performed for 1 h at 42°C in a 20-µl reaction mix containing 2 µg total RNA, lx RT buffer, 10 mM each dNTP (Promega, Madison, WI, USA), 100 U of reverse transcriptase (Promega) and 0.5 µM of Oligo dT primer. The first-strand cDNA (2 µl) was used as a PCR template in a 50-µl PCR reaction containing 5 µl 10× PCR buffer, 1 mM primers, 1 µl of 10 mM for each dNTP, and Taq polymerase (Promega). Amplification conditions for LaPDI52, LaPDI47, and LaPDI40 were 95°C for 5 min, followed by 40 cycles of 95°C for 1 min; 56°C for 1 min and 30 s; and 72°C for 2 min. For LaPDI15 amplification, the annealing temperature was 60°C.
Sequencing and sequence analysis
PCR products were directionally cloned into pET22b(+) plasmid (Novagen, Madison, WI, USA) with designed restriction enzymes, and recombinant plasmids were purified as a template for sequencing. Sequencing was conducted in the Protein Chemistry Lab at the University of Texas Medical Branch. Raw sequence data were assembled and edited with a DNASIS package (Version2.0). Homology analyses were performed using the DNA–DNA and protein–protein BLAST in the Leishmania genome databank (http://www.genedb.com/leishmania) and CLUSTAL W in the EMBL website (http://www.ebi.ac.uk/clustalw/). Potential signal peptide cleavage sites were identified with SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/).
Expression, purification and N-terminal sequencing of L. amazonensis PDI protein
The recombinant plasmid pET22b(+) containing the LaPDI gene was transformed into Escherichia coli BL21DE3 for protein expression. Bacteria with recombinant plasmid were cultured at 37°C until OD600 reached 0.6–0.8, and then 1 mM IPTG was added for an additional 4 h of cultivation. Proteins were purified using His-tag columns (Novagen), and purified proteins were verified by SDS-PAGE and Western blot. For protein purification, bacteria pellets were resuspended in lysis buffer containing 0.1 M Tris-Cl, pH 8.0, 0.15 M NaCl, 10 mM DTT, 2% Triton X-100, and a protease inhibitor cocktail (Sigma) for 30 min. Samples were freeze/thawed three times, sonicated on ice briefly, and centrifuged at 10,000×g for 30 min to remove insoluble debris. Clear lysates were loaded onto a His-tag column, and purified proteins were used for PDI activity assay and Western blot analysis. For Western blot analysis, proteins were resolved on 12% SDS-PAGE precasting gel and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After blocking with 1% casin for 1 h, membranes were incubated with either anti-His mAb (1:4,000; Novagen) or pooled mouse anti-L. amazonensis sera (1:100) in the blocking buffer for 1 h. Preimmune mouse serum was used as a control. After they were washed with TBS buffer (20 mM Tris-HCl and 137 mM NaCl, pH 7.4) and then with TBSTT buffer (TBS buffer with 0.1% Tween 20), membranes were incubated with HRP-conjugated goat anti-mouse IgG (1:10,000; Novagen) for 1 h. Specific proteins were visualized with the enhanced chemiluminescence detection system (Novagen). For N-terminal sequencing analysis, expressed PDI proteins were electrophoresed in 12% SDS-PAGE and then transferred to a PVDF membrane. After staining the membrane with Coomassie brilliant blue R-250, the expected protein bands were cut and stored in −80°C, and the N-terminal sequences were analyzed in an automated protein sequencer (Applied Biosystems, Foster, CA, USA) in the Protein Chemistry Lab at the University of Texas Medical Branch.
Parasite protein extraction and concentration
Freshly cultured L. amazonensis promastigotes (1×108/ml) were washed three times in PBS and suspended in lysis buffer containing 0.1 M Tris-Cl, pH 8.0, 0.15 M NaCl, 10 mM DTT, 2% Triton X-100, and a protease inhibitor cocktail (Sigma). Samples were freeze/thawed three times, sonicated on ice briefly and centrifuged at 10,000×g for 30 min to remove insoluble debris. Clear lysates were dialyzed against the AME buffer overnight, and aliquots from each preparation were diluted for measuring protein concentrations using a BCA protein assay (BioRad, Hercules, CA, USA). Lysates were immediately frozen down at −80°C until analysis.
PDI activity assays
PDIs are known to have isomerization activity, which can reduce scrambled RNase to native RNase, referred to as a PDI standard assay (Gilbert 1997). Some PDI isoforms also have oxidization and reduction activity, which can reduce oxidized glutathione (GSSG) to reduced glutathione (GSH) (Lambert and Freedman 1983). For the PDI standard assay, 8 µM scrambled RNAse was incubated with 1.4 µM PDI protein in a buffer containing 4.5 mM cytidine 2′,3′-cyclic monophosphate (cCMP), 1 mM reduced glutathione, 0.2 mM oxidized glutathione, 2 mM EDTA, and 100 mM Tris–HCl, pH 8.0. The cCMP reduced by active RNase into CMP was monitored by the absorbance of 296 nm for 20 min in a DU80 spectrophotometer (Beckman Instrument, Champlain, France). For the PDI reductive assay, the final reaction mixture contained 8 mM GSH, 120 µM NADPH, 1 U glutathione reductase, 5 mM EDTA, and PDI protein in 200 mM sodium phosphate buffer (pH 7.5) in 1-ml cuvettes. The mixture was equilibrated at 30°C for 2 min in an UA800 spectrophotometer, and then 30 µM insulin was added. After a stable, nonenzymic rate had been maintained for 2–3 min, PDI protein (20 µg) was added. After mixing, the ability of PDI to reduce the disulfide bonds between insulin chains was measured as a change in absorbance at 650 nm.
The growth of L. amazonensis promastigotes in the presence of PDI inhibitors
Log-phase L. amazonensis promastigotes (cultured in complete Schneider's medium for 3 days) were diluted into 1 × 106/ml and seeded in 24-well plates predisposed with PDI inhibitors, pCMBA (p-chloromercuribenzoic acid) or bacitracin (Sigma) at final concentrations of 5, 2, 1.5, 0.5, and 0.01 mM. At 24, 48, 72, 96, and 120 h of treatment, residual live parasites were counted daily under a microscope using trypan blue staining in which dead parasites were stained blue. Triplicate wells were done for each condition, and data were averaged for triple independent experiments.
Results
The PDI gene family in Leishmania spp. and cloning PDI genes for L. amazonensis
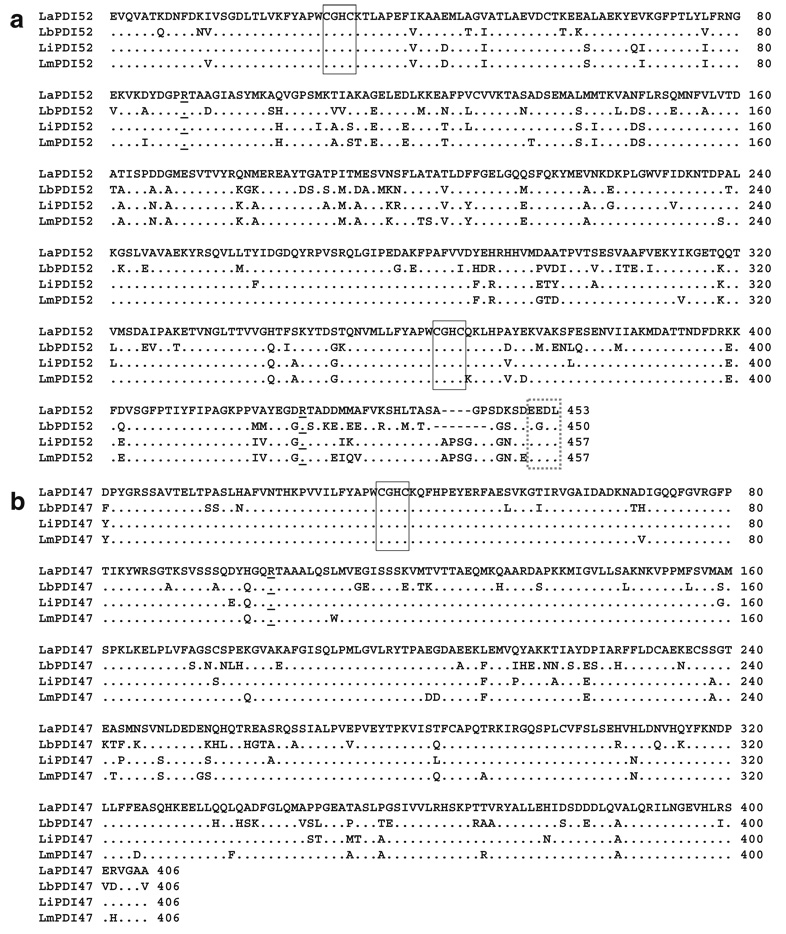
The activity sites of PDI have been reported to contain a “CGHC” motif (Ferrari and Soling 1999). After our protein motif BLAST search of the available Gene Data Bank (http://www.genedb.org/genedb/leish/), we found four expected PDI proteins in L. major, L. braziliensis, and L. infantum parasites, having molecular weights of 52-, 47-, 40-, and 15-kDa, respectively. Because the genome sequencing of L. amazonensis was incomplete, homology analysis of expected PDI sequences from these three Leishmania species was used to design pairs of universal primers for the amplification of L. amazonensis PDI genes. The resultant fragments were confirmed via digestion with internal restriction enzymes and directionally cloned into the pET-22b(+) plasmid. The complete amino acid sequences of LaPDI52 (Fig. 1a), LaPDI47 (Fig. 1b), LaPDI40 (Fig. 1c), and LaPDI15 (Fig. 1d) were compared with those of other Leishmania species, and the overall similarities between each PDI protein were found to be relatively high (75–99%). Of note, while the 40-kDa PDI was highly similar (99%) in both L. major and L. amazonensis, we were unable to find the homolog in L. braziliensis. In comparison to the PDI52 sequences for L. major and L. infantum, LaPDI52 missed four amino acids at the C-terminal end close to the ER-recognizing site (EEDL) (marked by dashed box in Fig. 1a).
Fig. 1.
Amino acid sequences of four PDI proteins among different Leishmania species. Amino acid sequence alignments for PDI52 (a), PDI47 (b), PDI40 (c), and PDI15 (d), among different New World species (L. amazonensis and L. braziliensis) and Old World species (L. major and L. infantum), are shown. Alignments and homology were analyzed with Clustral W bio-soft. The catalytic domains are denoted by solid boxes, and the ER recognizing site by a dashed box. The absent amino acids in the LaPDI52 and LbPDI52 sequences are indicated by dashes. The highly conservative arginine in the PDI family is underlined (Lappi et al. 2004). Gene Data Bank accession numbers for L. amazonensis PDIs are DQ886663, DQ886662, DQ886661, and DQ886660, respectively
Based upon the classification of PDI proteins in Trypanosoma brucei (Rubotham et al. 2005), we named the 52- and 40-kDa PDIs as class 1 and class 2, respectively, based upon their two “CGHC” redox-active motifs, and grouped the 47- and 15-kDa PDIs into the “single domain” class because they contained only one “CGHC” redox-active motif (Table 1). Potential isoelectrical points of each PDI appeared different between the New World and Old World species (Table 1).
Table 1.
Functionally inferred and cloned PDI proteins in different species of Leishmania
| Species/geneDB accession no. | Predicted size (kDa) | Isoelectric point | Redox-active motif | Class | Reference |
|---|---|---|---|---|---|
| L. amazonensis | |||||
| LaPDI52 | 52 | 5.0 | CGHC/CGHC | Class 1 | In this study |
| LaPDI40 | 40 | 6.1 | CGHC/CGHC | Class 2 | In this study |
| LaPDI47 | 47 | 6.5 | CGHC | Single domain 1 | In this study |
| LaPDI15 | 15 | 8.0 | CGHC | Single domain 2 | In this study |
| L. braziliensis | |||||
| LbrM35.6670 | 52 | 4.8 | CGHC/CGHC | Class 1 | / |
| LbrM20.3420 | 47 | 8.6 | CGHC | Single domain 1 | / |
| LbrM06.0880 | 15 | 9.7 | CGHC | Single domain 2 | / |
| L. major | |||||
| LmjF36.6940 | 52 | 5.0 | CGHC/CGHC | Class 1 | Ben Achour |
| LmjF26.0660 | 40 | 7.3 | CGHC/CGHC | Class 2 | / |
| LmjF34.2200 | 47 | 7.3 | CGHC | Single domain 1 | / |
| LmjF06.1050 | 15 | 8.3 | CGHC | Single domain 2 | / |
| L. infantum | |||||
| LinJ36.0630 | 52 | 5.3 | CGHC/CGHC | Class 1 | / |
| LinJ26.0630 | 40 | 6.9 | CGHC/CGHC | Class 2 | / |
| LinJ34.1760 | 47 | 7.2 | CGHC | Single domain 1 | / |
| LinJ06.1090 | 15 | 8.7 | CGHC | Single domain 2 | / |
Expression and purification of L. amazonensis PDI proteins
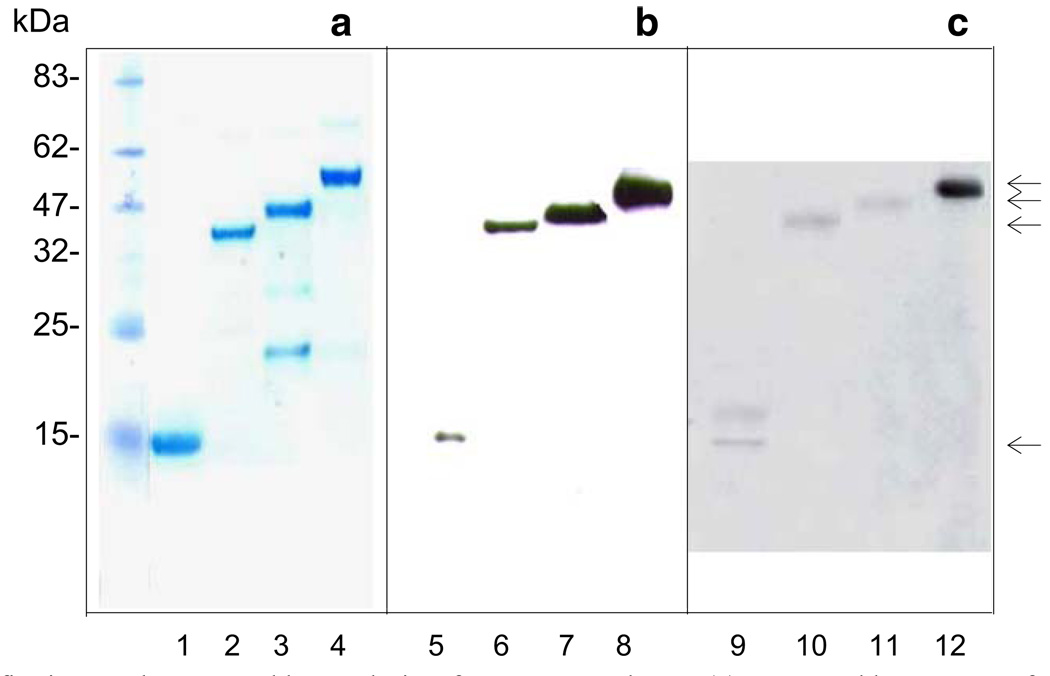
Recombinant plasmids were transformed into E. coli BL21(DE3) for the expression of recombinant proteins. LaPDI52, LaPDI47, and LaPDI15 had a comparable expression level at 37°C, while LaDPI40 had a relatively low expression level under the same conditions (data not shown). All of the purified LaPDI proteins (Fig. 2, lanes 1 to 4) were sequenced at the N- terminals, and their correct expression was confirmed. Through Western blots with an anti-His mAb, we detected strong bands of the expected size (Fig. 2, lanes 5 to 8). It appeared that only LaPDI52 (Fig. 2, lane 16) showed strong binding to sera derived from L. amazonensis-infected mice, whereas LaPDI15, LaPDI40, and LaPDI47 showed relatively weak reactions (Fig. 2, lanes 9 to 12). Recombinant PDI proteins showed no reactions to preimmune mouse serum (data not shown).
Fig. 2.
Expression, purification, and Western blot analysis of L. amazonensis PDI proteins. Recombinant plasmids containing the corresponding L. amazonensis PDI gene fragment (without the signal peptide) were transformed into BL21DE3 cells and induced for protein expression (1 mM IPTG for 4 h). Expressed PDI proteins were detected by electrophoresis in 12% SDS-PAGE and purified with His-tag columns (a). Western blots were performed for the detection of expressed PDI proteins by anti-His mAb (b) or pooled sera from L. amazonensis-infected mice (c). Lanes 1, 5, and 9 are for LaPDI15; lanes 2, 6, and 10 are for LaPDI40; lanes 3, 7, and 11 are for LaPDI47; and lanes 4, 8, and 12 are for LaPDI52. Arrows indicate the proteins with expected sizes
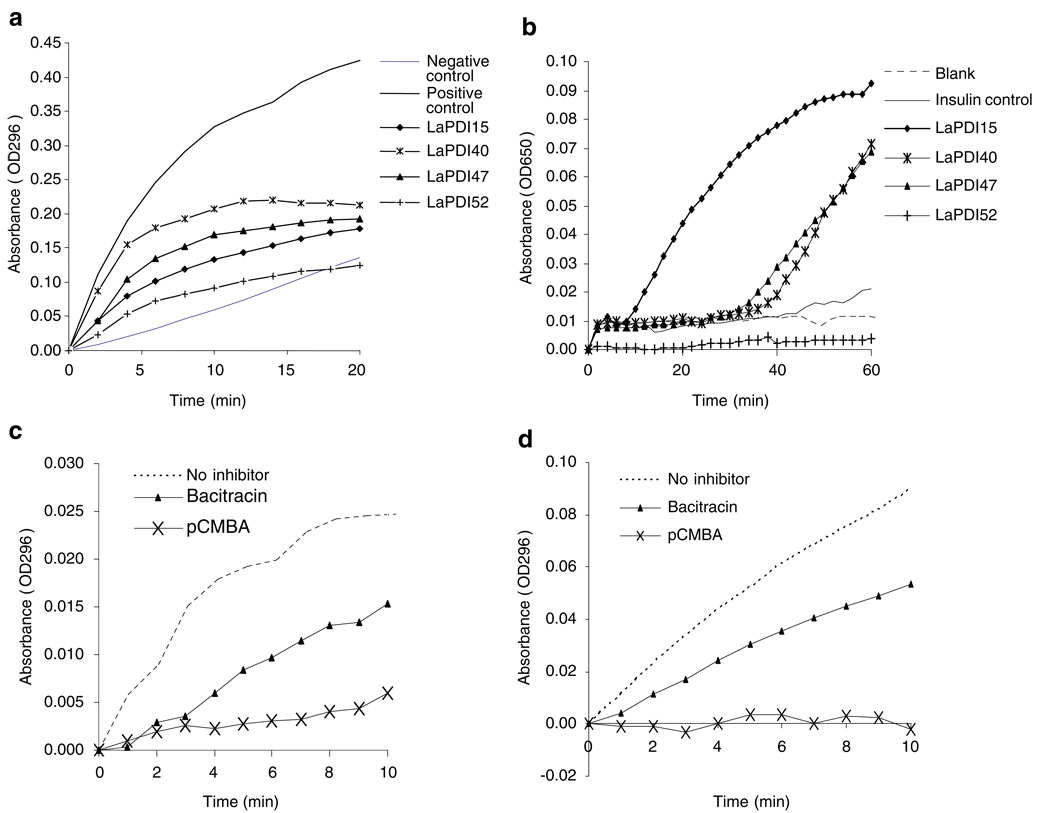
Enzymatic activities of LaPDI proteins and sensitivity to inhibitors
PDI activities are often assessed by a standard “scrambled RNase” assay, which monitors the conversion of cCMP to CMP at OD296, and, alternatively, by an insulin reduction assay, which measures the reduction of insulin at OD650. Although all four purified LaPDI recombinants displayed PDI activity, based upon the standard assay (Fig. 3a), their enzymatic levels were different with relative activity ranging in the following order: LaPDI40>LaPDI47>LaPDI15>LaPDI52. With regard to insulin reduction processes (Fig. 3b), LaPDI 15 had the highest level of activity, followed by LaPDI40 and LaPDI47. In contrast, LaPDI52 showed no insulin reduction activity. Because LaPDI40 protein has two ‘CGHC’ activity sites, whereas LaPDI47 protein has only one ‘CGHC’ activity site, we next examined their sensitivities to the PDI inhibitors pCMBA and bacitracin (Mandel et al. 1993). It was evident that the enzymatic activities of LaPDI40 (Fig. 3c) and LaPDI47 (Fig. 3d) were inhibited by 2 mM of pCMBA and bacitracin, respectively. The inhibitory effects of PDI inhibitors on LaPDI52 and LaPDI15 were similar to those on LaPD47 and LaPDI40 (data not shown). Together, these results confirm the functional activities of our recombinant proteins.
Fig. 3.
PDI activities in expressed LaPDI proteins and their responsiveness to PDI inhibitors. Representative results for the standard PDI assay (a) and reduction activity assay (b) are shown from one of three independent repeats. The reduction of cCMP to CMP by LaPDMO (c) and LaPDI47 (d) were assessed in the absence or presence of 2 mM bacitracin and 2 mM pCMBA, respectively, and representative results are shown from three independent repeats
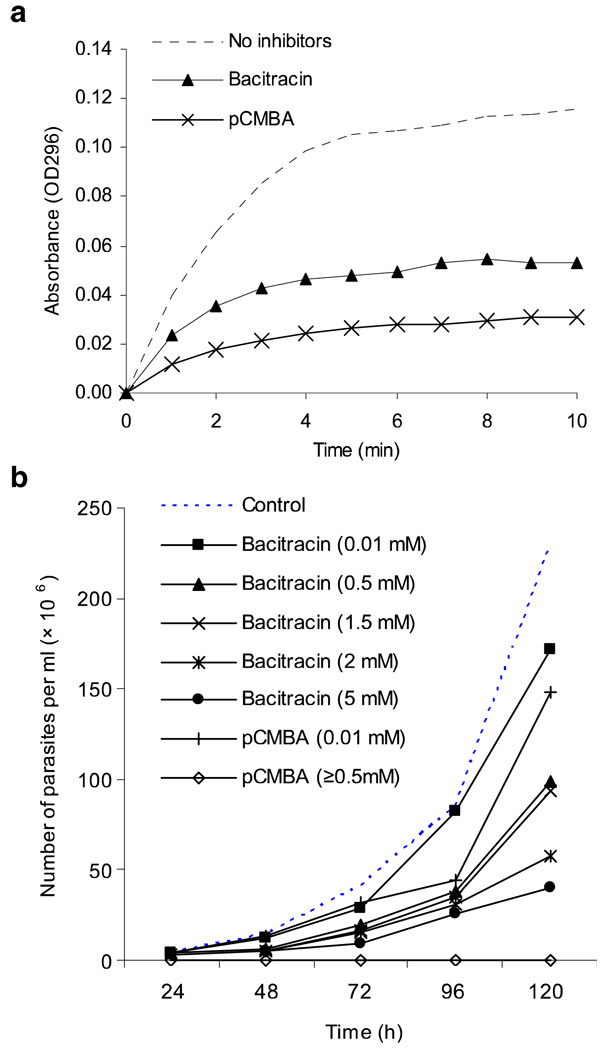
PDI activity in L. amazonensis promastigotes
Having demonstrated the enzymatic activities of PDI proteins and their sensitivities to PDI inhibitors, we next examined whether PDI activities can be detected in L. amazonensis. Parasite lysates were prepared from 1 × 109 promastigotes and tested for PDI activity using the “scrambled RNase” assay. As shown in Fig. 4a, promastigotes had appreciable levels of PDI activities, and the addition of 2 mM bacitracin and pCMBA resulted in a 50% and 70% reduction in enzymatic activities, respectively. We then examined whether PDI inhibitors could reduce parasite growth. Promastigotes (1×106 /ml) were cultured in 48-well plates in the presence or absence of indicated concentrations of pCMBA and bacitracin, and viable parasites were counted daily for 5 days. No viable parasites were detected in the presence of 0.5, 1.5, 2, or 5 mM of pCMBA; however, viable parasites (30×l06/ml) were still detected at day 5 after treatment with 5 mM bactericin (Fig. 4b). Together, these results indicate the presence of PDI activities in promastigotes and the potent effect of pCMBA for suppressing parasite growth.
Fig. 4.
PDI activity of L. amazonensis promastigotes and the effects of PDI inhibitors in parasite growth, a Promastigote lysates were prepared from 1 × 109 parasites and analyzed for PDI activity in the absence or presence of 2 mM bacitracin or 2 mM pCMBA. Results are presented as absorbance at OD296, and representative results are shown from three repeated experiments, b L. amazonensis promastigotes (l × l06/ml) were cultured in the absence or presence of indicated concentrations of PDI inhibitors for consecutive 5 days. Parasite numbers were counted microscopically, and results are presented as parasite numbers per ml (×106). Shown are representative results from three independent repeats
Discussion
PDI proteins are essential enzymes that catalyze thiol-disulfide interchange, ensuring the proper folding and conformation of proteins, acting as coreceptors of cell reorganization, and preventing cell toxicity associated with ER stress and protein misfolding (Kimura et al. 2005; Kimura et al. 2004; Tian et al. 2004). Most members of the PDI family can function both as oxidoreductases/isomerases and as molecular chaperones in the ER of eukaryotic cells (Ferrari and Soling 1999). PDI comprises two thio-redoxin-like catalytic domains that are separated by two noncatalytic domains. The catalytic domains contain a characteristic CXXC active-site motif with the two cysteine residues playing a major role in determining the redox potential of the enzyme and its function as a thiol-disulfide reductase, oxidase, or isomerase (Ellgaard and Ruddock 2005). Recently, a conservative arginine has been reported to modulate the pKa of the active-site cysteine residues by moving into and out of the active-site locale (Lappi et al. 2004).
Although much has been learned about PDIs in higher eukaryotes, limited information is available regarding PDIs in pathogens that are important for human infections. In this study, we have characterized and compared the PDI family in Leishmania spp. and have grouped Leishmania PDI proteins into two categories: one with two “CGHC” sites and another with only one “CGHC” activity site. Consistent with other known PDIs, all four Leishmania PDIs have a highly conserved arginine near their CGHC active site motifs (Fig. 1). Gene amplification of the L. amazonensis PDIs with specific primers allows us to perform homology analysis between New World and Old World species. Similar to the counterparts in other Leishmania species, LaPDI52 and LaPDI40 have two activity ‘CGHC’ sites, whereas LaPDI47 and LaPDI15 have only one active site. A 4- to 7- amino acid deletion was found in the 52-kDa PDI of New World species (L. amazonensis and L. braziliensis). Although the homology of the 40-kDa PDIs is very high (99%) in New World and Old World species, this gene appears to be absent in L. braziliensis, as judged by a sequence search and RT-PCR analysis (data not shown). Although the biological function of Leishmania PDIs remains unclear at this stage, the increased expression of LmPDI52 in highly virulent strains of L. major suggests their involvement in parasite pathogenesis (Ben Achour et al. 2002).
The enzymatic activity of PDI lies in its reduction oxidization and isomerase ability, and the standard assay for the activity utilizes refolding of the scrambled RNase. Other assays may employ reduction of insulin in some PDI proteins or the oxidation of reduced lysozyme or trypsin. We have found that although all of the four expressed PDI proteins are enzymatically active, their functional preference in parasites may differ significantly. In the standard PDI assay, LaPDI40 and LaPDI47 have a higher activity than do LaPDI52 and LaPDI15 (Fig. 3a). In the insulin reduction assay, LaPDI15 has the highest activity, while LaPDI40 and LaPDI47 show a relatively low and slow functional activity. In agreement with a report for the 52-kDa PDI of T. brucei (Rubotham et al. 2005), LaPDI52 possesses no oxidization/reduction activity (Fig. 3b). Our findings that PDI activities can be detected in L. amazonensis promastigote lysates and that bacitracin and pCMBA can decrease these activities by 50% to 70% further support the view that PDIs play an important role in Leishmania natural pathogenicity and may serve as new targets for anti-Leishmania chemotherapy (Ben Achour et al. 2002). Because a low concentration (0.5 mM) of pCMBA was sufficient to kill L. amazonensis promastigotes in culture, whereas relatively high (>5 mM) concentrations of bacitracin were required to reach a comparable effect, it would be interesting to examine the effect of these PDI inhibitors in macrophage infection in vitro.
TGases, a family of enzymes that catalyzes calcium-dependent, covalent cross-linking of cellular proteins, have been reported with PDI activity in several parasites, including Dirofilaria immitis (Chandrashekar et al. 1998), Brugia malayi (Singh and Mehta 1994), and Caenorhabditis elegans (Natsuka et al. 2001). We have recently demonstrated TGase activity in Leishmania promastigotes and amastigotes (Brobey and Soong 2006). Although we did not detect any TGase activity in any of the four purified L amazonensis PDI proteins (data not shown), it would be interesting to further evaluate the relationship of these two enzymes in other Leishmania species.
The members of the PDI family are often resident proteins in the lumen of the ER where they participate in the folding and maturation of newly synthesized proteins. Among the four Leishmania PDIs, only the 52-kDa PDI has the ER-retention signal (EEDL). Although the 15-kDa PDI of L. donovania does not have the ER-retention signal, it is located, at least in part, in the ER and plays a critical role in the leishmanial secretory pathway (Padilla et al. 2003). Given that a 55-kDa PDI identified in the salivary gland of the tick contains an ER-retention signal (KEEL) and that this protein is exclusively located in the ER (Knizetova et al. 2006), it is possible that PDIs without the ER-retention signal, such as the 45-kDa of PDI of T. brucei (Rubotham et al. 2005) and LaPDI47, LaPDI40, and LaPDI15, may have a relatively broad subcellular location and different functions. PDIs can be found on the surface of host cells, such as human B lymphocytes, and PDI-active sites may participate in the reducing environment of the cell exterior and/or be involved in cell adhesion (Turano et al. 2002).
PDIs may also be involved in the host mucosal immune system, inducing secretory IgA (Gregory and Filler 1987; Zierhut et al. 1998). For example, T. gondii PDIs are recognized by host IgA (Meek et al. 2002). Because L. braziliensis can cause mucosal leishmaniasis in humans, it will be interesting to further examine whether IgA from mucosal leishmaniasis patients can recognize leishmanial PDIs. Although we detected IgG specific to four LaPDIs (especially to the 52-kDa PDI) in L. amazonensis-infected mice (Fig. 2), additional studies are needed to assess the nature and extent of host immune responses to different parasite PDI isoforms. This is especially relevant, given the fact that the 52-kDa PDI appears to be linked to L. major virulence (Ben Achour et al. 2002) and that there is a correlation between B cell activation/Ab production and lesion progression in Leishmania-infected patients and experimental animals (Miles et al. 2005; Wanasen et al. 2007) Because three of the four PDI proteins apparently do not contain the ER-retention signal, further investigations are needed to address the intercellular location in Leishmania parasites and their possible function in host-parasite interaction.
In summary, our studies of L. amazonensis PDI proteins have indicated that these four PDI proteins have different oxidoreductases/isomerase activities and profiles of reaction with anti-Leishmania serum. Because PDI inhibitors can block expressed PDI protein activities and parasite growth in culture, we suggest that PDI proteins may involve different aspects of parasite biology and host–parasite interaction during Leishmania infection. Given the importance of PDI proteins in protein-protein interactions, additional studies would be needed to address the functional roles of these PDI proteins in Leishmania infection in vitro and in vivo.
Acknowledgements
We thank Reynolds Brobey for the helpful discussion and technical assistance during the course of this study and Mardelle Susman for the comments on the manuscript. This study was funded by the NIH grant AI43003 to L.S.
References
- Alvar J, Canavate C, Gutierrez-Solar B, Jimenez M, Laguna F, Lopez-Velez R, Molina R, Moreno J. Leishmania and human immunodeficiency virus coinfection: the first 10 years. Clin Microbiol Rev. 1997;10:298–319. doi: 10.1128/cmr.10.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbouche R, Lortat-Jacob H, Jones IM, Fenouillet E. Glycosaminoglycans and protein disulfide isomerase-mediated reduction of HIV Env. Mol Pharmacol. 2005;67:1111–1118. doi: 10.1124/mol.104.008276. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Mendez S, Lira R, Kadambi N, Milon G, Sacks D. A natural model of Leishmania major infection reveals a prolonged “silent” phase of parasite amplification in the skin before the onset of lesion formation and immunity. J Immunol. 2000;165:969–977. doi: 10.4049/jimmunol.165.2.969. [DOI] [PubMed] [Google Scholar]
- Ben Achour Y, Chenik M, Louzir H, Dellagi K. Identification of a disulfide isomerase protein of Leishmania major as a putative virulence factor. Infect Immun. 2002;70:3576–3585. doi: 10.1128/IAI.70.7.3576-3585.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brobey RK, Soong L. Leishmania species: evidence for transglutaminase activity and its role in parasite proliferation. Exp Parasitol. 2006;114:94–102. doi: 10.1016/j.exppara.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Chandrashekar R, Tsuji N, Morales T, Ozols V, Mehta K. An ERp60-like protein from the filarial parasite Dirofilaria immitis has both transglutaminase and protein disulfide isomerase activity. Proc Natl Acad Sci USA. 1998;95:531–536. doi: 10.1073/pnas.95.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant CG, Stephens RS. Chlamydia attachment to mammalian cells requires protein disulfide isomerase. Cell Microbiol. 2007;9:222–232. doi: 10.1111/j.1462-5822.2006.00783.x. [DOI] [PubMed] [Google Scholar]
- Desjeux P. Leishmaniasis: current situation and new perspectives. Comp Immunol Microbiol Infect Dis. 2004;27:305–318. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Ellerman DA, Myles DG, Primakoff P. A role for sperm surface protein disulfide isomerase activity in gamete fusion: evidence for the participation of ERp57. Dev Cell. 2006;10:831–837. doi: 10.1016/j.devcel.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engwerda CR, Ato M, Kaye PM. Macrophages, pathology and parasite persistence in experimental visceral leishmaniasis. Trends Parasitol. 2004;20:524–530. doi: 10.1016/j.pt.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Eschenlauer SC, Page AP. The Caenorhabditis elegans ERp60 homolog protein disulfide isomerase-3 has disulfide isomerase and transglutaminase-like cross-linking activity and is involved in the maintenance of body morphology. J Biol Chem. 2003;278:4227–4237. doi: 10.1074/jbc.M210510200. [DOI] [PubMed] [Google Scholar]
- Ferrari DM, Soling HD. The protein disulphide-isomerase family: unravelling a string of folds. Biochem J. 1999;339(Pt 1):1–10. [PMC free article] [PubMed] [Google Scholar]
- Gilbert HF. Protein disulfide isomerase and assisted protein folding. J Biol Chem. 1997;272:29399–29402. doi: 10.1074/jbc.272.47.29399. [DOI] [PubMed] [Google Scholar]
- Gregory RL, Filler SJ. Protective secretory immunoglobulin A antibodies in humans following oral immunization with Streptococcus mutans. Infect Immun. 1987;55:2409–2415. doi: 10.1128/iai.55.10.2409-2415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handman E. Leishmaniasis: current status of vaccine development. Clin Microbiol Rev. 2001;14:229–243. doi: 10.1128/CMR.14.2.229-243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Hosoda Y, Sato Y, Kitamura Y, Ikeda T, Horibe T, Kikuchi M. Interactions among yeast protein-disulfide isomerase proteins and endoplasmic reticulum chaperone proteins influence their activities. J Biol Chem. 2005;280:31438–31441. doi: 10.1074/jbc.M503377200. [DOI] [PubMed] [Google Scholar]
- Kimura T, Nishida A, Ohara N, Yamagishi D, Horibe T, Kikuchi M. Functional analysis of the CXXC motif using phage antibodies that cross-react with protein disulphide-isomerase family proteins. Biochem J. 2004;382:169–176. doi: 10.1042/BJ20040116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knizetova P, Vancova I, Kocakova P, Slovak M, Proost P, Kopacek J. New member of the protein disulfide isomerase (PDI) family identified in Amblyomma variegatum tick. Insect Biochem Mol Biol. 2006;36:943–953. doi: 10.1016/j.ibmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Kuhls K, Mauricio IL, Pratlong F, Presber W, Schonian G. Analysis of ribosomal DNA internal transcribed spacer sequences of the Leishmania donovani complex. Microbes Infect. 2005;7:1224–1234. doi: 10.1016/j.micinf.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Lambert N, Freedman RB. Kinetics and specificity of homogeneous protein disulphide-isomerase in protein disulphide isomerization and in thiol-protein-disulphide oxidoreduction. Biochem J. 1983;213:235–243. doi: 10.1042/bj2130235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappi AK, Lensink MF, Alanen HI, Salo KE, Lobell M, Juffer AH, Ruddock LW. A conserved arginine plays a role in the catalytic cycle of the protein disulphide isomerases. J Mol Biol. 2004;335:283–295. doi: 10.1016/j.jmb.2003.10.051. [DOI] [PubMed] [Google Scholar]
- Liao M, Ma L, Bannai H, Lee EG, Xie Z, Tang X, Zhang H, Xuan X, Fujisaki K. Identification of a protein disulfide isomerase of Neospora caninum in excretory-secretory products and its IgA binding and enzymatic activities. Vet Parasitol. 2006;139:47–56. doi: 10.1016/j.vetpar.2006.02.029. [DOI] [PubMed] [Google Scholar]
- Mandel R, Ryser HJ, Ghani F, Wu M, Peak D. Inhibition of a reductive function of the plasma membrane by bacitracin and antibodies against protein disulfide-isomerase. Proc Natl Acad Sci USA. 1993;90:4112–4116. doi: 10.1073/pnas.90.9.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio IL, Yeo M, Baghaei M, Doto D, Pratlong F, Zemanova E, Dedet JP, Lukes J, Miles MA. Towards multilocus sequence typing of the Leishmania donovani complex: resolving genotypes and haplotypes for five polymorphic metabolic enzymes (ASAT, GPI, NH1, NH2, PGD) Int J Parasitol. 2006;36:757–769. doi: 10.1016/j.ijpara.2006.03.006. [DOI] [PubMed] [Google Scholar]
- McMahon-Pratt D, Alexander J. Does the Leishmania major paradigm of pathogenesis and protection hold for New World cutaneous leishmaniases or the visceral disease. Immunol Rev. 2004;201:206–224. doi: 10.1111/j.0105-2896.2004.00190.x. [DOI] [PubMed] [Google Scholar]
- Meek B, Back JW, Klaren VN, Speijer D, Peek R. Protein disulfide isomerase of Toxoplasma gondii is targeted by mucosal IgA antibodies in humans. FEBS Lett. 2002;522:104–108. doi: 10.1016/s0014-5793(02)02911-3. [DOI] [PubMed] [Google Scholar]
- Miles SA, Conrad SM, Alves RG, Jeronimo SM, Mosser DM. A role for IgG immune complexes during infection with intracellular pathogen Leishmania. J Exp Med. 2005;201:747–754. doi: 10.1084/jem.20041470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguleswaran A, Alaeddine F, Guionaud C, Vonlaufen N, Sonda S, Jenoe P, Mevissen M, Hemphill A. Neospora caninum protein disulfide isomerase is involved in tachyzoite-host cell interaction. Int J Parasitol. 2005;35:1459–1472. doi: 10.1016/j.ijpara.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Natsuka S, Takubo R, Seki R, Ikura K. Molecular cloning and expression of Caenorhabditis elegans ERp57-homologue with transglutaminase activity. J Biochem (Tokyo) 2001;130:731–735. doi: 10.1093/oxfordjournals.jbchem.a003042. [DOI] [PubMed] [Google Scholar]
- Padilla A, Noiva R, Lee N, Mohan KV, Nakhasi HL, Debrabant A. An atypical protein disulfide isomerase from the protozoan parasite Leishmania containing a single thioredoxin-like domain. J Biol Chem. 2003;278:1872–1878. doi: 10.1074/jbc.M210322200. [DOI] [PubMed] [Google Scholar]
- Papp E, Szaraz P, Korcsmaros T, Csermely P. Changes of endoplasmic reticulum chaperone complexes, redox state, and impaired protein disulfide reductase activity in misfolding alphal-antitrypsin transgenic mice. Faseb J. 2006;20:1018–1020. doi: 10.1096/fj.05-5065fje. [DOI] [PubMed] [Google Scholar]
- Robinson CG, Roy CR. Attachment and fusion of endoplasmic reticulum with vacuoles containing Legionella pneumophila. Cell Microbiol. 2006;8:793–805. doi: 10.1111/j.1462-5822.2005.00666.x. [DOI] [PubMed] [Google Scholar]
- Rubotham J, Woods K, Garcia-Salcedo JA, Pays E, Nolan DP. Characterization of two protein disulfide isomerases from the endocytic pathway of bloodstream forms of Trypanosoma brucei. J Biol Chem. 2005;280:10410–10418. doi: 10.1074/jbc.M409375200. [DOI] [PubMed] [Google Scholar]
- Sacks D, Sher A. Evasion of innate immunity by parasitic protozoa. Nat Immunol. 2002;3:1041–1047. doi: 10.1038/ni1102-1041. [DOI] [PubMed] [Google Scholar]
- Singh RN, Mehta K. Purification and characterization of a novel transglutaminase from filarial nematode Brugia malayi. Eur J Biochem. 1994;225:625–634. doi: 10.1111/j.1432-1033.1994.00625.x. [DOI] [PubMed] [Google Scholar]
- Tian R, Li SJ, Wang DL, Zhao Z, Liu Y, He RQ. The acidic C-terminal domain stabilizes the chaperone function of protein disulfide isomerase. J Biol Chem. 2004;279:48830–48835. doi: 10.1074/jbc.M407076200. [DOI] [PubMed] [Google Scholar]
- Tinsley CR, Voulhoux R, Beretti JL, Tommassen J, Nassif X. Three homologues, including two membrane-bound proteins, of the disulfide oxidoreductase DsbA in Neisseria meningitidis: effects on bacterial growth and biogenesis of functional type IV pili. J Biol Chem. 2004;279:27078–27087. doi: 10.1074/jbc.M313404200. [DOI] [PubMed] [Google Scholar]
- Turano C, Coppari S, Altieri F, Ferraro A. Proteins of the PDI family: unpredicted non-ER locations and functions. J Cell Physiol. 2002;193:154–163. doi: 10.1002/jcp.10172. [DOI] [PubMed] [Google Scholar]
- Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- Wanasen N, Xin L, Soong L. Pathogenic role of B cells and antibodies in murine Leishmania amazonensis infection. Int J Parasitol. 2007 doi: 10.1016/j.ijpara.2007.08.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut M, Elson CO, Forrester JV, Kijlstra A, Kraehenbuhl JP, Sullivan DA. Mucosal immunology and the eye. Immunol Today. 1998;19:148–150. doi: 10.1016/s0167-5699(97)01229-2. [DOI] [PubMed] [Google Scholar]