Abstract
Intracerebral hemorrhage (ICH) produces a high rate of death and disability. The molecular mechanisms of damage in perihematomal tissue in humans have not been systematically characterized. This study determines the gene expression profile and molecular networks that are induced in human perihematomal tissue through molecular analysis of tissue obtained from endoscopic clot evacuation. Differentially expressed genes and their cellular origin were confirmed in a mouse model of ICH. A total of 624 genes showed altered regulation in human ICH. Bioinformatic analysis shows that these genes form interconnected networks of proinflammatory, anti-inflammatory, and neuronal signaling cascades. Intracerebral hemorrhage evokes coordinated upregulation of proinflammatory signaling through specific cytokines and chemokines and their downstream molecular pathways. Anti-inflammatory networks are also induced by ICH, including annexins A1 and A2 and transforming growth factor beta (TGF β) and their intracellular cascades. Intracerebral hemorrhage downregulates many neuronal signaling systems, including the N-methyl-d-aspartic acid (NMDA) receptor complex and membrane ion channels. Select portions of these molecular networks were confirmed in the mouse, and the proteins in a subset of these networks localized to subsets of neurons, oligodendrocytes, or leukocytes. These inflammatory and anti-inflammatory networks interact at several key points in neutrophil signaling, apoptotic cell death, and protease responses, and indicate that secondary damage in ICH activates opposing molecular systems.
Keywords: annexin, anti-inflammatory, chemokine, microarray, neuroprotection, perihematomal
Introduction
Intracerebral hemorrhage (ICH) is a common and lethal stroke subtype. Intracerebral hemorrhage constitutes 15% to 20% of all strokes, with only 10% of victims independent at 1 month and 38% survival after 1 year (Qureshi et al, 2001). Intracerebral hemorrhage produces primary damage from the initial hemorrhage and dissection through tissue planes, and an evolving secondary phase of injury in the surrounding tissue. Apoptotic cell death and tissue necrosis occur adjacent to the hemorrhage (Qureshi et al, 2001; Xi et al, 2006). In humans, ICH induces secondary damage in the brain through the induction of cerebral edema and perihematomal injury (Qureshi et al, 2001; Xi et al, 2006).
Perihematomal edema with mass effect is an almost universal complication of the ICH, elaborated over several days after the initial insult. Perihematomal blood flow is reduced, but not to ischemic levels (Miller et al, 2007). Nonetheless, several additional pathologic processes occur in human perihematomal tissue, including glutamate release, bioenergetic failure, inflammation, and apoptosis. Microdialysis of perihematomal regions shows massive local glutamate release; blood glutamate levels correlate with hemorrhage size and reduced functional outcome after stroke (Kidwell et al, 2001; Castillo et al, 2002; Miller et al, 2007). Animal studies support these findings in showing elevated glutamate levels, abnormal glutamate signaling, and increased lactate production in a region with preserved blood flow in perihematomal tissue (Ardizzone et al, 2004; Wagner, 2007). Diffusion magnetic resonance imaging studies show that one-quarter of the patients exhibit a perihematomal region of diffusion abnormality most likely because of advanced bioenergetic failure (Kidwell et al, 2001). Unlike acute ischemic stroke, these processes in perihematomal tissue occur at a delay after ICH—when patients are in the hospital and under direct medical observation. This creates a frustrating clinical scenario because patients’ condition worsens at the time in which there is maximal potential for medical intervention. This pattern of clinical worsening in perihematomal tissue highlights the significance of studies that may provide basic insights into the cellular mechanisms of perihematomal damage and edema after ICH. However, there have been no systematic studies of the molecular pathways in human perihematomal tissue that mediate this secondary brain damage.
Molecular and cellular studies in animal models of ICH indicate that hemorrhage induces inflammation, apoptotic cell death, and progressive tissue destruction in perihematomal tissue. Intracerebral hemorrhage produces a prompt infiltration of white blood cells, inflammatory cytokine production, and secondary inflammatory gene activation during a period of apoptotic cell death (Felberg et al, 2002; Lu et al, 2006; Xi et al, 2006; Wagner, 2007; Wasserman et al, 2007). Gene expression profiling in rat models of ICH indicate a robust upregulation of inflammatory genes in perihematomal tissue (Lu et al, 2006; Wasserman et al, 2007). Studies of human white blood cell gene expression patterns also indicate an inflammatory reaction in blood after ICH (Tang et al, 2006). However, these same studies have shown a poor correlation between genomic expression patterns in peripheral blood in rat ICH compared with peripheral blood in human ICH (Tang et al, 2001, 2006).
The present studies were undertaken to determine the gene expression profile in human perihematomal tissue in the acute period after ICH. On the basis of experimental animal studies, we hypothesized that ICH would induce inflammatory cytokine signaling centered around tumor necrosis factor-α (TNFα), interleukin (IL)1β, and IL6; that ICH induces glial edema-associated genes including aquaporin 4 and vascular endothelial growth factor; and that ICH triggers the expression of matrix metalloproteinase 9 (MMP9) and serum proteases (plasminogen) as mediators of secondary extracellular matrix damage. Human brain samples were obtained during neuroendoscopic evacuation of hemorrhage clots within the first 24 h after ICH. The gene expression profile of perihematomal tissue was determined with microarray analysis compared with control samples from the same brain regions. Bioinformatics analyses and molecular pathway modeling show that ICH induces both inflammatory and anti-inflammatory pathways and a downregulation of neuronal signaling systems. These expression profiles were further studied in a mouse model of ICH, where this pattern of human ICH gene expression was localized to specific cell types for a select number of gene products. These data indicate that intricate proinflammatory and anti-inflammatory systems are activated in human perihematomal tissue and that these gene networks localize within neurons, astrocytes, and oligodendrocytes in a region-specific manner.
Materials and methods
Human Subjects
Tissue was obtained from patients and index cases in a clinical trial (Hemorrhage Evacuation using MR-guided Endoscopy Surgery—HEME Surgery trial). The HEME Surgery trial was a study to evaluate the feasibility, safety, and potential efficacy of minimally invasive, magnetic resonance-guided endoscopic hematoma evacuation after ICH. Patients were randomized to receive clot evacuation or maximal medical care within 24 h of the hemorrhage. No patient received intracranial pressure (ICP) monitoring or activated factor VIIa before clot removal and tissue sampling. The UCLA Institutional Review Board approved the study protocol and consent form.
Genomic studies were performed on perihematomal parenchymal tissue adherent to removed clot. No specific perihematomal wall biopsy was performed; rather, the adherent tissue typically incidentally removed during hematoma evacuation was carefully isolated. After clot evacuation, the evacuated material was immediately rinsed of blood in ice-cold buffered saline and adherent neural parenchymal tissue microdissected off the clot. One sample was not included because of insufficient tissue adherent to the clot. The typical size of the aliquot of microdissected brain parenchyma was 150 to 200 mg. Two-thirds of the tissue was sent to UCLA Neuropathology for diagnostic evaluation and one-third was immediately frozen in liquid nitrogen. Control tissue was obtained from five autopsy cases within 4 h of death from the same brain areas as the sites of the ICH specimens: frontal and parietal cortex; putamen and subcortical white matter (Table 1). Death in these cases was from cardiopulmonary arrest in the setting of advanced but healthy aging (Table 1) and did not occur after prolonged illness. Previous studies have shown that RNA isolation from autopsy cases up to 40 h after death in this clinical situation produces good RNA quality without evidence of systematic changes in gene expression patterns (Li et al, 2004; Stan et al, 2006). This lack of a systematic effect of this postmortem interval on the relevant genes in this study was confirmed with mouse gene expression analysis (Supplementary Figure 1).
Table 1.
Cases
| Case | Age (years) |
Gender | Race | Time (h) |
Hematoma (mL) |
Site | Treatment |
|---|---|---|---|---|---|---|---|
| HS3 | 55 | F | Viet | 6 | 35 | Putamen | Anti-HTN, DPH, insulin |
| HS4 | 79 | M | Cauc | 24 | 40 | Lobar | Anti-HTN, insulin, DPH, platelets, DDAVP |
| HS5 | 71 | M | Cauc | 12 | 70 | Lobar | Anti-HTN, insulin, DPH, platelets, DDAVP |
| HS6 | 51 | M | Hisp | 24 | 30 | Putamen | Anti-HTN, DPH |
| HS8 | 71 | F | Pers | 24 | 45 | Lobar | Anti-HTN, insulin, DPH |
| HS10 | 84 | M | Cauc | 60 | Lobar | Anti-HTN, insulin, DPH | |
| Control 1 | 76 | F | Cauc | 3 | Frontal (C.FC1), putamenal (C.Put), white matter (C.WM1) | ||
| Control 2 | 80 | F | Cauc | 2 | Frontal (C.FC2), white matter (WM2) | ||
| Control 3 | 84 | F | Cauc | 4 | White matter (C.WM3) | ||
| Control 4 | 75 | M | Cauc | 2 | Parietal (C.B_1) | ||
| Control 5 | 78 | M | Cauc | 2 | Parietal (C.B_1877) |
F, female; M, male.
Terms in parentheses correspond to symbols for control specimens in Figure 1. anti-HTN, antihypertensive medicines; Cauc, Caucasian; DDAVP, desmopressin acetate; DPH, dilantin; Hisp, Hispanic; Pers, Persian; Time, time to autopsy or hemorrhage evacuation; Viet, Vietnamese.
RNA Isolation and Array Hybridization
Total RNA was isolated with TRIzol reagent (Carmichael et al, 2005) and RNA integrity assayed (Nanochip, Agilent Biotechnologies, Santa Clara, CA, USA). Three samples were rejected because of insufficient RNA quality (28s:18s ribosomal RNA ratio < 1.7). These samples were micro-dissected and frozen in excess of 10 mins from the time of endoscopic clot retrieval. The remaining six samples and all control samples had excellent RNA quality (ribosomal RNA 28s:18s > 1.7; Supplementary Figure 1). Total RNA was used to generate probes for microarrays following the manufacturer’s guidelines (performed in conjunction with the UCLA Microarray Core facility).
Array Analysis and Bioinformatics
The data set included 14 arrays (8 controls on the u133a platform, 3 perihematomal cases on the u133a platform, and 3 perihematomal on the u133plus platform). Raw data from CEL files were loaded in R statistical software using the ‘affy’ package. RNA degradation was estimated using 3′/5′ ratios in RNA degradation plots and was not significant (Supplementary Figure 2). Intensity values for individual probes were (1) log2-transformed and background corrected, then (2) normalized using quantile normalization, and (3) summarized in individual probe sets using median polish. Data from the two platforms were merged after normalization (MergeMaid package; http://www.bioconductor.org), and the shared probe sets (22,277) were used for further analyses. Quality assessment was performed looking at interarray Pearson’s correlation, and clustering based on top variant genes was used to assess overall data coherence (Figure 1 and Supplementary Figure 2). Contrast analysis of differential expression was performed using the LIMMA package (http://www.bioconductor.org). After linear model fitting, a Bayesian estimate of differential expression was calculated using a false discovery rate (FDR) of 0.001%. Differentially expressed genes were also ranked according to their B score (Figure 2), which ranks genes according to the highest probability of being differentially expressed (Huber et al, 2005). A differentially expressed gene in this FDR of 0.001 data set was accepted only with a B score ≥1.5.
Figure 1.
Microarray analysis of human perihematomal brain samples. (A) Standard deviation of array signals across control and experimental cases after RMA normalization. Lines in each column indicate mean and boxes indicated 2 standard deviations. Red boxes are U133a arrays; green are U133plus arrays. See Table 1 for×axis abbreviations. (B) Scatter plot of the significance of expression of all genes in perihematomal tissue versus control. Y axis is the B score, a statistic derived from Bayesian analysis that reflects the probability of being differentially expressed. Red dots are genes that are significantly expressed in perihematomal versus control, having a B score ≥1.5 (C). Array clustering according to the 1,000 most highly differentially expressed genes. The colors represent a ‘heat map’ and show the MAD (median absolute deviation) score for each array relative to each other array in the entire group. The color key for the MAD score is in the top left. A lower MAD score indicates less difference between arrays in the matrix, and a red score indicates a greater statistical difference in gene expression between arrays.
Figure 2.
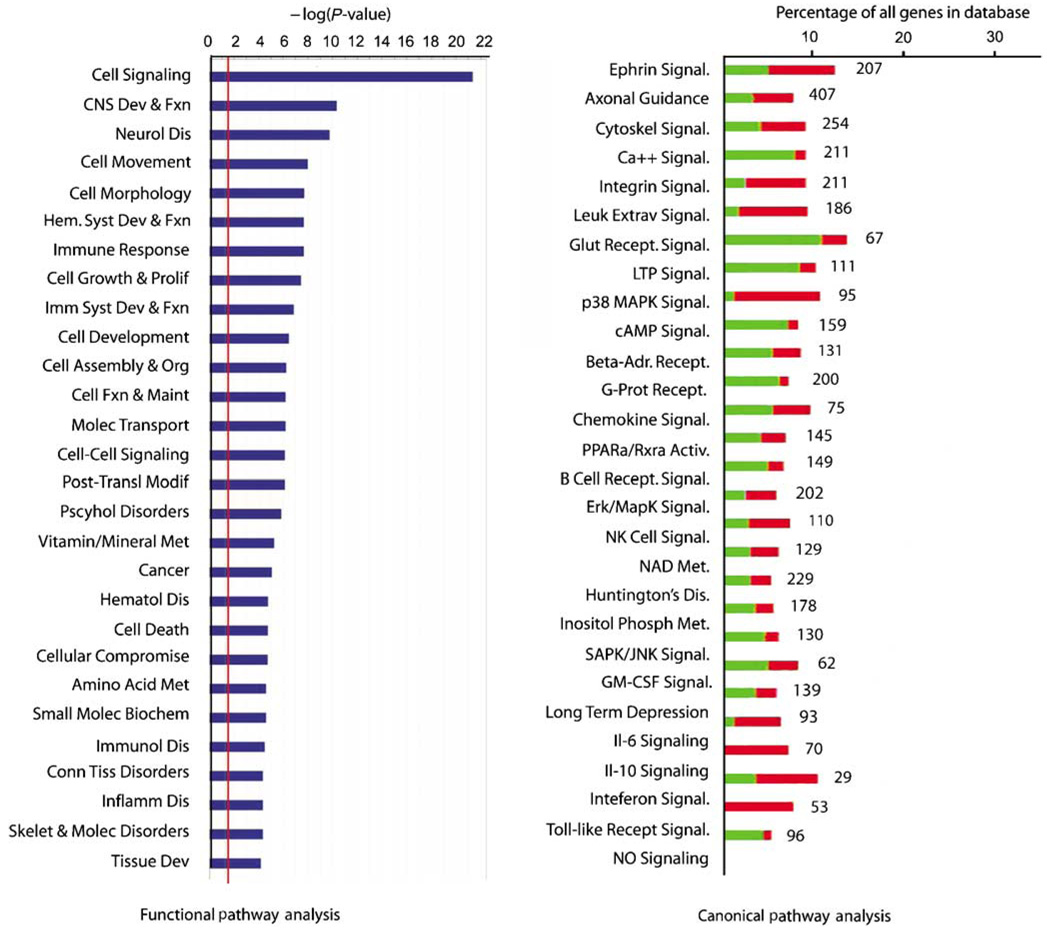
Functional grouping of differentially expressed genes. The red line in the left graph indicates the threshold for a significant association, the −log(0.05). The right plot shows the percentage of genes within a canonical signaling pathway that are differentially regulated by ICH versus the total number of genes in the database for that pathway. Green indicates the number that is downregulated; red is upregulated. The number at the right of the column indicates the total number of genes for that category in the database. All of the association in the canonical signaling pathway are significant (Fisher’s exact t-test): the probability of finding the number of genes listed for each pathway as regulated in ICH out of all of the genes in the database for that pathway is greater than 0.05 for each class.
Gene Ontology and Pathway Analysis
This set of differentially regulated genes was submitted to gene ontology (DAVID/EASE, http://david.niaid.nih.gov/david/ease.htm) and molecular pathway analysis (Ingenuity Systems, Redwood City, CA, USA; www.ingenuity.com). Molecular pathway analysis was used to classify the significantly regulated genes into coherent biologic systems, and was carried out at two levels. In the first level, the genes differentially expressed after ICH at the FDR of 0.001 level were tested for their association into known functional pathways and canonical receptor/cell signaling pathways. In this analysis, genes were tested for significant association in specific cell functional or signaling pathways versus random chance association in a total curated database of gene interactions of over 23,900 human, rat, and mouse genes by right-tailed Fisher’s exact test (Ingenuity Systems). Significance was assessed by testing the number of genes that were regulated by ICH in a specific pathway versus total number of genes in this database for that pathway. Data were plotted both as the significance value and as the ratio of ICH-regulated genes versus total genes (Figure 2). In the second level of analysis, the FDR of 0.001 genes were tested for molecular interactions within this database that forms specific gene networks. This analysis determines individual biologic pathways in which the genes that are induced in perihematomal tissue participate (Figure 3).
Figure 3.
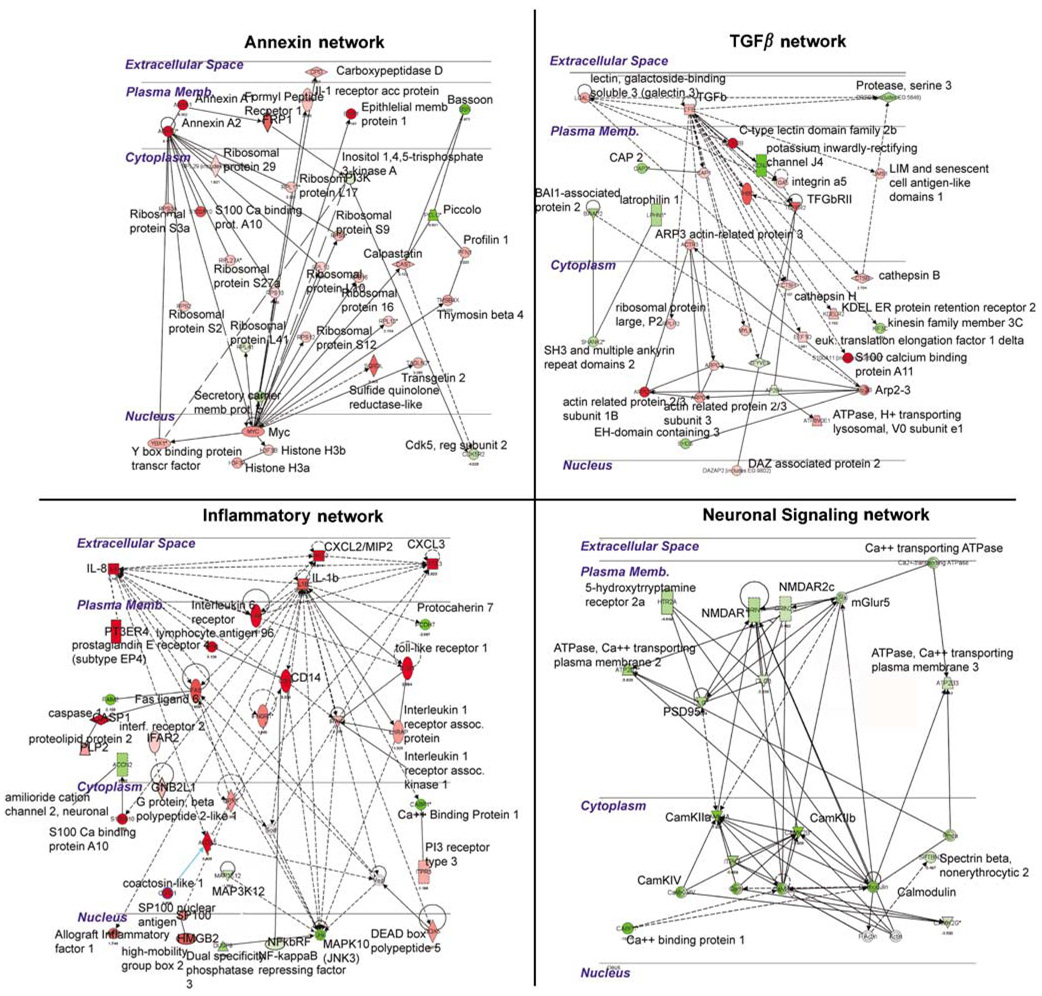
Molecular networks regulated by ICH in perihematomal tissue. Each quadrant shows a network of interacting genes and gene products whose expression is differentially regulated in response to ICH. Genes colored red are upregulated; genes colored green are downregulated. Solid lines depict a direct physical interaction between two proteins, such as annexin A1 directly binding the formyl peptide receptor 1. Dotted lines indicate an indirect interaction, such as an alteration in expression levels, posttranslational modification, or changes in protein localization.
Mouse Intracerebral Hemorrhage Model
One-third of the human brain sample was available for gene expression analysis (two-thirds of each sample being submitted for neuropathological diagnosis), limiting the amount of RNA left over for confirmatory studies. For this reason, a mouse model of ICH was used to study gene expression levels with an independent method, quantitative reverse-transcription polymerase chain reaction (qRT-PCR), and to localize select differentially regulated genes in specific cell types with tissue immunohistochemistry. Mouse ICH was produced using a well-characterized method of a single injection of nonclotted autologous blood (Nakamura et al, 2004; Xue et al, 2006) in a procedure approved by the UCLA Chancellor’s Animal Research Committee. Briefly, adult male C57Bl6 mice were anesthetized with isoflurane in N2O/O2, the scalp incised, and a small burr hole opened at A/P 0.86, M/L 2.0 (Paxinos and Franklin, 2001). A volume of 10 µL of blood, drawn from the tail tip, was injected at 0.6 µL/min through a 30 gauge needle attached to a Hamilton syringe that was placed 2.5 mm deep to the surface of the brain. Sham ICH animals received saline injection at the same rate. Control animals underwent skin incision and burr hole drilling but no blood injection. One cohort of animals (n = 6 ICH, n = 6 sham ICH, and n = 7 controls) was killed at 1 day for qRT-PCR studies. The brains were dissected on an ice block and the tissue surrounding the hematoma was rapidly frozen. A second cohort of animals (n = 6 ICH and n = 6 control) was perfused at 1 day with 4% paraformaldehyde, cryoprotected in 30% sucrose, and the brains frozen and then sectioned for tissue immunohistochemistry. The volume of the ICH was measured in these brains (Ohab et al, 2006) in a series of sections spaced 200 µm apart and stained for Nissl substance. The volume of the ICH is 1.76 ± 0.35 mm3.
Quantitative Reverse-Transcription Polymerase Chain Reaction
Total RNA was isolated from mouse perihematomal tissue (RNeasy minikit, Qiagen, Valencia, CA, USA) and assayed for integrity (Nanochip, Agilent Technologies, Santa Clara, CA, USA). 28s:18s rRNA ratios were consistently greater than 1.75 (Supplementary Figure 1). Quantitative reverse-transcription polymerase chain reaction was performed as described (Carmichael et al, 2005). A total of 2 µg of total RNA was converted to cDNA in a single-step reverse transcription reaction (Omniscript, Qiagen). This product was used for a PCR reaction using SYBYR green with fluorescence quantification software and compared with mRNA levels of GAPDH (glyceraldehyde 3-phosphate dehydrogenase; Roche Lightcycler, Roche Diagnostics, Indianapolis, IN, USA). The level of GAPDH message is not influenced by ischemia (Harrison et al, 2000). Primers for the genes of interest (Supplementary Table 1) were designed in unique mRNA segments for each gene, verified by BLAST search (NCBI), and each sequence was confirmed by end product sequencing (UCLA Sequencing Core). The expression values for each gene in control (n=7) and ICH conditions (n = 6) were compared using a two-tailed t-test.
Immunohistochemistry
Fluorescent immunohistochemistry was performed as described (Ohab et al, 2006). A mouse-on-mouse staining protocol was used according to the manufacturer’s guidelines (Vector Labs, Burlingame, CA, USA) except that streptavidin cy3 (Molecular Probes/Invitrogen, Carlsbad, CA, USA) was used to minimize background staining with mouse primary antibodies. In all experiments, no-primary and no-secondary antibody controls were run in parallel. These showed no specific staining. The primary antibodies were rabbit anti-CCR1 (CC chemokine receptor 1, 1:200; Abcam, Cambridge, MA, USA), mouse anti-IL1R1 (1:100; Abcam), rabbit anti-annexin A1 (1:500; Abcam), mouse anti-annexin A2 (1:100; Zymed, San Francisco, CA, USA), rat anti-CD68 (1:100; AbD Serotec, Raleigh, NC, USA), rat anti-glial fibrillary acidic protein (GFAP, 1:500; Zymed/Invitrogen, Carlsbad, CA, USA), mouse anti-NeuN (1:500; Chemicon, Temecula, CA, USA), chicken anti-tissue inhibitor of metalloproteinase 1 (TIMP-1; 1:200; R&D Systems, Madison, WI, USA), goat anti-aquaporin 9 (1:200; Chemicon), and sheep anti-carbonic anhydrase II (CAII, 1:300; Abcam). Tissue sections for each animal were also stained for Nissl substance using standard protocols.
Results
Gene Expression Characteristics of Human Perihematomal Tissue
Neuroendoscopic clot removal was performed within 24 h of ICH. The evacuated clot tissue contained both gray and white matter components from the region of the hemorrhages: putamen, frontal and parietal cortex, and subcortical white matter. The gene expression profile of perihematomal tissue in this study was obtained from six subjects, ranging in age from 51 to 84 years (Table 1). Control tissue was obtained from autopsy specimens in brain areas that matched the sites of primary hemorrhage in the ICH cases. The arrays showed minimal RNA degradation and a close correlation of signal intensity (Figure 1A, Supplementary Figure 1 and Supplementary Figure 2). Interarray Pearson’s correlation showed no effects of array hybridization batch, array platform, sample accrual time, or postmortem interval on array signals (data not shown). A Bayesian estimate of differential expression was calculated using an FDR of 0.001%. This is a highly stringent statistical cutoff for significance in gene expression levels that controls for multiple comparisons and means that 1 out of 1,000 genes would be expected as false positive in this comparison. At this statistical level, most genes are not significantly different in perihematomal versus control tissue (Supplementary Figure 2B) and show an expected normal distribution around a ‘0’-fold change in expression level in perihematomal versus control (Figure 1B). The experimental and control tissues contained a potentially large degree of genetic variation in subject characteristics (sex, age, and race), hemorrhage type (lobar versus hypertensive), and tissue origin (putamen, parietal/frontal cortex, and white matter). Unsupervised cluster analysis of arrays was performed to determine whether these genetic variations contribute to gene expression differences that would obscure those related to perihematomal versus control differences. Unsupervised clustering of the 1,000 most variably expressed genes showed a very tight segregation of perihematomal arrays versus control arrays, as assessed by both heat map plots (Figure 1C) and dendrogram plots (Supplementary Figure 2C). This segregation indicates that, despite genetic variation in the subjects, in the types of hemorrhage and in the site of origin of the brain samples, the gene expression profile in human perihematomal tissue is very distinct from the control nonhemorrhagic brain.
Gene Expression Profile of Human Perihematomal Tissue
A total of 624 genes (331 downregulated and 293 upregulated) are differentially expressed in perihematomal tissue versus control (FDR < 0.001, see Supplementary Table 2 for list). This gene set was further analyzed in its organization into functional cellular pathways. Ranking these by significance reveals that most of the genes induced in perihematomal tissue belong to pathways functionally associated with central nervous system development and disease, immune responses and general immune system function, and the cell biology of signaling, cell movement, development, and metabolism (Figure 2, Supplementary Figure 2D). To further break down this gene set, genes were grouped according to their statistical association with known canonical molecular pathways. Intracerebral hemorrhage induces a pattern of gene expression in 27 canonical pathways (Figure 2). A large number of these relate to neuronal or cellular signaling functions, including glutamate receptor signaling, synaptic signaling (long-term depression and potentiation), calcium signaling, and ephrin pathways. Similar to the functional pathway analysis, canonical pathway analysis shows that ICH induces or downregulates genes in several inflammatory pathways, including leukocyte extravasation, chemokine signaling, Toll-like receptor signaling, and the cytokine pathways of GM-CSF (granulocyte macrophage colony stimulating factor), IL6, and interferons (Figure 2). Genes within neuronal signaling pathways of long-term potentiation and both depression and axonal structure are induced and downregulated after ICH. Overrepresentation of cellular signaling and intracellular signal transduction systems was confirmed with hierarchical gene ontology analysis (DAVID, Supplementary Figure 2D).
To better define specific members of these functional molecular groups in human perihematomal tissue, the genes regulated by ICH at FDR of 0.001 (Supplementary Table 2) were assessed individually for their molecular associations (Table 2). A large number of inflammatory genes are induced in human perihematomal tissue. These include the chemokines CXCL2/MIP2, CXCL3, CCR1, and IL8; IL1β and its receptor complex, IL6 receptor, IL13 receptor, interferon receptor 2, Toll-like receptor-1; and several downstream effector molecules related to these signaling molecules. Tumor necrosis factor-α is not significantly regulated after ICH (P = 0.498). This set of molecules is clearly proinflammatory in cerebrovascular disease (Wang and Dore, 2007) and, in most cases, is highly upregulated in human ICH (Table 2). A set of genes that functions in an anti-inflammatory role is also upregulated in perihematomal tissue, including annexins A1 and A2, transforming growth factor beta (TGFβ), and the IL10 receptor. Annexins A1 and A2, TGFβ, and IL10 have been implicated in a anti-inflammatory response in cerebrovascular disease (Gavins et al, 2007) and in systemic illness (Parente and Solito, 2004). Glial or extracellular matrix-associated genes are also regulated by ICH, including inductions in TIMP-1, aquaporin 9, and several proteoglycans; vascular endothelial growth factor and aquaporin 4 expression are not significantly changed. Interestingly, the expression of a wide range of genes associated with neuronal excitability is significantly downregulated after ICH, including those for synaptic proteins, glutamate receptor and other neurotransmitter proteins, ion channels, and calcium second messenger cascades (Table 2). This finding reinforces the functional pathway results noted above, which showed a significant regulation of neuronal signaling genes by ICH (Figure 2). This downregulation of neuronal signaling genes is not simply a reflection of neuronal loss in the tissue sample, as most neuron-specific or neuron-enriched genes on the array are not differentially regulated by ICH, for example neurotransmitter receptors (GABARa1 to a5; metabotropic glutamate receptors 1 to 4, 6 to 8; alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunits; dopamine receptor subunits), voltage-dependent calcium channels (L and P/Q type calcium channels), and axonal transport proteins (kinesin family members). Intracerebral hemorrhage appears to coordinately downregulate aspects of N-methyl-d-aspartic acid (NMDA) receptor and synaptic signaling across many levels of this process, from specific receptors subclasses and ion channels to their intracellular scaffolding proteins and second messenger systems (Table 2).
Table 2.
Differential gene expression in perihematomal tissue
| Gene | Fold |
|---|---|
| Anti-inflammatory | |
| Annexin A1 | 9.302 |
| Annexin A2 | 9.475 |
| Interleukin 10 receptor beta | 4.005 |
| Transforming growth factor beta induced 68 kDa | 7.568 |
| Transforming growth factor beta receptor 2 | 5.478 |
| FPR1-formyl peptide receptor | 6.667 |
| S100 calcium binding protein A10 | 5.867 |
| Transforming growth factor, beta 1 | 2.848 |
| Neurotransmitter/glutamate signaling | |
| Cholinergic receptor, muscarinic 3 | 0.115 |
| 5-HT receptor 2A | 0.215 |
| GABAb receptor 1 | 0.218 |
| Glutamate receptor, metabotropic 5 | 0.116 |
| NMDAR1 | 0.215 |
| NMDAR2c | 0.286 |
| Synaptic proteins | |
| Synaptophysin | 0.152 |
| Piccolo | 0.114 |
| Bassoon | 0.1 |
| Neurexin 1 | 0.131 |
| Neurexin 2 | 0.356 |
| EphA4 | 0.267 |
| Synaptic vesicle glycoprotein 2a | 0.163 |
| Dendrin | 0.151 |
| Synaptobrevin 2 | 0.218 |
| Synpatic vessicle glycoprotein 2a | 0.163 |
| Synaptobrevin 2 | 0.218 |
| PSD95 | 0.196 |
| Liprin α4 | 0.173 |
| Calcium signaling | |
| Calcium binding protein 1 (calbrain) | 0.073 |
| CaM kinase II alpha | 0.125 |
| CaM kinase II beta | 0.126 |
| CaM kinase II gamma | 0.286 |
| Calmod. 3 (phosphorylase kinase, delta) | 0.149 |
| Calmod. regulated spectrin-associated protein 1 | 0.396 |
| Neurocalcin delta | 0.131 |
| ATPase, Ca2+ transporting, plasma membrane 2 | 0.177 |
| ATPase, Ca2+ transporting, plasma membrane 1 | 0.244 |
| ATPase, Ca2+ transporting, plasma membrane 3 | 0.293 |
| Ca2+ channel, voltage-dependent, beta 1 | 0.38 |
| Ca2+ channel, voltage-dependent, alpha 2/delta | 0.468 |
| Ca2+ channel, voltage-dependent, gamma | 0.072 |
| Ca2+ channel, voltage-dependent, beta 3 subunit | 0.203 |
| Ca2+ channel, voltage-dependent, P/Q type, alpha 1A | 0.298 |
| Inflammatory | |
| CXCL2 | 8.079 |
| Leuk. immunoglob.-like receptor, subfamily B, member 1 | 21.14 |
| Complement component 5a receptor 1 | 5.24 |
| CCR1 | 4.118 |
| Chemokine-like factor | 3.76 |
| IL-13 receptor alpha1 | 3.6 |
| Interleukin receptor associated kinase 1 | 2.23 |
| Interferon regulatory factor 8 | 2.245 |
| Interleukin 1 accessory protein | 0.406 |
| IL-8 | 8.827 |
| Interleukin 1 receptor 1 | 5.994 |
| Interferon gamma inducible protein 16 | 7.802 |
| Toll-like receptor 1 | 5.5 |
| NF-kappaB repressing factor | 0.518 |
| Complement component 3a receptor 1 | 1.595 |
| Interferon (alpha, beta and omega) receptor 2 | 1.967 |
| Fc fragment of IgG, high affinity Ia, receptor (CD64) | 6.206 |
| CXCL3 | 7.59 |
| Il-6 receptor | 4.95 |
| Il-1 beta | 3.8 |
| TNFα | NS |
| ECM/glial | |
| Versican | 5.986 |
| Brevican | 0.23 |
| Decorin | 3.074 |
| Galectin-1 | 2.739 |
| Integrin alpha 5 | 2.376 |
| Neuroglycan C | 0.276 |
| Aquaporin 9 | 9.98 |
| Aquaporin 4 | NS |
| TMP-1 | 10.26 |
| VEGF | NS |
| Ion channels and transporters transporter | |
| Na+ channel, voltage-gated, type III, beta | 0.057 |
| Na+-dep. inorganic phosphate cotransporter, member 7 | 0.043 |
| Na+/Ca+ exchanger 8, member 2 | 0.117 |
| Na+/H+ exchanger 9, member 6 | 0.331 |
| FXYD domain containing ion transport regulator 7 | 0.158 |
| Solute carrier organic anion transporter family, member 1C1 | 0.285 |
| Organic cation transporter 22, member 17 | 0.291 |
| Amiloride-sens. cation channel 2, neuronal | 0.329 |
| K+ channel tetramerisation domain containing 13 | 0.365 |
| Voltage-gated K+ channel, Shab-related subfamily, member 1 | 0.15 |
| Inwardly-rectifying K+ channel, subfamily J, member 4 | 0.065 |
| Voltage-gated K+ channel, KQT-like subfamily, member 2 | 0.083 |
| K+ channel, subfamily V, member 1 | 0.292 |
NS, not significant.
Fold is fold change in perihematomal tissue versus control tissue. CaM kinase is calcium/calmodulin-dependent kinase.
To identify specific relationships between genes that are coordinately regulated by ICH in perihematomal tissues, the differentially regulated genes were submitted to an unsupervised molecular pathway analysis. Seventeen different networks contain 15 or more genes that are significantly regulated by ICH in perihematomal tissue and form specific cellular signaling pathways. Four pathways contain distinct, multilevel, and interrelated gene systems with greater than 25 genes regulated by ICH: Annexin, TGFβ, IL/chemokine, and NMDA/neuronal signaling systems (Figure 3). These will be described in detail. Six additional networks contain several interconnected gene systems with hubs at genes that are regulated by the ICH/fibroblast growth factor (FGF) network, CDC42/TIMP-11 network, PTEN network, G-protein network, mitogen activated protein kinase (MAPK)/RhoA network, and an ID2 network (Supplementary Figure 3).
In the four networks with the most highly interconnected genes, ICH regulates inflammatory, anti-inflammatory (annexin and TGFβ), and neuronal glutamate receptor interactions in perihematomal tissue (Figure 3). In these pathways, ICH induces specific genes that function in signaling from the cell surface through cytoplasmic proteins to nuclear transcription factors. Intracerebral hemorrhage induces a highly integrated network of chemokines CXCL2/MIP2, CXCL3, and IL8 with IL1β into signaling cascades that link the fas ligand, Toll-like receptor 1, and caspase 1/ICE with specific nuclear regulatory genes. In the TGFβ network, ICH induces TGFβ, its receptor and several downstream nuclear and cytoplasmic genes directly linked to TGFβ signaling. In the annexin pathway, ICH induces annexins A1 and A2, the annexin A1 receptor FPR, and a series of ribosomal regulatory proteins and myc and Y box-binding protein transcription factors. A fourth major network involves glutamate receptor signaling (Figure 3), confirming the functional and canonical pathway analysis described previously. Intracerebral hemorrhage coordinately downregulates neuronal signaling genes in human perihematomal tissue, extending from the NMDA receptor through its linked PSD95 and calcium calmodulin kinase II (CamKII) cytoplasmic cascades.
In summary, a relatively small number of all genes tested (624 out of over 14,500) are differentially regulated by ICH after a stringent analysis. Networks of proinflammatory and anti-inflammatory genes are coinduced in human perihematomal tissue. In contrast, major aspects of neuronal synaptic function (membrane receptors and ion channels and their protein kinase second messenger cascades) are downregulated.
Mouse Intracerebral Hemorrhage: Quantitative Reverse-Transcription Polymerase Chain Reaction
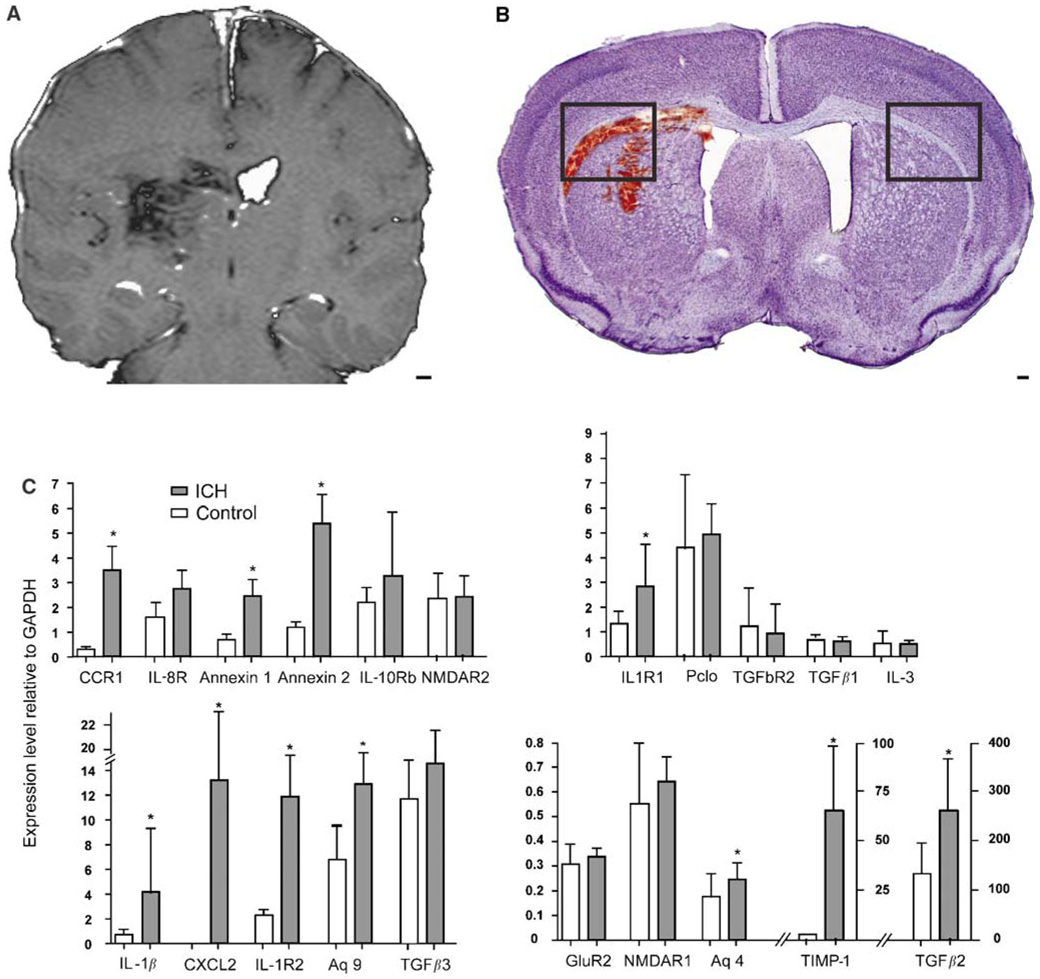
Perihematomal gene expression was studied with qRT-PCR in a mouse model of ICH (Figure 3) to test the array findings with an independent technique and to extend the human findings in an experimental brain hemorrhage model. In this model, autologous blood injection into the striatum produces inflammatory cell infiltration, edema, and neuronal cell death within the first 24 h (Nakamura et al, 2004; Xue et al, 2006). Perihematomal tissue from the mouse striatum at 1 day after ICH was compared with sham ICH and with control striatum/subcortical white matter. There was no significant difference in gene expression between sham ICH and control so these two groups were combined. Intracerebral hemorrhage significantly upregulates IL1β, CCR1, IL1R1, IL1R2, annexin A1, annexin A2, CXCL2, TIMP-11, TGFβ2, and aquaporins 4 and 9 in mouse perihematomal tissue at 1 day after hemorrhage (Figure 3C). Tissue inhibitor of metallproteinase 1 is the most differentially regulated gene in mouse perihematomal tissue, with a 217-fold induction. TGFβ1 is not induced in mouse ICH, whereas TGFβ2 is upregulated in mouse perihematomal tissue (Figure 4C). Piccolo and NMDAR1 and 2c are downregulated in human perihematomal tissue but are not altered at day 1 after ICH in the mouse.
Figure 4.
Mouse ICH model and qRT-PCT testing of differentially regulated genes in human perihematomal tissue. (A) T1-weighted MRI image of human ICH case, HS6 (Table 1). (B) Nissl photomicrograph of mouse ICH model. Boxes in mouse ICH model indicate regions shown in Figures 5A and 5B. Note that the human ICH image has been contrast-enhanced to highlight the location of the hemorrhage. (C) Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) results of differentially regulated genes in mouse perihematomal tissue based on human microarray experiments. *P≤0.05. Scale bar =3 mm (A); 100 µm (B).
Mouse Intracerebral Hemorrhage: Cellular Localization of Induced Genes
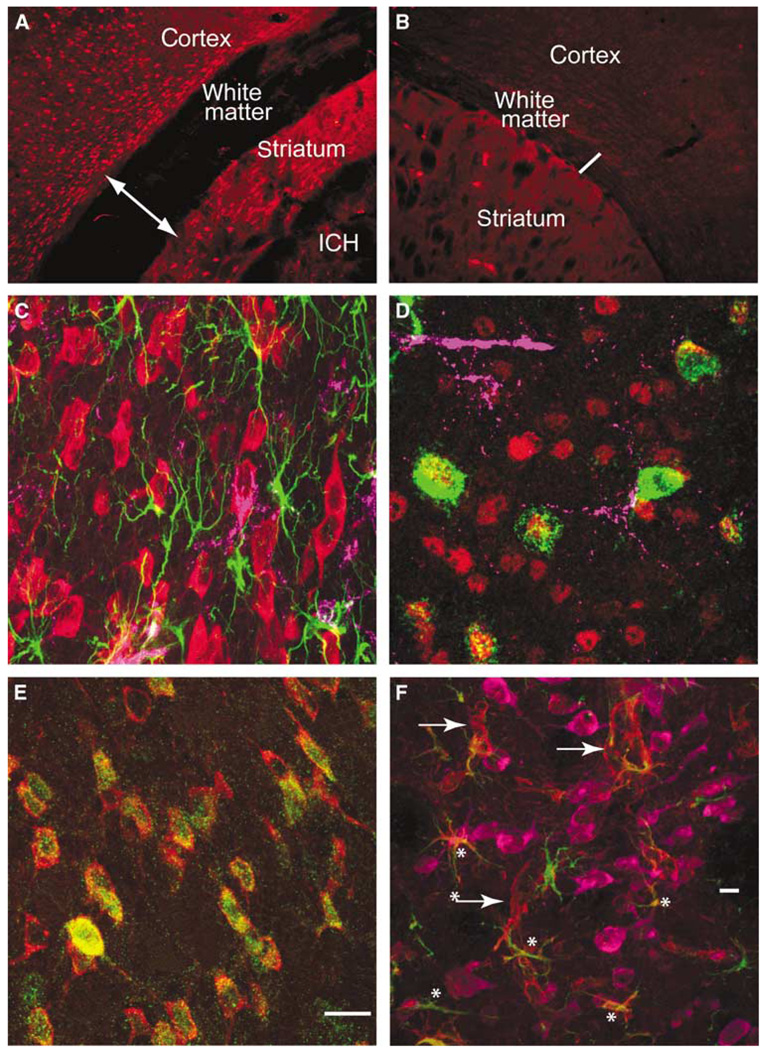
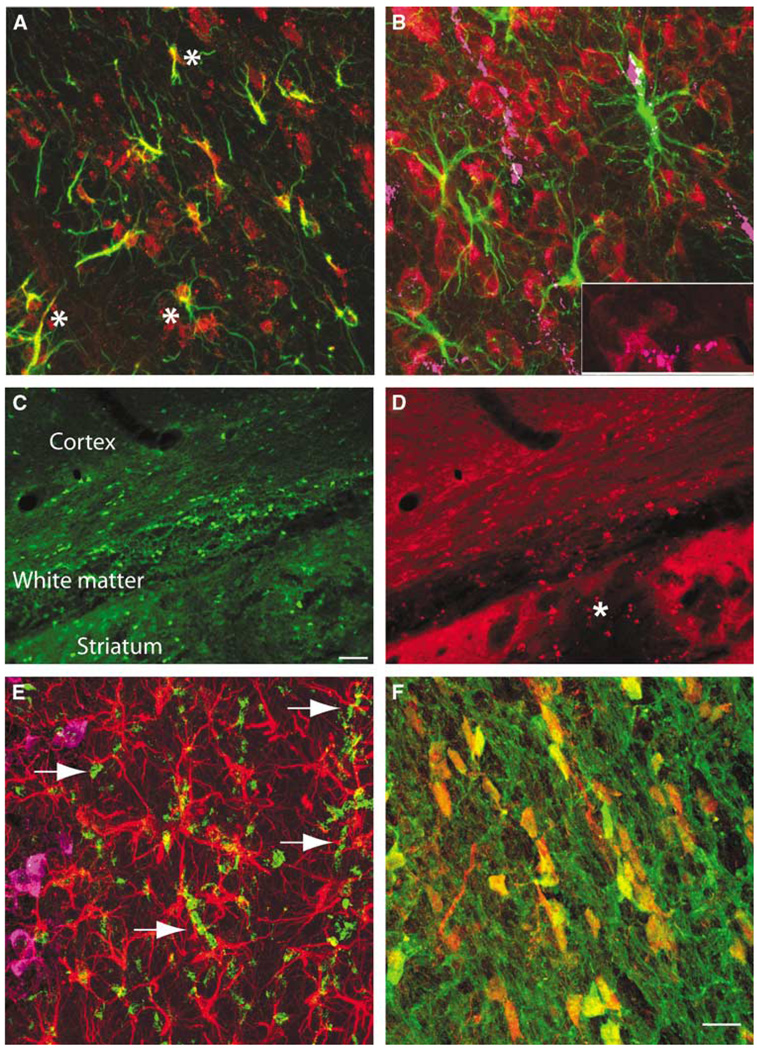
Multilabel immunofluorescent studies were used to localize protein products of select genes at 1 day after ICH in the mouse. Both annexins A1 and A2 are induced in cells that border the hemorrhage site in the striatum, as well as in the cortex that borders the edematous white matter above the striatum (Figures 5A and 5C). Annexin A2 immunoreactivity is present in cells that are also positive for the mature neuronal marker NeuN (Figure 5D). Annexin A1 staining colocalizes with annexin A2 staining in these same cells (Figure 5E). Baseline annexin staining in the hemisphere contralateral to the ICH (Figure 5A) and in control brains (data not shown) is not present in neurons. Thus, ICH induces annexins A1 and A2 expression in perihematomal neurons in the striatum and the cortex. Tissue inhibitor of metallproteinase 1 immunoreactivity is normally present in endothelial cells and neurons in the striatum and white matter. After ICH, this protein is induced in reactive astrocytes (Figure 5F, Supplementary Figure 4D). CD68-positive inflammatory cells infiltrate the hemorrhage site and both blood-borne macrophages and microglia are present in perihematomal tissue. These cells express IL1R1 (Supplementary Figure 4). Interleukin-1R1 localizes with annexin A2-positive cells and is also present in cells with the neuronal morphology of after ICH (Figure 6B, Supplementary Figure 4B). Aquaporin 9 is weakly present in control tissue in neurons and astrocytes. After ICH, there is increased staining in reactive astrocytes (Figure 5F; Supplementary Figure 4E). The chemokine CCR1 is present in cells that form chains in white matter that is adjacent to ICH and in inflammatory cells that infiltrate the ICH and perihematomal tissue (Figure 6D). These chains of CCR1-immunoreactive cells also stain for markers of mature oligodendrocytes, including myelin basic protein (data not shown) and carbonic anhydrase II (Figures 6C and 6F). Neurons and astrocytes do not express CCR1 (Figure 6E). These data suggest a cell type-specific response in perihematomal tissue, with neurons upregulating annexins, oligodendrocytes upregulating CCR1, and astrocytes upregulating TIMP-1 and aquaporin 9.
Figure 5.
Cellular pattern of annexins and inflammatory cells in perihematomal tissue. Annexin A2 immunoreactivity in the cortex, white matter, and striatum adjacent to ICH (A) and in the contralateral hemisphere (B). The area shown is indicated in the two boxes in Figure 3. Line in panel A is through edematous white matter and is 380 µm across, as compared with the line in through the contralateral white matter, which is 130 µm. (C) Cortex adjacent to hemorrhage stained for annexin A2 (red), GFAP (green), and CD68 (purple). (D) Perihematomal cortex stained for annexin A1 (green), NeuN (red), and CD68 purple. Many annexin A1-positive cells also stain for NeuN. (E) Annexin A1 (green) and A2 (red) staining in same cells in the perihematomal striatum. (F) Tissue inhibitor of metallproteinase 1 (TIMP-1, red), NeuN (purple), and GFAP (green) immunoreactivity in perihematomal tissue. Tissue inhibitor of metallproteinase 1 is present in neurons and astrocytes (asterisks) and cells with the appearance of endothelial cells (arrows). All neurons stained for TMP1 and NeuN, rendering the composite color a red/purple. Scale bar = 15 µm in panel E corresponds to panels C to E. Scale bar = 60 µm (E).
Figure 6.
Cellular Expression Pattern of Aquaporin 9, CCR1, and IL1R1 expression in perihematomal tissue. (A) Aquaporin 9 (red) and GFAP (green) immunostaining in periinfarct tissue. Colocalized staining is seen as yellow (asterisks). (B) GFAP (green), IL1R1 (red), and CD68 (purple) immunohistochemical staining in the perihematomal cortex. Arrows denote double labeling of CD68 and IL1R1. The inset in the bottom right is an enlargement of the bottom left corner of this panel showing overlap of staining of IL1R1 and CD68. (C) Low-power photomicrograph of CAII immunoreactivity in the perihematomal white matter and striatum. CAII is a maker for oligodendrocytes. (D) Same tissue stained for CCR1. Asterisk denotes region of hemorrhage. Area depicted in panel A is in the same region as in Figure 5A. (E) Perihematomal tissue stained for CCR1 (green), GFAP (red), and NeuN (purple). Arrows show CCR1-positive chains of cells in subcortical white matter. (F) CCR1 (red) and CAII (oligodendrocyte marker) (green) in perihematomal white matter. Note cell body colocalization of these two immunomarkers. Scale bar in panel C = 60 µm also applies to panel D. Scale bar = 10 µm in panel F and applies to panels B, E, and F.
Discussion
Intracerebral hemorrhage regulates coordinated networks of gene expression in inflammatory, anti-inflammatory, and neuronal signaling systems in human perihematomal tissue within the first day of the hemorrhage. A molecular network of proinflammatory signaling starts with the cytokines/chemokines IL1β, IL8, IL6 receptor, CCR1, CXCL2/MIP2, and CXCL3 and signals through IL and Toll-like receptors to activate a signaling cascade that involves the Fas ligand, NF-κB (nuclear factor-κB), and a MEKK/JNK pathway. Anti-inflammatory signaling is activated in ICH in a cascade that progresses from annexins A1 and A2, IL10 and TGFβ, through downstream calcium-binding, cytoskeletal and ribosomal proteins, and c-Myc. Intracerebral hemorrhage downregulates parallel neuronal signaling systems in human perihematomal tissue. These downregulated neuronal genes include molecules that function in glutamate signaling, pre- and postsynaptic structure, and a number of ion channels and calcium signaling proteins. At the cellular level, specific cell types respond to ICH with altered expression of these genes. Astrocytes in the perihematomal rim express aquaporin 9 and TIMP-1. Annexin A2 is induced in inflammatory cells in the hemorrhage and in neurons that border the hemorrhage site. Oligodendrocytes in injured white matter express the inflammatory chemokine CCR1. Inflammatory and endothelial cells in the immediate perihematomal rim and hemorrhage site express the IL receptor IL1R1. Perihematomal tissue includes gray and white matter structures. At the protein level, select members of the molecular cascades induced in perihematomal tissue after ICH localize within different glial and neuronal cell types in these two tissue compartments.
Inflammatory signaling is initiated by cytokine and chemokine signaling in both human studies and animal models of ICH. Elevated serum levels of the cytokines IL6 and TNFα correlate with edema and functional deficits in patients (Castillo et al, 2002; Dziedzic et al, 2002; Qureshi et al, 2003a). The IL6 receptor is upregulated in human perihematomal tissue. Tumor necrosis factor-α was not upregulated in the present human perihematomal data set. Tumor necrosis factor-α mRNA is induced within 1 day in the rat after ICH (Mayne et al, 2001; Wasserman et al, 2007; Wasserman and Schlichter, 2007), but TNFα protein levels are induced only within hours and return to baseline by 1 day in the mouse after ICH (Hua et al, 2006). These differences in the TNFα response across species may reflect the fact that much of the TNFα released in the brain after cerebral insults is in the neutrophils and macrophages (Mayne et al, 2001), which may release stored TNFα, and variations in the initial inflammatory response would then contribute to different levels of this cytokine across species.
Many of the upregulated inflammatory genes in rat perihematomal tissue (Wasserman et al, 2007; Lu et al, 2006) are also increased in this data set in humans after ICH, including CXCL2, IL1β, and its converting enzyme caspase 1/ICE, CD14, and CD44. In a systematic microarray study of genes regulated in the striatum and cortex 24 h after ICH in the rat, Lu et al report in detail on 336 genes excluding ESTs that are regulated in these perihematomal tissue and belong to distinct functional subclasses. Although there are substantial differences between the present human data and this published rat study, including tissue of origin, timing of tissue harvest, array type, and statistical and bioinformatic processing, it is useful to note that 13.7% of the human genes are also regulated in the rat and that these genes are regulated in the same direction (induced or down-regulated). The common genes altered by ICH in both species include similar classes of these inflammatory genes or gene families: IL-related: rat: IL1β, IL2r2, IL1ra, and IL18; human: IL13r, IL6r, IL8, and IL1r1, IL1rap. Additionally, annexins A1 and A2, gamma-aminobutyric acid (GABA) type B receptor, NMDAR1, and HTR2c are commonly regulated in rat and human ICH, as well as genes in the fibroblast growth family: FGF2b rat; FGF9, 12, 13 human.
Intracerebral hemorrhage also induces several chemokine systems in human perihematomal tissue, including CCR1, IL8, CXCL2, and CXCL3. CCR1 has been consistently shown to play a role in leukocyte infiltration in the brain in inflammatory disorders, such as in experimental autoimmune encephalomyelitis (EAE) (Trebst et al, 2003), and was upregulated in oligodendrocytes in perihematomal white matter in this study. The ligands for CCR1, CCL2, 3, 5, 7, 8, 14 to 16 and 23 are not regulated by ICH in humans. Interleukin-8, CXCL2, and CXCL3 are members of the Glutamic acid-Leucine-Arginine (ELR) class of chemokines and promote neutrophil localization (Bajetto et al, 2001). With their prominent early expression, ELR chemokine signaling in human ICH is positioned to initiate neutrophil homing and diapedesis (Rakonjac et al, 2006) during an initial inflammatory response in this disease.
A novel finding in the present data set is that this initial signaling system of cytokines and chemokines is at the front end of an inflammatory molecular pathway that is coordinately activated in human perihematomal tissue. Interleukin-1β, IL8, IL6, CXCL2, and CXLC3 are part of a network of genes that form a link from cell surface receptors to cytosolic effector molecules and transcriptional regulators (Figure 2, Table 2), including Toll-like receptor 1, fas ligand 6, allograft inflammatory factor 1, HMGB2, SP100a, cathepsins B, H, L, coactosin-like protein 1, and a downregulation of NF-κB repressing factor. These molecules mediate many of the deleterious downstream effects of inflammatory signaling, including microglial activation (Postler et al, 2000; Gan et al, 2004), leukotriene signaling (Rakonjac et al, 2006), p53 activity and apoptosis (Stros et al, 2002), and oligodendrocyte cell death (D’Souza et al, 1996). Past studies have shown that apoptotic cell death can be mitigated by treatments that address initiation points in inflammatory signaling, such as TNFα, ILβ, and IL6 (Wagner, 2007; Wang and Dore, 2007). It is possible that future approaches to the control of inflammatory cascades in ICH may target the downstream mediators of damage that are seen in these inflammatory networks.
In addition to these proinflammatory cascades, ICH induces coordinated upregulation of several networks of anti-inflammatory genes. Annexin A1, annexin A2 (also termed annexins I and II), the annexin A1 receptor FPR, the annexin A2 binding protein S100A10, TGFβ1, its receptor TGBR2, the IL10 receptor, galectin-1, and specific intracellular downstream genes from these anti-inflammatory molecules are upregulated (Figure 3, Table 2). Annexins A1 and annexin A2 block inflammatory second messenger systems and recruitment of neutrophils (Lim and Pervaiz, 2007). Annexin A1 inhibits interferon-γ and endotoxin-induced inflammatory cellular damage and inhibits inducible nitric oxide synthetase (iNOS) and cyclooxygenase 2 (COX2) (Parente and Solito, 2004; Gavins et al, 2007; Lim and Pervaiz, 2007). Nonhemorrhagic stroke induces annexin A1 and the blockade of annexin A1 levels or function is neuroprotective (Gavins et al, 2007). Interleukin- 10 inhibits leukocyte production of COX2, iNOS, and IL12,and is also neuroprotective in stroke (Spera et al, 1998; Grilli et al, 2002). Transforming growth factorβ has long been implicated in anti-inflammatory actions and is neuroprotective in stroke (Buisson et al, 2003). Thus these ICH-activated anti-inflammatory networks act to dampen the immune response to brain hemorrhage and, in many cases, directly oppose the inflammatory gene networks that are also activated by ICH. Many of these pro- and anti-inflammatory signaling systems converge on neutrophil recruitment and activation.
Intracerebral hemorrhage induces a downregulation of neuronal signaling systems, including pre- and postsynaptic, ion channel and calcium signaling networks, and the NMDA receptor complex. PSD95, spectrin, NMDA receptor 2 subunits, CamKII, and calmodulin, all downregulated in human perihematomal tissue, form a complex that mediates many aspects of normal postsynaptic glutamate responses in neurons (Schoch and Gundelfinger, 2006). The excitotoxic effects of ischemic glutamate release are also mediated by NMDA receptor/PSD95 interactions through nitric oxide production (Aarts et al, 2002). Piccolo, bassoon, synaptophysin, neurexins 1 and 2, and liprinα4 are presynaptic proteins, linked to the active zone, that interact in synaptic vesicle fusion, membrane recycling, and proper pre- and post-synaptic alignment (Schoch and Gundelfinger, 2006) and are downregulated in human perihematomal tissue. It is remarkable that after human ICH, the expression level of such a broad array of molecules in aspects of NMDA receptor pre- and postsynaptic functions is coordinately downregulated. On top of this downregulation in synaptic function, ICH induces a substantial downregulation of cellular ion channel and calcium signaling pathways, including 15 calcium signaling proteins and 13 cellular ion channels (Table 2). This coordinated downregulation of neuronal signaling systems does not appear to be due to a general loss of neurons in perihematomal tissue, as other neuron-specific or neuron-enriched molecules (Cahoy et al, 2008) are induced (Figure 2) or without significant change, such as EphA7, Nov, SLA, AMPA, and GABA receptor subunits.
Intracerebral hemorrhage induces a massive and early glutamate release (Castillo et al, 2002; Qureshi et al, 2003a, b; Miller et al, 2007) and elevated serum glutamate in humans after ICH correlates with reduced functional outcome and larger hemorrhage cavity (Castillo et al, 2002). The coordinated down-regulation of neuronal excitability genes may serve an endogenous neuroprotective mechanism in the tissue immediately adjacent to and adherent to the hemorrhage, the target tissue obtained in these human studies. Because further adjacent tissue is at-risk for progressive apoptotic cell death (Felberg et al, 2002; Qureshi et al, 2003b; Wasserman et al, 2007), white matter loss, and tissue atrophy (Felberg et al, 2002), future neuropathological studies in humans will need to determine the expression profile of neuronal signaling genes in these adjacent areas.
Intracerebral hemorrhage alters extracellular matrix, glial, and vascular structure in perihematomal tissue during the initiation of cell death and edema. In human perihematomal tissue, ICH induces aquaporin 9 but does not significantly upregulate matrix metalloproteinases or serine proteases (such as, prothrombin mRNA), vascular endothelial growth factor, or aquaporin 4. Aquaporin 4 and vascular endothelial growth factor are induced in animal models of ICH and several are elevated in the blood in humans after ICH (Xi et al, 2006; Wang and Dore, 2007). These discrepancies may relate to actual molecular differences between human and rodent ICH with, for example, glial aquaporin 9 instead of aquaporin 4 implicated in human perihematomal edema formation. The tissue sampling differences in this study, compared with rodent studies, may also partially account for these gene expression differences. In this study, gene expression analyses were drawn from tissue adherent to clot, which is different from the more extensive periinfarct tissue sampled in published rodent experiments (Lu et al, 2006) and in those rodent experiments in this study. Rodent studies thus include both immediate periinfarct tissue and more distant sites and likely include both injured and normal glia, neurons and endothelial cells; conversely, tissue adherent to clot in human studies may be incorporating into a perihematomal scar and solely include already damaged cells.
From human ICH material to confirmatory studies in a mouse ICH model, this data set provides important conclusions but has limitations. First, this data set has a limited number of experimental samples. A stringent statistical cutoff established a robust data set from these samples but many genes that are known to be upregulated in periinfarct tissue in animal models of ICH and stroke, such as HSP70, heme oxygenase 1, MMP9, and MMP2 (Lu et al, 2006; Wang and Dore, 2007), were not significantly altered in this data set. Future human studies with larger sample sizes may identify these genes as being significantly regulated after ICH. Second, this data set relates to biologic inferences from gene expression data. Molecular pathway analysis does identify distinct receptor-ligand signaling systems and their downstream signaling cascades that are induced in human perihematomal tissue. But future experiments will need to use gain and loss of function studies in these systems to identify their causal role in perihematomal damage. The mouse model of ICH was used to study gene expression changes identified in the human and localize select proteins within specific cell types after ICH. However, the timing of neuroinflammation, apoptotic cell death, and anti-inflammatory processes may differ between the mouse and human, and future time course studies in both species are necessary. The strengths of this data set are that it is the first description of the molecules that are induced or altered in human perihematomal tissue, the target region for much of the clinical pathology in this disease. With rigorous bioinformatics approaches, the study takes a potentially large and complex set of genes and narrows these to several key molecular signaling pathways. The regulation of many of these pathways in human ICH is then confirmed in an experimental model and this model indicates distinct neuronal, oligodendrocyte, and astrocyte roles in ICH. Pro- and anti-inflammatory signaling involves ligands, such as the annexins, galectin-1, and CCR1, that are positioned to interface at key cellular points in ICH damage. These and other molecules in human ICH molecular pathways may serve as targets for future mechanistic studies to interdict the progressive perihematomal damage in this disease.
Supplementary Material
Acknowledgements
We thank Michal Machniki, Ellen Walker, and Martin Rutkoswski for excellent technical assistance in qRT-PCR and immunohistochemistry, and Jill Haines for patient care coordination.
These studies were supported by NIH NS044378 and by the Dr Miriam and Sheldon G Adelson Medical Research Foundation.
Footnotes
Conflict of interest
The authors state no conflict of interest.
References
- Aarts M, Liu Y, Liu L, Besshoh S, Arundine M, Gurd JW, Wang YT, Salter MW, Tymianski M. Treatment of ischemic brain damage by perturbing NMDA receptor-PSD–95 protein interactions. Science. 2002;298:846–850. doi: 10.1126/science.1072873. [DOI] [PubMed] [Google Scholar]
- Ardizzone TD, Lu A, Wagner KR, Tang Y, Ran R, Sharp FR. Glutamate receptor blockade attenuates glucose hypermetabolism in perihematomal brain after experimental intracerebral hemorrhage in rat. Stroke. 2004;35:2587–2591. doi: 10.1161/01.STR.0000143451.14228.ff. [DOI] [PubMed] [Google Scholar]
- Bajetto A, Bonavia R, Barbero S, Pirani P, Florio T, Schettini G. Chemokines and their receptors in the central nervous system. Front Neuroendocrinol. 2001;22:147–184. doi: 10.1006/frne.2001.0214. [DOI] [PubMed] [Google Scholar]
- Buisson A, Lesne S, Docagne F, Ali C, Nicole O, MacKenzie ET, Vivien D. Transforming growth factor-beta and ischemic brain injury. Cell Mol Neurobiol. 2003;23:539–550. doi: 10.1023/A:1025072013107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael ST, Archibeque I, Luke L, Nolan T, Momiy J, Li S. Growth-associated gene expression after stroke: evidence for a growth-promoting region in peri-infarct cortex. Expt Neurol. 2005;193:291–311. doi: 10.1016/j.expneurol.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Castillo J, Davalos A, Alvarez-Sabin J, Pumar JM, Leira R, Silva Y, Montaner J, Kase CS. Molecular signatures of brain injury after intracerebral hemorrhage. Neurology. 2002;58:624–629. doi: 10.1212/wnl.58.4.624. [DOI] [PubMed] [Google Scholar]
- D’Souza SD, Bonetti B, Balasingam V, Cashman NR, Barker PA, Troutt AB, Raine CS, Antel JP. Multiple sclerosis: Fas signaling in oligodendrocyte cell death. J Exp Med. 1996;184:2361–2370. doi: 10.1084/jem.184.6.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziedzic T, Bartus S, Klimkowicz A, Motyl M, Slowik A, Szczudlik A. Intracerebral hemorrhage triggers interleukin-6 and interleukin-10 release in blood. Stroke. 2002;33:2334–2335. doi: 10.1161/01.str.0000027211.73567.fa. [DOI] [PubMed] [Google Scholar]
- Felberg RA, Grotta JC, Shirzadi AL, Strong R, Narayana P, Hill-Felberg SJ, Aronowski J. Cell death in experimental intracerebral hemorrhage: the ‘black hole’ model of hemorrhagic damage. Ann Neurol. 2002;51:517–524. doi: 10.1002/ana.10160. [DOI] [PubMed] [Google Scholar]
- Gan L, Ye S, Chu A, Anton K, Yi S, Vincent VA, von Schack D, Chin D, Murray J, Lohr S, Patthy L, Gonzalez-Zulueta M, Nikolich K, Urfer R. Identification of cathepsin B as a mediator of neuronal death induced by Abeta-activated microglial cells using a functional genomics approach. J Biol Chem. 2004;279:5565–5572. doi: 10.1074/jbc.M306183200. [DOI] [PubMed] [Google Scholar]
- Gavins FN, Dalli J, Flower RJ, Granger DN, Perretti M. Activation of the annexin 1 counter-regulatory circuit affords protection in the mouse brain microcirculation. FASEB J. 2007;21:1751–1758. doi: 10.1096/fj.06-7842com. [DOI] [PubMed] [Google Scholar]
- Grilli M, Barbieri I, Basudev H, Brusa R, Casati C, Lozza G, Ongini E. Interleukin-10 modulates neuronal threshold of vulnerability to ischaemic damage. Eur J Neurosci. 2002;12:2265–2272. doi: 10.1046/j.1460-9568.2000.00090.x. [DOI] [PubMed] [Google Scholar]
- Harrison DC, Medhurst AD, Bond BC, Campbell CA, Davis RP, Philpott KL. The use of quantitative RT-PCR to measure mRNA expression in a rat model of focal ischemia—caspase-3 as a case study. Brain Res Mol Brain Res. 2000;75:143–149. doi: 10.1016/s0169-328x(99)00305-8. [DOI] [PubMed] [Google Scholar]
- Hua Y, Wu J, Keep RF, Nakamura T, Hoff JT, Xi G. Tumor necrosis factor-alpha increases in the brain after intracerebral hemorrhage and thrombin stimulation. Neurosurgery. 2006;58:542–550. doi: 10.1227/01.NEU.0000197333.55473.AD. [DOI] [PubMed] [Google Scholar]
- Huber W, Li X, Gentleman R. Visualizing data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and computational biology solutions using R and bioconductor. NY, New York: Springer; 2005. [Google Scholar]
- Kidwell CS, Saver JL, Mattiello J, Starkman S, Vinuela F, Duckwiler G, Gobin YP, Jahan R, Vespa P, Villablanca JP, Liebeskind DS, Woods RP, Alger JR. Diffusion–perfusion MR evaluation of perihematomal injury in hyperacute intracerebral hemorrhage. Neurology. 2001;57:1611–1617. doi: 10.1212/wnl.57.9.1611. [DOI] [PubMed] [Google Scholar]
- Li JZ, Vawter MP, Walsh DM, Tomita H, Evans SJ, Choudary PV, Lopez JF, Avelar A, Shokoohi V, Chung T, Mesarwi O, Jones EG, Watson SJ, Akil H, Bunney WE, Jr, Myers RM. Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum Mol Genet. 2004;13:609–616. doi: 10.1093/hmg/ddh065. [DOI] [PubMed] [Google Scholar]
- Lu A, Tang Y, Ran R, Ardizzone TL, Wagner KR, Sharp FR. Brain genomics of intracerebral hemorrhage. J Cereb Blood Flow Metab. 2006;26:230–252. doi: 10.1038/sj.jcbfm.9600183. [DOI] [PubMed] [Google Scholar]
- Lim LH, Pervaiz S. Annexin 1: the new face of an old molecule. FASEB J. 2007;21:968–975. doi: 10.1096/fj.06-7464rev. [DOI] [PubMed] [Google Scholar]
- Mayne M, Ni W, Yan HJ, Xue M, Johnston JB, Del Bigio MR, Peeling J, Power C. Antisense oligodeoxynucleotide inhibition of tumor necrosis factor-alpha expression is neuroprotective after intracerebral hemorrhage. Stroke. 2001;32:240–248. doi: 10.1161/01.str.32.1.240. [DOI] [PubMed] [Google Scholar]
- Miller CM, Vespa PM, McArthur DL, Hirt D, Etchepare M. Frameless stereotactic aspiration and thrombolysis of deep intracerebral hemorrhage is associated with reduced levels of extracellular cerebral glutamate and unchanged lactate pyruvate ratios. Neurocrit Care. 2007;6:22–29. doi: 10.1385/NCC:6:1:22. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Xi G, Hua Y, Schallert T, Hoff JT, Keep RF. Intracerebral hemorrhage in mice: model characterization and application for genetically modified mice. J Cereb Blood Flow Metab. 2004;24:487–494. doi: 10.1097/00004647-200405000-00002. [DOI] [PubMed] [Google Scholar]
- Ohab JJ, Fleming S, Blesch A, Carmichael ST. A neurovascular niche for neurogenesis after stroke. J Neurosci. 2006;26:13007–13016. doi: 10.1523/JNEUROSCI.4323-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente L, Solito E. Annexin 1: more than an anti-phospholipase protein. Inflamm Res. 2004;53:125–132. doi: 10.1007/s00011-003-1235-z. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. Mouse brain in stereotaxic coordinates. San Diego: Academic Press; 2001. [Google Scholar]
- Postler E, Rimner A, Beschorner R, Schluesener HJ, Meyermann R. Allograft-inflammatory-factor-1 is upregulated in microglial cells in human cerebral infarctions. J Neuroimmunol. 2000;108:244–250. doi: 10.1016/s0165-5728(00)00283-6. [DOI] [PubMed] [Google Scholar]
- Qureshi AI, Ali Z, Suri MF, Shuaib A, Baker G, Todd K, Guterman LR, Hopkins LN. Extracellular glutamate and other amino acids in experimental intracerebral hemorrhage: an in vivo microdialysis study. Crit Care Med. 2003a;31:1482–1489. doi: 10.1097/01.CCM.0000063047.63862.99. [DOI] [PubMed] [Google Scholar]
- Qureshi AI, Suri MF, Ostrow PT, Kim SH, Ali Z, Shatla AA, Guterman LR, Hopkins LN. Apoptosis as a form of cell death in intracerebral hemorrhage. Neurosurgery. 2003b;52:1041–1047. [PubMed] [Google Scholar]
- Qureshi AI, Tuhrim S, Broderick JP, Batjer HH, Hondo H, Hanley DF. Spontaneous intracerebral hemorrhage. N Engl J Med. 2001;344:1450–1460. doi: 10.1056/NEJM200105103441907. [DOI] [PubMed] [Google Scholar]
- Rakonjac M, Fischer L, Provost P, Werz O, Steinhilber D, Samuelsson B, Rådmark O. Coactosin-like protein supports 5-lipoxygenase enzyme activity and up-regulates leukotriene A4 production. Proc Natl Acad Sci USA. 2006;103:13150–13155. doi: 10.1073/pnas.0605150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch S, Gundelfinger ED. Molecular organization of the presynaptic active zone. Cell Tissue Res. 2006;326:379–391. doi: 10.1007/s00441-006-0244-y. [DOI] [PubMed] [Google Scholar]
- Spera PA, Ellison JA, Feuerstein GZ, Barone FC. IL10 reduces rat brain injury following focal stroke. Neurosci Lett. 1998;251:189–192. doi: 10.1016/s0304-3940(98)00537-0. [DOI] [PubMed] [Google Scholar]
- Stan AD, Ghose S, Gao XM, Roberts RC, Lewis-Amezcua K, Hatanpaa KJ, Tamminga CA. Human postmortem tissue: what quality markers matter? Brain Res. 2006;1123:1–11. doi: 10.1016/j.brainres.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stros M, Ozaki T, Bacikova A, Kageyama H, Nakagawara A. HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J Biol Chem. 2002;277:7157–7164. doi: 10.1074/jbc.M110233200. [DOI] [PubMed] [Google Scholar]
- Tang Y, Lu A, Aronow BJ, Sharp FR. Blood genomic responses differ after stroke, seizures, hypoglycemia, and hypoxia: blood genomic fingerprints of disease. Ann Neurol. 2001;50:699–707. doi: 10.1002/ana.10042. [DOI] [PubMed] [Google Scholar]
- Tang Y, Xu H, Du X, Lit L, Walker W, Lu A, Ran R, Gregg JP, Reilly M, Pancioli A, Khoury JC, Sauerbeck LR, Carrozzella JA, Spilker J, Clark J, Wagner KR, Jauch EC, Chang DJ, Verro P, Broderick JP, Sharp FR. Gene expression in blood changes rapidly in neutrophils and monocytes after ischemic stroke in humans: a microarray study. J Cereb Blood Flow Metab. 2006;26:1089–1102. doi: 10.1038/sj.jcbfm.9600264. [DOI] [PubMed] [Google Scholar]
- Trebst C, Staugaitis SM, Tucky B, Wei T, Suzuki K, Aldape KD, Pardo CA, Troncoso J, Lassmann H, Ransohoff RM. Chemokine receptors on infiltrating leucocytes in inflammatory pathologies of the central nervous system (CNS) Neuropathol Appl Neurobiol. 2003;29:584–595. doi: 10.1046/j.0305-1846.2003.00507.x. [DOI] [PubMed] [Google Scholar]
- Wagner KR. Modeling intracerebral hemorrhage: glutamate, nuclear factor-kappa B signaling and cytokines. Stroke. 2007;38:753–758. doi: 10.1161/01.STR.0000255033.02904.db. [DOI] [PubMed] [Google Scholar]
- Wang J, Dore S. Inflammation after intracerebral hemorrhage. J Cereb Blood Flow Metab. 2007;27:894–908. doi: 10.1038/sj.jcbfm.9600403. [DOI] [PubMed] [Google Scholar]
- Wasserman JK, Schlichter LC. Neuron death and inflammation in a rat model of intracerebral hemorrhage: effects of delayed minocycline treatment. Brain Res. 2007;1136:208–218. doi: 10.1016/j.brainres.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Wasserman JK, Zhu X, Schlichter LC. Evolution of the inflammatory response in the brain following intracerebral hemorrhage and effects of delayed minocycline treatment. Brain Res. 2007;1180:140–154. doi: 10.1016/j.brainres.2007.08.058. [DOI] [PubMed] [Google Scholar]
- Xi G, Keep RF, Hoff JT. Mechanisms of brain injury after intracerebral haemorrhage. Lancet Neurol. 2006;5:53–63. doi: 10.1016/S1474-4422(05)70283-0. [DOI] [PubMed] [Google Scholar]
- Xue M, Hollenberg MD, Yong VW. Combination of thrombin and matrix metalloproteinase-9 exacerbates neurotoxicity in cell culture and intracerebral hemorrhage in mice. J Neurosci. 2006;26:10281–10291. doi: 10.1523/JNEUROSCI.2806-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.