Abstract
Oleanolic acid is a plant-derived triterpenoid, which protects against various hepatotoxicants in rodents. In order to determine whether oleanolic acid activates nuclear factor erythroid-2 related factor 2 (Nrf2), a transcription factor known to induce various antioxidant and cytoprotective genes, wild-type and Nrf2-null mice were treated with oleanolic acid (90 mg/kg, i.p.) once daily for three days. Oleanolic acid increased nuclear accumulation of Nrf2 in wild-type but not Nrf2-null mice, as determined by Western blot and immunofluorescence. Oleanolic acid-treated wild-type mice had increased hepatic mRNA expression of the Nrf2 target genes NAD(P)H:quinone oxidoreductase 1 (Nqo1); glutamate-cysteine ligase, catalytic subunit (Gclc); heme oxygenase-1 (Ho-1); as well as Nrf2 itself. In addition, oleanolic acid increased protein expression and enzyme activity of the prototypical Nrf2 target gene, Nqo1, in wild-type, but not in Nrf2-null mice. Oleanolic acid protected against acetaminophen hepatotoxicity in wild-type mice but to a lesser extent in Nrf2-null mice. Oleanolic acid-mediated Nrf2-independent protection from acetaminophen is, in part, due to induction of Nrf2-independent cytoprotective genes, such as metallothionein. Collectively, the present study demonstrates that oleanolic acid facilitates Nrf2 nuclear accumulation, causing induction of Nrf2-dependent genes, which contributes to protection from acetaminophen hepatotoxicity.
Keywords: Nrf2, oleanolic acid, hepatoprotection, oxidative stress, acetaminophen
1. INTRODUCTION
Oleanolic acid is a natural triterpenoid that is a constituent of the leaves of Olea europaea, Viscum album L., and other plants, and is an aglycone of many saponins. Oleanolic acid is used in Chinese medicine for the treatment of liver disorders, such as viral hepatitis, and has been shown to protect mice from various hepatotoxicants that cause oxidative and electrophilic stress, including carbon tetrachloride, acetaminophen, bromobenzene, and thioacetamide. In mice pretreated with oleanolic acid, and then exposed to one of the above chemical hepatotoxicants, serum alanine transaminase concentrations and centrilobular necrosis were diminished [1-4].
Acetaminophen is a widely-used anitpyretic drug that causes hepatotoxicity at high doses. At therapeutic doses, most acetaminophen is conjugated with either glucuronic acid in a reaction with UDP-glucuronosyltransferases or sulfate in a reaction with sulfotransferases. However, at higher doses, UDP-glucuronosyltransferases and sulfotransferases become saturated and more of the toxic metabolite, N-acetyl-p-benzoquinoneimine (NAPQI), is formed via a reaction with cytochrome p450s. If NAPQI is not detoxified via conjugation with glutathione or if glutathione becomes depleted, NAPQI can bind to cellular proteins and form adducts, which leads to hepatotoxicity [5].
Nuclear factor erythroid 2-related factor 2 (Nrf2), and its repressor kelch-like ECH-associated protein 1 (Keap1), have been characterized over the past decade as an important endogenous cellular mechanism for coping with oxidative stress [6]. Nrf2 is a transcription factor that binds to antioxidant response elements (AREs) in upstream promoter regions of various cytoprotective enzymes, the induction of which helps restore the intracellular balance between oxidants and antioxidants. Under conditions when oxidative/electrophilic stress is low, Keap1 sequesters Nrf2 in the cytosol, by acting as an adaptor for Cullin (Cul) 3-based E3 ligase, to target Nrf2 for proteasomal degradation [7]. Cul3 and other Cullin-based E3 ligases regulate the turnover of important transcription factors by acting as scaffold proteins to help facilitate ubiquitination, marking proteins for proteasomal degradation [8]. When oxidative/electrophilic stress is increased in the cell, Nrf2 is able to avoid Keap1-mediated proteasomal degradation by Cul3, and translocate into the nucleus [9]. Once in the nucleus, Nrf2 heterodimerizes with a small musculo-aponeurotic fibrosarcoma (Maf) protein, c-jun, or activating transcription factor-4 (ATF-4) and binds to AREs, promoting transcription of various cytoprotective genes [10-12].
Nrf2 target genes include, but are not limited to, glutathione-S-transferases, NAD(P)H:quinone oxidoreductase 1 (Nqo1), glutamate-cysteine ligase catalytic subunit (Gclc), and heme oxygenase-1 (Ho-1). Nqo1 catalyzes the reduction of quinones, protecting cells against redox cycling and oxidative stress [13]. In addition, Nqo1 can reduce the toxic metabolite of acetaminophen back to the parent compound in vitro [14]. Gclc is the rate-limiting enzyme in the synthesis of glutathione, a major antioxidant molecule in cells, which is critical for the maintenance of cellular redox homeostasis and detoxification of electrophiles, including NAPQI [15]. Ho-1 catalyzes the breakdown of heme into bilirubin, carbon monoxide, and iron. Bilirubin is an antioxidant, and its generation following Ho-1 induction reduces cytotoxicity caused by oxidative stress [16]. In addition, small quantities of carbon monoxide are cytoprotective, anti-apoptotic, vasorelaxant, and anti-inflammatory [17]. Also, hemin-mediated induction of Ho-1 or pretreatment with biliverdin (a bilirubin precursor) protects rats against acetaminophen-induced hepatotoxicity [18].
The group of enzymes under the transcription control of Nrf2 are referred to as the “Nrf2 regulon” and provide antioxidative protection [19]. Nrf2-null mice have compromised antioxidant protection and are highly susceptible to target-organ injury from acetaminophen [20], benzo[a]pyrene [21], diesel exhaust [22], hyperoxia [23], hydrogen peroxide [24], and many other oxidative-type pathologies. In contrast, mice with a hepatocyte-specific deletion of Keap1 have marked increases in Nrf2 target genes and are protected from acetaminophen hepatotoxicity [25].
Previous studies have suggested that oleanolic acid is unable to activate the Nrf2-Keap1 pathway because oleanolic acid is a natural compound and lacks necessary functional groups (Michael acceptors) possessed by its synthetic derivatives, including CDDO-Im [26, 27]. Nqo1 is a prototypical target gene of Nrf2 activation; therefore, Nqo1 mRNA expression and enzyme activity are used as biomarkers of Nrf2 activation [28]. In previous studies, low doses of oleanolic acid were unable to induce Nqo1 activity in vitro in Hepa1c1c7 murine hepatoma cells [26]. In addition, a low, single, oral dose of oleanolic acid (9 mg/kg) failed to induce hepatic Nqo1 mRNA expression in mice [27].
However, in a recent study, dosing oleanolic acid (~25 mg/kg, s.c.) once daily for four days, induced many cytoprotective genes, such as the Nrf2-dependent genes Nqo1, Gclc, and Ho-1 (3-to 5-fold over vehicle) [29]. However, in this study by Liu et al. nuclear translocation of Nrf2 protein was implied but not actually shown. Furthermore, Nrf2-null mice were not used by Liu et al. to demonstrate definitively whether induction of Nqo1, Gclc, and Ho-1 was caused by Nrf2 activation, as other transcription factors and mechanisms are capable of inducing these genes [30-33]. Therefore, the purpose of the present study was to investigate whether oleanolic acid (90 mg/kg, i.p.), administered once daily for three days, as was used previously in hepatoprotection studies [34], activates the Nrf2-Keap1 pathway using wild-type and Nrf2-null mice, and whether activation of Nrf2 is responsible for oleanolic acid-mediated protection against acetaminophen hepatotoxicity.
2. METHODS
2.1 Reagents
Oleanolic acid was a gift from Dr. Jie Liu (NIEHS, Research Triangle Park, NC). Nqo1 and β-actin antibodies were purchased from Abcam (Ab2346, Ab8227, respectively, Cambridge, MA). Nrf2 antibody was purchased from Santa Cruz Biotechnology (sc-30915, Santa Cruz, CA). All other chemicals, unless otherwise specified, were purchased from Sigma-Aldrich (St. Louis, MO).
2.2 Animals and Husbandry
Eight-week-old male C57BL/6 mice were purchased from Charles River Laboratories, Inc (Wilmington, MA). Nrf2-null mice on a mixed C57BL/6 and AKR background were obtained from Dr. Jefferson Chan (University of California, Irvine, Irvine, CA) and backcrossed seven generations into C57BL/6 mice to >99% congenicity, as was determined by Jackson Laboratories (Bar Harbor, ME). Animals were housed in a temperature-, light-, and humidity-controlled environment and fed Teklad Rodent Diet #8604 (Harlan Laboratories, Madison, WI) ad libitum. The housing facility is an American Animal Associations Laboratory Animal Care-accredited facility at the University of Kansas Medical Center, and all procedures were preapproved in accordance with the Institutional Animal Care and Use Committee guidelines.
2.3 Animal Treatment
Male wild-type and Nrf2-null mice were dosed with oleanolic acid (90 mg/kg, i.p.) or vehicle (2% Tween 80 in sterile saline, 10 mL/kg, i.p) in the morning, once daily for three days. After cervical dislocation on the morning of the fourth day, livers were removed, cut into small pieces, frozen in liquid nitrogen, and stored at -80°C for later use for quantification and analysis of mRNA, protein, and enzyme activity. For immunofluorescent staining of Nrf2, a section of liver was frozen in isopentane and stored at -80°C.
For the hepatoprotection study, wild-type and Nrf2-null mice were dosed with oleanolic acid (90 mg/kg, i.p.) or vehicle (2% Tween 80, 10 mL/kg, i.p) in the morning, once daily for three days. On the fourth day, mice were administered acetaminophen (500 mg/kg, i.p.) in saline (pH 8) in a volume of 20 mL/kg. Blood and livers were collected 8 h later. Portions of the liver were fixed in 10% zinc formalin for 48 h.
2.4 Total RNA Isolation
Total liver RNA was isolated using RNA-Bee reagent (Tel-Test, Inc., Friendswood, TX) according to the manufacturer’s protocol. Total RNA concentrations were determined spectrophotometrically at 260 nm. The RNA samples were diluted to 500 ng/μL with diethyl pyrocarbonate-treated deionized water.
2.5 Branched DNA Signal Amplification (bDNA) Analysis
The mRNA expression of Nqo1, Gclc, and Ho-1 in mouse livers was quantified using the bDNA assay (Quantigene 1.0 bDNA signal amplification kit; Panomics, Inc., Fremont, CA) with modifications [35]. Gene sequences of interest were accessed from GenBank. Target sequences were analyzed using ProbeDesigner software v1.0 (Chiron Diagnostics Corp., Walpole, MA) to design oligonucleotide probe sets (capture, label, and blocker probes). All probes were designed with a melting temperature of 63°C, enabling hybridization conditions to be held constant (i.e., 53°C) during each hybridization step. Probe sets for Ho-1 and Nqo1 were previously described [36]. Probe sets for Gclc and metallothionein (MT) are listed in Table 1. Total RNA was added to each well of a 96-well plate containing 50 μL of each diluted probe set. RNA was allowed to hybridize with the probe sets overnight at 53°C. Subsequent hybridization steps were carried out according to the manufacturer’s protocol, and luminescence was quantified with a Synergy 2 Multi-Detection Microplate Reader interfaced with Gen5 Reader Control and Data Analysis Software (Biotek, Winoosky, VT). Data are presented as relative light units (RLU) normalized to control.
Table 1.
Oligonucleotide probes used for analysis of mouse gene expression by bDNA signal amplification assay.
| Gene | Accession # | Targeta | Functionb | Probe Sequence |
|---|---|---|---|---|
| Gclc | NM_012815 | 1548-1566 | CE | atggctcggagctggtctgTTTTTctcttggaaagaaagt |
| 1996-1724 | CE | cttaattagcttcaggtagttcagaataTTTTTctcttggaaagaaagt | ||
| 1725-1749 | CE | tcattagttctccagatgctctcttTTTTTctcttggaaagaaagt | ||
| 1848-1870 | CE | tcattagttctccagatgctctcttTTTTTctcttggaaagaaagt | ||
| 1896-1917 | CE | acttcgcttttctaaagcctgaTTTTTctcttggaaagaaagt | ||
| 1528-1547 | LE | ggccttgctacacccatccaTTTTTaggcataggacccgtgtct | ||
| 1567-1588 | LE | atgagcgtgtactcctctgcagTTTTTaggcataggacccgtgtct | ||
| 1589-1612 | LE | ccattgatgatggtgtctatgctcTTTTTaggcataggacccgtgtct | ||
| 1656-1678 | LE | tcgacttccatgttttcaaggtaTTTTTaggcataggacccgtgtct | ||
| 1679-1696 | LE | ctgcatcgggtgtccacgTTTTTaggcataggacccgtgtct | ||
| 1750-1770 | LE | ctctcatccacctggcaacagTTTTTaggcataggacccgtgtct | ||
| 1771-1794 | LE | agtcaggatggtttgcaataaactTTTTTaggcataggacccgtgtct | ||
| 1822-1847 | LE | tttcaaaatgaggctatagttgatctTTTTTaggcataggacccgtgtct | ||
| 1918-1939 | LE | gggtcgcttttacctccactgtTTTTTaggcataggacccgtgtct | ||
| 1613-1632 | BL | caggaaacacgccttccttc | ||
| 1633-1655 | BL | ggagttcagaatggggatgagtc | ||
| 1795-1821 | BL | catcagttattacactgtcttgcttgt | ||
| 1871-1895 | BL | tccaagtaactctggacattcacac | ||
| MT | NM_013602 | 72-92 | CE | ggtccattccgagatctggtgTTTTTctcttggaaagaaagt |
| 170-190 | CE | ggagcagcagctcttcttgcaTTTTTctcttggaaagaaagt | ||
| 130-147 | LE | caggcgcaggagctggtgTTTTTaggcataggacccgtgtct | ||
| 191-207 | LE | cagcccacgggacagcaTTTTTaggcataggacccgtgtct | ||
| 227-244 | LE | ggcgcctttgcagacacaTTTTTaggcataggacccgtgtct | ||
| 245-262 | LE | gcacgtgcacttgtccgcTTTTTaggcataggacccgtgtct | ||
| 263-283 | LE | ctgttcgtcacatcaggcacaTTTTTaggcataggacccgtgtct | ||
| 284-302 | LE | tttacacgtggtggcagcgTTTTTaggcataggacccgtgtct | ||
| 303-323 | LE | cgctgggttggtccgatactaTTTTTaggcataggacccgtgtct | ||
| 93-112 | BL | ggtggagcaggagcagttgg | ||
| 113-129 | BL | caagtgcaggagccgcc | ||
| 148-169 | BL | ggaggtgcacttgcagttcttg | ||
| 208-226 | BL | gccctgggcacatttggag |
Target refers to the sequence of the mRNA transcript as enumerated in the GenBank file.
Function refers to the use of the oligonucleotide probe in the assay (CE, capture extender; LE, label extender; BL, blocker probe).
2.6 Nrf2 and Nqo1 Protein Expression
Livers were homogenized in sucrose-Tris buffer (0.25 mol/L sucrose, 10 mmol/L Tris–HCl, pH 7.4) and centrifuged at 100,000 g for 60 min at 4°C. The resulting supernatant, containing the cytosolic fraction, was used to assay for Nqo1 protein. Nuclear extracts were prepared with the NE-PER nuclear extraction kit according to the manufacturer’s directions (Pierce Biotechnology, Rockford, IL). Nuclear extracts were used for Nrf2 immunoblotting. Protein concentrations were determined with BCA Assay Kit from Pierce Biotechnology (Rockford, IL).
Cytosolic and nuclear proteins (40 μg protein/lane) were electrophoretically resolved using polyacrylamide gels (4% stacking and 12% resolving for Nqo1; 4% stacking and 10% resolving for Nrf2). Gels were transblotted overnight at 4°C onto a nitrocellulose membrane for Nqo1, and a polyvinylidene fluoride membrane for Nrf2. Membranes were then washed with PBS–buffered saline containing 0.05% Tween-20 (PBS-T). Membranes were blocked for 1 h at room temperature with 5% non-fat milk in PBS–T. Blots were then incubated with primary antibody (1:1000 dilution for both Nqo1 and Nrf2, in 2% non-fat milk in PBS-T) for 3 h at room temperature. Blots were then washed in PBS-T and incubated with secondary antibody conjugated with horseradish peroxidase (1:2000 dilution for Nqo1 and Nrf2) in 2% non-fat milk in PBS-T buffer for 1 hr at room temperature. Blots were then washed with PBS-T. Protein-antibody complexes were detected using an enhanced chemiluminescent kit (Pierce Biotechnology, Rockford, IL) and exposed to X-ray film (Denville Scientific, Metuchen, NJ). Intensity of protein bands was quantified using the Discovery Series Quantity One 1-D Analysis software (Bio-Rad Laboratories, Hercules, CA). Intensity values were normalized to β-actin and expressed as relative protein expression.
2.7 Immunofluorescence
Nrf2 was detected as previously described [37]. Briefly, isopentane frozen liver was embedded in Optimal Cutting Temperature (OCT) compound prior to cyrosectioning. Cryosections were air-dried at room temperature for 5 min and fixed with 4% paraformaldehyde. The sections were blocked at room temperature for 30 min with 5% donkey serum/phosphate-buffered saline with 0.2% Triton X-100 (PBS-Tx), and then incubated overnight with Nrf2 antibody diluted 1:50 in 5% donkey serum/PBS-Tx. A fluorescein isothiocyanate-labeled secondary antibody (Jackson Immunoresearch Laboratories, West Grove, PA) diluted 1:200 was used, along with rhodamine-labeled phalloidin (Invitrogen, Carlsbad, CA) diluted 1:200 in 5% donkey serum/PBS-Tx. 4,6-diamidino-2-phenylindole (Invitrogen, Carlsbad, CA) staining was performed to define nuclear regions. Antibody solutions were filtered through 0.22 μm membrane syringe-driven filter units (Osmonics Inc., Minnetonka, MN) prior to use. Frozen liver sections were stained and imaged under uniform conditions for each antibody. Negative controls without primary antibody were also included in the analysis (data not shown). Images were captured on an Olympus BX41 fluorescent microscope with a DP70 camera and DP Controller software (Olympus, Melville, NY).
2.8 Nqo1 activity assay
Cytosolic Nqo1 enzyme activity was determined by quantifying the reduction of 2,6-dichlorophenol-indophenol (DCPIP) as described previously [38, 39].
2.9 Serum alanine transaminase (ALT) concentrations
Serum ALT concentrations were determined as a biochemical indicator of hepatocellular injury using Pointe Scientific Liquid ALT Reagent Set (Canton, MI) according to the manufacturer’s protocol.
2.10 Histopathology
Liver samples were fixed in 10% neutral-buffered zinc formalin prior to routine processing and paraffin embedding. Liver sections (5 μm) were stained with hematoxylin and eosin. Liver sections were analyzed and assigned one of six scores of severity of necrosis: 0, none; 1, minimal (>2 foci of single cell necrosis per section); 2, mild (at least 5 areas of focal necrosis per section); 3, moderate (at least five foci of zonal necrosis per section); 4, severe (lobular damage, with many viable lobules per section); and 5, global (severe lobular damage, few areas of viability per section). A section was defined as a single viewpoint at low power (×100) magnification.
2.11 Statistical Analysis
An n=5 for all groups was used. Data were analyzed using one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test (p ≤ 0.05). Histopathological data were rank ordered prior to ANOVA analysis, which was followed by Newman-Keuls multiple range test (p ≤ 0.05).
3. RESULTS
3.1 Nrf2 nuclear translocation in oleanolic acid-treated wild-type and Nrf2-null mice
Oleanolic acid (90 mg/kg, i.p.), given once daily for three days, increased Nrf2 protein 432% in nuclear extracts from livers of wild-type mice (Fig 1, upper). Nrf2-null mice treated with vehicle or oleanolic acid did not have detectable levels of Nrf2 protein in the nuclear fractions. Indirect immunofluorescent analysis revealed minimal Nrf2 staining (green) in frozen sections from wild-type mice (Fig 1, lower). The actin cytoskeleton was stained in red. Nrf2 staining of liver sections from oleanolic acid-treated wild-type mice was strong and localized to the nucleus (blue). No Nrf2 staining was observed in vehicle-or oleanolic acid-treated Nrf2-null mice.
Fig 1.
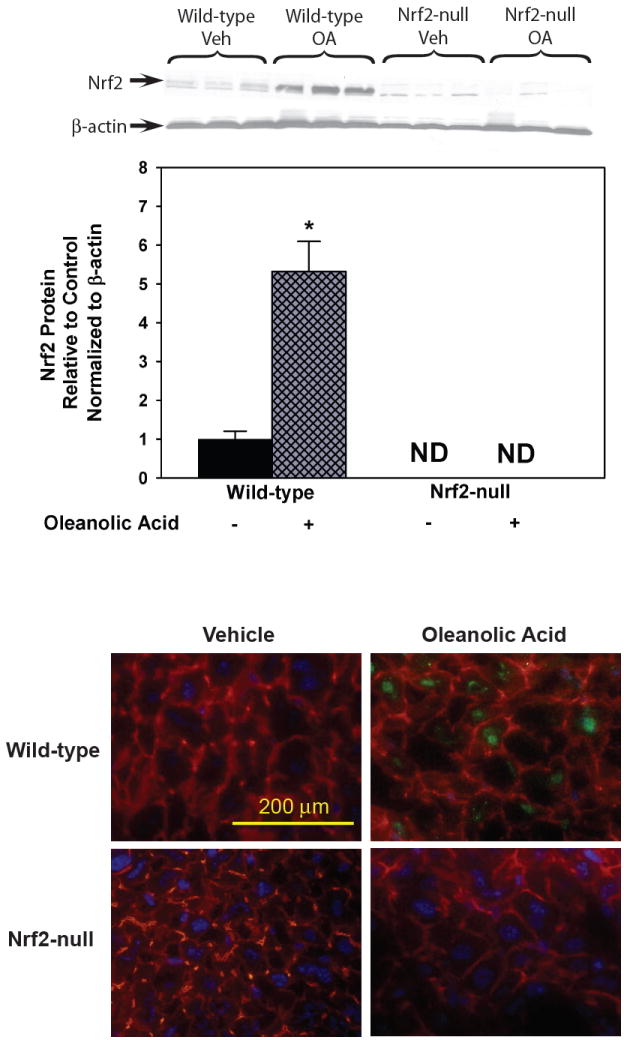
Upper. Western blot of liver nuclear fractions for Nrf2 after treatment with oleanolic acid (90 mg/kg, i.p.) once daily for three days. Also shown is the quantification of specific band intensity, normalized to β-actin, and expressed relative to control as mean ± S.E.M. Abbreviations: ND, Not Detected; Veh, Vehicle; OA, oleanolic acid; WT, wild-type; null, Nrf2-null. Asterisks (*) indicate a statistically significant difference from wild-type mice treated with vehicle (p ≤ 0.05). Lower. Immunofluorescent localization of Nrf2 in livers from wild-type mice after vehicle or oleanolic acid. Indirect immunofluorescence to detect Nrf2 (green) and actin (red) was performed on liver cryosections (5 μm) from wild-type mice after oleanolic acid (90 mg/kg, i.p.) treatment once daily for three days. Sections were mounted in Prolong Gold containing DAPI for nuclear staining (blue). Representative images are shown at high-power magnification (×400). Bar represents 200 μm.
3.2 Nrf2, Nqo1, Gclc, and Ho-1 mRNA expression in oleanolic acid-treated wild-type and Nrf2-null mice
Oleanolic acid increased mRNA expression of Nrf2 (347%), Nqo1 (1108%), Gclc (521%), and Ho-1 (955%) in livers of wild-type mice (Fig 2). Nrf2-null mice treated with oleanolic acid showed no increase in Nrf2, Nqo1, Gclc, or Ho-1 mRNA expression.
Fig 2.
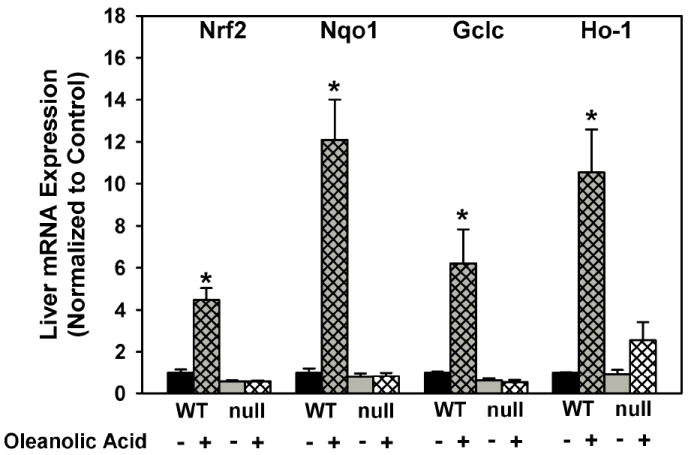
Messenger RNA expression of Nqo1, Gclc, and Ho-1 in livers of wild-type and Nrf2-null mice after treatment with oleanolic acid (90 mg/kg, i.p.) once daily for three days. Messenger RNA was quantified by the bDNA assay. Data is expressed relative to wild-type controls as mean ± S.E.M. Abbreviations: WT, wild-type; null, Nrf2-null. Asterisks (*) indicate a statistically significant difference from wild-type mice receiving vehicle (p ≤ 0.05).
3.3 Nqo1 protein expression and enzyme activity in oleanolic acid-treated wild-type and Nrf2-null mice
Because Nqo1 is considered the prototypical Nrf2-target gene [40-43], Nqo1 protein and enzyme activity were evaluated. Wild-type mice treated with oleanolic acid had increased Nqo1 protein expression (79%) as determined by western blot (Fig 3, upper). Vehicle treated Nrf2-null mice had much lower Nqo1 protein expression (-73%) than wild-type mice. Furthermore, oleanolic acid did not change Nqo1 protein expression in Nrf2-null mice. Oleanolic acid induced Nqo1 enzyme activity 154% in wild-type mice (Fig 3, lower). Nqo1 activity is less in Nrf2-null mice (-47%) than in wild-type mice and is not induced by oleanolic acid. Of note, in wild-type mice treated with oleanolic acid, even though Nqo1 mRNA expression was induced over 1100%, Nqo1 protein expression and enzyme activity did not increase more than 150%.
Fig 3.
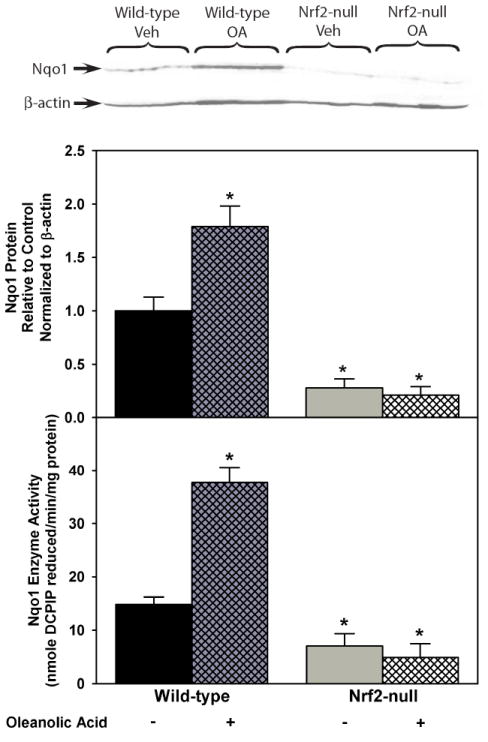
Upper. Protein expression of Nqo1 determined by western blot in livers of wild-type and Nrf2-null mice after treatment with oleanolic acid (90 mg/kg, i.p.) once daily for three days. Also shown is the quantification of specific band intensity, normalized to β-actin, and expressed relative to control as mean ± S.E.M. Lower. Nqo1 enzyme activity in livers of wild-type and Nrf2-null mice after treatment with oleanolic acid (90 mg/kg, i.p.) once daily for three days. Units are in nmole DCPIP reduced per min per mg of protein expressed as mean ± S.E.M. Abbreviations: Veh, Vehicle; OA, oleanolic acid; WT, wild-type; null, Nrf2-null. Asterisks (*) indicate a statistically significant difference from wild-type mice receiving vehicle (p ≤ 0.05).
3.4 Oleanolic acid-mediated protection against acetaminophen in wild-type and Nrf2-null mice
In this study, serum ALT concentrations corresponded with histopathological necrosis grading. Oleanolic acid did not alter serum ALTs (baseline of 20-30 IU/L) or liver histology in wild-type or Nrf2-null mice (data not shown). Oleanolic acid-pretreatment decreased acetaminophen-induced hepatotoxicity in wild-type mice, as indicated by an 82% reduction in serum ALTs (Fig 4) and reduction in necrosis grade (Table 2). Acetaminophen produced more severe hepatotoxicity in Nrf2-null mice (increases in serum ALTs [250%] and necrosis grade) than wild-type mice, similar to a previous report of increased sensitivity of Nrf2-null mice [20]. Oleanolic acid-pretreatment of Nrf2-null mice protected against acetaminophen hepatotoxicity (reduction in serum ALTs [65%] and necrosis grade), but less so than in wild-type mice. Of note, one Nrf2-null mouse pretreated with vehicle and challenged with acetaminophen died before completion of the study (8 h).
Fig 4.
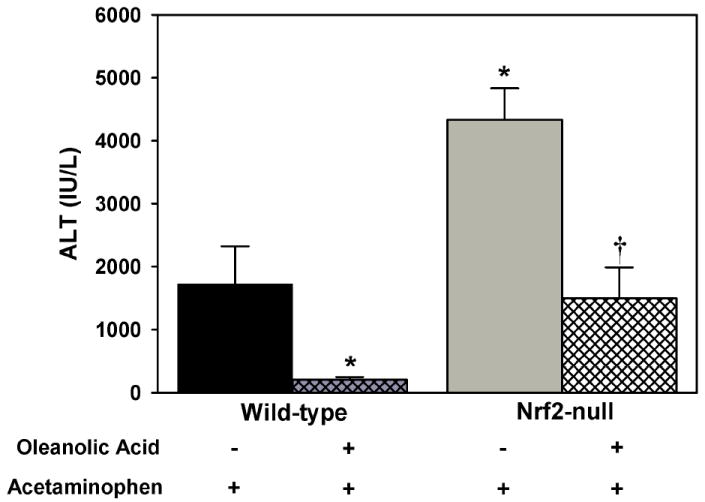
Serum alanine transaminase (ALT) levels in wild-type and Nrf2-null mice pretreated with oleanolic acid (90 mg/kg, i.p.) once daily for three days (n=5 for all groups except Nrf2-null mice receiving vehicle, n=4), administered acetaminophen (500 mg/kg, i.p.) on the fourth day, and sacrificed 8-h later. Units are in International Units/Liter (IU/L) expressed as mean ± S.E.M. A minus sign (-) represents that the animal was administered the appropriate vehicle for the compound specified. A plus sign (+) represents that the animal was administered the compound specified. Asterisks (*) indicate a statistically significant difference from wild-type mice receiving vehicle (p ≤ 0.05). Daggers (†) indicate a statistically significant difference from Nrf2-null mice receiving vehicle (p ≤ 0.05).
Table 2.
Histological Analysis of Livers from Vehicle-and OA-pretreated Wild-type and Nrf2-null mice after AA Challenge.
| Histological Grade | |||||||
|---|---|---|---|---|---|---|---|
| Treatment Group | 0 | 1 | 2 | 3 | 4 | 5 | p≤0.05 |
| Wild-type VC-AA | 0 | 0 | 0 | 2 | 3 | 0 | |
| Wild-type OA-AA | 0 | 3 | 2 | 0 | 0 | 0 | * |
| Nrf2-null VC-AA | 0 | 0 | 0 | 0 | 0 | 5 | * |
| Nrf2-null OA-AA | 0 | 0 | 0 | 1 | 4 | 0 | † |
Grade of liver injury in wild-type and Nrf2-null mice pretreated with OA (90 mg/kg, i.p.) once daily for three days (n=5 for all groups except Nrf2-null mice receiving vehicle, n=4), administered acetaminophen (500 mg/kg, i.p.) on the fourth day, and sacrificed 8-h later. Wild-type and Nrf2-null mice administered OA or vehicle only did not have any histological abnormalities. Liver sections were analyzed at low power (×100) with six scores of severity: 0, none; 1, minimal (>2 foci of single cell necrosis per section); 2, mild (at least 5 areas of focal necrosis per section); 3, moderate (at least five foci of zonal necrosis per section); 4, severe (lobular damage, with many viable lobules per section); and 5, global (severe lobular damage, few areas of viability per section). Values are expressed as mean ± S.E.M. Abbreviations: WT, wild-type; null, Nrf2-null; VC, Vehicle Control; OA, oleanolic acid; AA, Acetaminophen.
Asterisks indicate a statistically significant difference from wild-type mice receiving vehicle (VC-AA) (p ≤ 0.05).
Daggers indicate a statistically significant difference from Nrf2-null mice receiving vehicle (VC-AA) (p ≤ 0.05).
3.5 MT mRNA expression in oleanolic acid-treated wild-type and Nrf2-null mice
Because oleanolic acid produced some protection from acetaminophen hepatotoxicity in Nrf2-null mice, mRNA expression of MT, a Nrf2-independent gene [44] that has been shown to protect against acetaminophen hepatotoxicity [45], was quantified in oleanolic acid-treated wild-type and Nrf2-null mice (Fig 5). It has been hypothesized that the high sulfhydryl content of MT can scavenge the electrophiles and free radicals formed during the pathogenesis of acetaminophen hepatotoxicity [45]. Indeed, oleanolic acid induced hepatic MT mRNA expression in both wild-type (31-fold) and Nrf2-null mice (22-fold).
Fig 5.
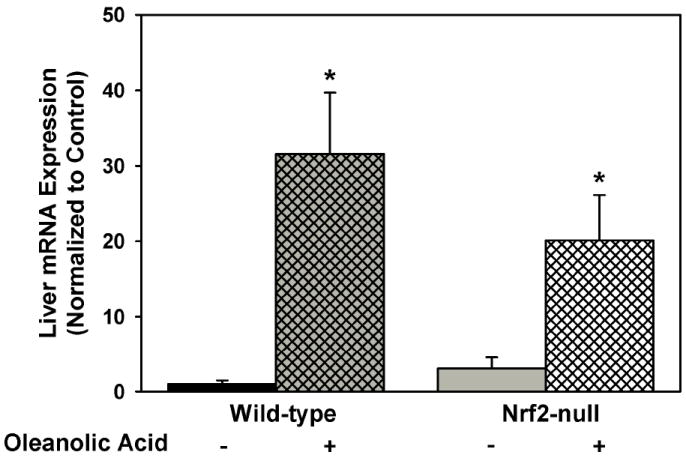
Messenger RNA expression of MT from livers of wild-type and Nrf2-null mice after treatment with oleanolic acid (90 mg/kg, i.p.) once daily for three days. Messenger RNA was quantified by the bDNA assay. Data is expressed relative to wild-type controls as mean ± S.E.M. Asterisks (*) indicate a statistically significant difference from wild-type mice receiving vehicle (p ≤ 0.05).
4. DISCUSSION
Because it has previously been shown that oleanolic acid (90 mg/kg, i.p.) given once daily for three days is protective against various hepatotoxicants, which cause toxicity, in part, through oxidative or electrophilic stress mechanisms [34], the present studies were undertaken to establish whether oleanolic acid is an activator of the Nrf2-Keap1 pathway. Oleanolic acid (90 mg/kg, i.p.), given once daily for three days, increases nuclear accumulation of Nrf2 in wild-type, but not in Nrf2-null mice, as determined by both western blot of nuclear fractions and immunofluorescence. Also, the oleanolic acid-dosing regimen used in the current studies, increases mRNA of Nrf2, as well as three characteristic Nrf2-target genes (Nqo1, Gclc, and Ho-1) in wild-type, but not in Nrf2-null mice.
In addition to mRNA expression, Nqo1 protein expression and activity were quantified because Nqo1 is a prototypical Nrf2-target gene [40-43]. Oleanolic acid increased protein expression of Nqo1 in wild-type mice (Fig 4). As expected, Nrf2-null mice have lower Nqo1 protein expression than their wild-type counterparts, as reported previously [46]. Oleanolic acid was not able to increase Nqo1 protein expression in Nrf2-null mice, which correlates with the mRNA expression data. In addition, oleanolic acid increased Nqo1 enzyme activity in wild-type, but not Nrf2-null mice.
The functional involvement of Nrf2 in oleanolic acid-mediated hepatoprotection was investigated using acetaminophen as a model hepatotoxicant. Acetaminophen produced liver injury in both genotypes, but the injury was more severe in Nrf2-null mice. Enhanced susceptibility of Nrf2-null mice to acetaminophen is consistent with previous reports [20]. Oleanolic acid-pretreatment of wild-type mice decreased acetaminophen hepatotoxicity. Oleanolic acid-pretreatment also decreased acetaminophen hepatotoxicity in Nrf2-null mice, although the reduction was less than in wild-type mice.
Because oleanolic acid also had some hepatoprotective effects against acetaminophen toxicity in Nrf2-null mice, the possibility that oleanolic acid might induce MT in Nrf2-null mice and produce protection by this mechanism was evaluated. MT is a low-molecular-weight, cysteine-rich, metal-binding protein that is suggested to be capable of trapping reactive oxygen and nitrogen species, as well as electrophiles [47], which all contribute to the pathogenesis of acetaminophen hepatotoxicity [48]. MT has at least one ARE in its promoter region and is induced by Nrf1 (a transcription factor in the same family as Nrf2), but not Nrf2 [44]. Oleanolic acid induces MT in both mice and rats [29], and oleanolic acid-mediated MT induction protects against cadmium toxicity [49]. In addition, MT-null mice are more susceptible than wild-type mice to acetaminophen-mediated hepatotoxicity [49]. Oleanolic acid induced hepatic MT mRNA expression in both wild-type and Nrf2-null mice (Fig 5), which suggests that that other mechanisms, especially MT induction, in addition to Nrf2 activation might contribute to protection from oleanolic acid-mediated hepatoprotection from acetaminophen.
Other genes that may contribute to Nrf2-independent hepatoprotection by oleanolic acid include a 70 kDa heat shock protein (Hsp70) and the G protein-coupled bile acid receptor 1 (Gpbar1 or Tgr5). Hsp70 mRNA is induced by oleanolic acid [29] and acts to protect cells by reducing the ability of oxidized proteins to aggregate, permitting them time to refold and return to a functional conformation. Furthermore, Hsp70-null mice are more sensitive to acetaminophen hepatotoxicity [50]. TGR5 mRNA is also induced by oleanolic acid [51] and is a bile acid responsive and cell surface G protein-coupled receptor that when activated suppresses macrophage proinflammatory cytokine production, particularly interleukin-1α (IL-1α), IL-1β, and tumor necrosis factor α (TNFα) [52, 53]. Induction of IL-1 and TNFα lead to induction of inducible nitric oxide synthase (iNos) [54, 55]. iNos contributes to acetaminophen hepatotoxicity by leading to increased production of nitric oxide and subsequent peroxynitrite formation. Thus, iNos suppression, possibly mediated by oleanolic acid, may contribute to the observed hepatoprotection from acetaminophen in Nrf2-null mice [5, 56]. Thus, oleanolic acid has Nrf2-dependent and Nrf2-independent effects, both of which likely contribute to oleanolic acid-mediated hepatoprotection from acetaminophen.
Collectively, increased nuclear accumulation of Nrf2, increased mRNA expression of Nrf2-target genes, and up-regulation of Nqo1 protein and activity provide strong evidence that oleanolic acid activates the Nrf2-Keap1 pathway. In addition oleanolic acid protected from acetaminophen hepatotoxicty in wild-type but less so in Nrf2-null mice, suggesting that oleanolic acid also activates Nrf2-independent cytoprotective mechanisms. Thus, this study establishes oleanolic acid as a Nrf2 activator; however, Nrf2-independent mechanisms that contribute to oleanolic acid-mediated hepatoprotection from acetaminophen require further study.
Acknowledgments
The authors would like to thank Xiaohong Lei and the rest of the Klaassen laboratory for technical and manuscript revision assistance. Nrf2-null mice were provided by Dr. Jefferson Chan (University of California-Irvine, Irvine, CA). This research was financially supported by NIH Grants ES07079, ES09649, ES09716, ES013714, and RR021940.
Non-Standard Abbreviations
- ALT
alanine transaminase
- ARE
antioxidant response element
- ATF-4
activating transcription factor-4
- Cul3
Cullin3
- Gclc
glutamate cysteine ligase catalytic subunit
- Ho-1
heme oxygenase-1
- MT
metallothionein
- Nrf2
nuclear factor-erythroid 2-related factor 2
- Keap1
kelch-like ECH associated protein 1
- Maf
musculo-aponeurotic fibrosarcoma
- Nqo1
NAD(P)H: quinone oxidoreductase 1
- NAPQI
N-acetyl-p-benzoquinoneimine
- OA
oleanolic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu J, Liu Y, Parkinson A, Klaassen CD. Effect of oleanolic acid on hepatic toxicant-activating and detoxifying systems in mice. J Pharmacol Exp Ther. 1995;275:768–74. [PubMed] [Google Scholar]
- 2.Liu J, Liu Y, Klaassen CD. The effect of Chinese hepatoprotective medicines on experimental liver injury in mice. J Ethnopharmacol. 1994;42:183–91. doi: 10.1016/0378-8741(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Liu Y, Mao Q, Klaassen CD. The effects of 10 triterpenoid compounds on experimental liver injury in mice. Fundam Appl Toxicol. 1994;22:34–40. doi: 10.1006/faat.1994.1005. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Liu Y, Madhu C, Klaassen CD. Protective effects of oleanolic acid on acetaminophen-induced hepatotoxicity in mice. J Pharmacol Exp Ther. 1993;266:1607–13. [PubMed] [Google Scholar]
- 5.James LP, Mayeux PR, Hinson JA. Acetaminophen-induced hepatotoxicity. Drug Metab Dispos. 2003;31:1499–506. doi: 10.1124/dmd.31.12.1499. [DOI] [PubMed] [Google Scholar]
- 6.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 7.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006;387:1311–20. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 8.Wolf DA, Zhou C, Wee S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol. 2003;5:1029–33. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- 9.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 10.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–22. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 11.He CH, Gong P, Hu B, Stewart D, Choi ME, Choi AM, et al. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem. 2001;276:20858–65. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 12.Venugopal R, Jaiswal AK. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17:3145–56. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- 13.Riley RJ, Workman P. DT-diaphorase and cancer chemotherapy. Biochem Pharmacol. 1992;43:1657–69. doi: 10.1016/0006-2952(92)90694-e. [DOI] [PubMed] [Google Scholar]
- 14.Moffit JS, Aleksunes LM, Kardas MJ, Slitt AL, Klaassen CD, Manautou JE. Role of NAD(P)H:quinone oxidoreductase 1 in clofibrate-mediated hepatoprotection from acetaminophen. Toxicology. 2007;230:197–206. doi: 10.1016/j.tox.2006.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalton TP, Dieter MZ, Yang Y, Shertzer HG, Nebert DW. Knockout of the mouse glutamate cysteine ligase catalytic subunit (Gclc) gene: embryonic lethal when homozygous, and proposed model for moderate glutathione deficiency when heterozygous. Biochem Biophys Res Commun. 2000;279:324–9. doi: 10.1006/bbrc.2000.3930. [DOI] [PubMed] [Google Scholar]
- 16.Clark JE, Foresti R, Green CJ, Motterlini R. Dynamics of haem oxygenase-1 expression and bilirubin production in cellular protection against oxidative stress. Biochem J. 2000;348(Pt 3):615–9. [PMC free article] [PubMed] [Google Scholar]
- 17.Ryter SW, Otterbein LE. Carbon monoxide in biology and medicine. Bioessays. 2004;26:270–80. doi: 10.1002/bies.20005. [DOI] [PubMed] [Google Scholar]
- 18.Chiu H, Brittingham JA, Laskin DL. Differential induction of heme oxygenase-1 in macrophages and hepatocytes during acetaminophen-induced hepatotoxicity in the rat: effects of hemin and biliverdin. Toxicol Appl Pharmacol. 2002;181:106–15. doi: 10.1006/taap.2002.9409. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–40. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O’Connor T, et al. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol Sci. 2001;59:169–77. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- 21.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98:3410–5. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aoki Y, Sato H, Nishimura N, Takahashi S, Itoh K, Yamamoto M. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol Appl Pharmacol. 2001;173:154–60. doi: 10.1006/taap.2001.9176. [DOI] [PubMed] [Google Scholar]
- 23.Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, et al. Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol. 2002;26:175–82. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 24.Kraft AD, Johnson DA, Johnson JA. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. J Neurosci. 2004;24:1101–12. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 26.Dinkova-Kostova AT, Liby KT, Stephenson KK, Holtzclaw WD, Gao X, Suh N, et al. Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proc Natl Acad Sci U S A. 2005;102:4584–9. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yates MS, Tauchi M, Katsuoka F, Flanders KC, Liby KT, Honda T, et al. Pharmacodynamic characterization of chemopreventive triterpenoids as exceptionally potent inducers of Nrf2-regulated genes. Mol Cancer Ther. 2007;6:154–62. doi: 10.1158/1535-7163.MCT-06-0516. [DOI] [PubMed] [Google Scholar]
- 28.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 2003;374:337–48. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Wu Q, Lu YF, Pi J. New insights into generalized hepatoprotective effects of oleanolic acid: key roles of metallothionein and Nrf2 induction. Biochem Pharmacol. 2008;76:922–8. doi: 10.1016/j.bcp.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Dahl EL, Mulcahy RT. Cell-type specific differences in glutamate cysteine ligase transcriptional regulation demonstrate independent subunit control. Toxicol Sci. 2001;61:265–72. doi: 10.1093/toxsci/61.2.265. [DOI] [PubMed] [Google Scholar]
- 31.Ferrandiz ML, Devesa I. Inducers of heme oxygenase-1. Curr Pharm Des. 2008;14:473–86. doi: 10.2174/138161208783597399. [DOI] [PubMed] [Google Scholar]
- 32.Knight TR, Choudhuri S, Klaassen CD. Induction of Hepatic Glutathione S-Transferases in Male Mice By Prototypes of Various Classes of Microsomal Enzyme Inducers. Toxicol Sci. 2008;106:329–38. doi: 10.1093/toxsci/kfn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okey AB, Franc MA, Moffat ID, Tijet N, Boutros PC, Korkalainen M, et al. Toxicological implications of polymorphisms in receptors for xenobiotic chemicals: the case of the aryl hydrocarbon receptor. Toxicol Appl Pharmacol. 2005;207:43–51. doi: 10.1016/j.taap.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Liu Y, Klaassen CD. Protective effect of oleanolic acid against chemical-induced acute necrotic liver injury in mice. Zhongguo Yao Li Xue Bao. 1995;16:97–102. [PubMed] [Google Scholar]
- 35.Hartley DP, Klaassen CD. Detection of chemical-induced differential expression of rat hepatic cytochrome P450 mRNA transcripts using branched DNA signal amplification technology. Drug Metab Dispos. 2000;28:608–16. [PubMed] [Google Scholar]
- 36.Aleksunes LM, Slitt AM, Cherrington NJ, Thibodeau MS, Klaassen CD, Manautou JE. Differential expression of mouse hepatic transporter genes in response to acetaminophen and carbon tetrachloride. Toxicol Sci. 2005;83:44–52. doi: 10.1093/toxsci/kfi013. [DOI] [PubMed] [Google Scholar]
- 37.Slitt AL, Cherrington NJ, Dieter MZ, Aleksunes LM, Scheffer GL, Huang W, et al. trans-Stilbene oxide induces expression of genes involved in metabolism and transport in mouse liver via CAR and Nrf2 transcription factors. Mol Pharmacol. 2006;69:1554–63. doi: 10.1124/mol.105.014571. [DOI] [PubMed] [Google Scholar]
- 38.Benson AM, Hunkeler MJ, Talalay P. Increase of NAD(P)H:quinone reductase by dietary antioxidants: possible role in protection against carcinogenesis and toxicity. Proc Natl Acad Sci U S A. 1980;77:5216–20. doi: 10.1073/pnas.77.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ernster L. Methods Enzymol. 1967;10:309–317. [Google Scholar]
- 40.Dhakshinamoorthy S, Jaiswal AK. Small maf (MafG and MafK) proteins negatively regulate antioxidant response element-mediated expression and antioxidant induction of the NAD(P)H:Quinone oxidoreductase1 gene. J Biol Chem. 2000;275:40134–41. doi: 10.1074/jbc.M003531200. [DOI] [PubMed] [Google Scholar]
- 41.Dhakshinamoorthy S, Jaiswal AK. Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 2001;20:3906–17. doi: 10.1038/sj.onc.1204506. [DOI] [PubMed] [Google Scholar]
- 42.Jaiswal AK. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic Biol Med. 2000;29:254–62. doi: 10.1016/s0891-5849(00)00306-3. [DOI] [PubMed] [Google Scholar]
- 43.Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93:14960–5. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. NRF1 and NRF2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem. 2008 doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Liu Y, Hartley D, Klaassen CD, Shehin-Johnson SE, Lucas A, et al. Metallothionein-I/II knockout mice are sensitive to acetaminophen-induced hepatotoxicity. J Pharmacol Exp Ther. 1999;289:580–6. [PubMed] [Google Scholar]
- 46.Aleksunes LM, Slitt AL, Maher JM, Dieter MZ, Knight TR, Goedken M, et al. Nuclear factor-E2-related factor 2 expression in liver is critical for induction of NAD(P)H:quinone oxidoreductase 1 during cholestasis. Cell Stress Chaperones. 2006;11:356–63. doi: 10.1379/CSC-217.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klaassen CD, Liu J, Choudhuri S. Metallothionein: an intracellular protein to protect against cadmium toxicity. Annu Rev Pharmacol Toxicol. 1999;39:267–94. doi: 10.1146/annurev.pharmtox.39.1.267. [DOI] [PubMed] [Google Scholar]
- 48.Michael SL, Pumford NR, Mayeux PR, Niesman MR, Hinson JA. Pretreatment of mice with macrophage inactivators decreases acetaminophen hepatotoxicity and the formation of reactive oxygen and nitrogen species. Hepatology. 1999;30:186–95. doi: 10.1002/hep.510300104. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Kreppel H, Liu J, Choudhuri S, Klaassen CD. Oleanolic acid protects against cadmium hepatotoxicity by inducing metallothionein. J Pharmacol Exp Ther. 1993;266:400–6. [PubMed] [Google Scholar]
- 50.Tolson JK, Dix DJ, Voellmy RW, Roberts SM. Increased hepatotoxicity of acetaminophen in Hsp70i knockout mice. Toxicol Appl Pharmacol. 2006;210:157–62. doi: 10.1016/j.taap.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Sato H, Genet C, Strehle A, Thomas C, Lobstein A, Wagner A, et al. Anti-hyperglycemic activity of a TGR5 agonist isolated from Olea europaea. Biochem Biophys Res Commun. 2007;362:793–8. doi: 10.1016/j.bbrc.2007.06.130. [DOI] [PubMed] [Google Scholar]
- 52.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–40. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 53.Keitel V, Donner M, Winandy S, Kubitz R, Haussinger D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun. 2008;372:78–84. doi: 10.1016/j.bbrc.2008.04.171. [DOI] [PubMed] [Google Scholar]
- 54.Busse R, Mulsch A. Induction of nitric oxide synthase by cytokines in vascular smooth muscle cells. FEBS Lett. 1990;275:87–90. doi: 10.1016/0014-5793(90)81445-t. [DOI] [PubMed] [Google Scholar]
- 55.Kilbourn RG, Gross SS, Jubran A, Adams J, Griffith OW, Levi R, et al. NG-methyl-L-arginine inhibits tumor necrosis factor-induced hypotension: implications for the involvement of nitric oxide. Proc Natl Acad Sci U S A. 1990;87:3629–32. doi: 10.1073/pnas.87.9.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bourdi M, Masubuchi Y, Reilly TP, Amouzadeh HR, Martin JL, George JW, et al. Protection against acetaminophen-induced liver injury and lethality by interleukin 10: role of inducible nitric oxide synthase. Hepatology. 2002;35:289–98. doi: 10.1053/jhep.2002.30956. [DOI] [PubMed] [Google Scholar]


