Abstract
Thioredoxins are oxido-reductase enzymes present in all organisms, catalyzing the reduction of disulfide bonds in proteins. By applying a calibrated force to a substrate disulfide, the chemical mechanisms of Trx catalysis can be examined in detail at the single molecule level. Here we use single molecule force-clamp spectroscopy to explore the chemical evolution of Trx catalysis by probing the chemistry of eight different thioredoxin enzymes. While all Trxs show a characteristic Michaelis-Menten mechanism detected when the disulfide bond is stretched at low forces, two different chemical behaviors distinguish bacterial from eukaryotic-origin Trxs at high forces. Eukaryotic-origin Trxs reduce disulfide bonds through a single-electron transfer reaction (SET) whereas bacterial-origin Trxs exhibit both nucleophilic substitution (SN2) and SET reactions. A computational analysis of Trx structures identifies the evolution of the binding groove as an important factor controlling the chemistry of Trx catalysis.
Enzymes are exceptional catalysts able to accelerate reaction rates by several orders of magnitude1. However, little is known about how enzymes developed their chemical mechanisms to obtain high reaction rates and specificity. The mechanisms of numerous enzymatic reactions have been studied using protein biochemistry and structural biological techniques such as X-ray and NMR2,3. These studies have been useful in identifying many structural features and conformational changes necessary for the catalytic activity of enzymes. Nonetheless, the dynamic subÅngström scale rearrangements of the participating atoms necessary for catalysis cannot be detected by these techniques. Recently developed single molecule techniques have shown promise in uncovering the dynamics of enzymatic activity at a length scale that was previously impossible to observe. For example, both optical tweezers and fluorescence techniques have been used extensively to detect the motions of molecular motors, a large class of ATP-consuming force generating enzymes4,5.
In this work, we use single molecule force-clamp spectroscopy techniques to investigate the chemical mechanisms of catalysis of disulfide reducing enzymes called thioredoxins (Trx). These oxido-reductases are present in all known organisms from bacteria to human. Trxs posses a highly conserved active site (CXXC) that catalyzes the reduction of target disulfide bonds being involved in a multitude of cellular processes6,7. Several methods based on spectrophotometry have been widely used to determine the activity of Trx such as monitoring of oxidation of NADPH in the presence of Trx reductase or ribonucleotide reductase8,9; the observation of the turbidity of solutions containing insulin, which readily aggregates after reduction of its disulfide bonds9 or the use of Ellman's reagent (DTNB), where upon reduction by thiol groups generates products that can be easily detected with a spectrophotometer6. The change in tryptophan fluorescence has also been used to measure rates of Trx oxidation and reduction10. While highly effective in monitoring the overall activity of Trx enzymes, these methods do not probe the chemical mechanisms underlying the catalytic activity of these enzymes.
Recent single molecule force-spectroscopy experiments have demonstrated that the application of a mechanical force to a substrate disulfide bond can regulate the catalytic activity of thioredoxins11, revealing different chemical mechanisms of reduction that could be readily distinguished by their sensitivity to an applied force. A simple form of catalysis in E. coli Trx corresponded to a straightforward SN2 type chemical reaction characterized by an exponential increase of the rate of reduction with the pulling force12,13. This SN2 type reaction was absent in human Trx. Instead, a force-independent disulfide reduction mechanism was observed. An additional chemical mechanism of reduction was characterized by its rapid inhibition by a force applied to the substrate disulfide bond. This chemical mechanism is unique to disulfide bond reduction catalyzed by Trx enzymes and was explained by a Michaelis-Menten type reaction where the binding of the enzyme to the stretched polyprotein and the subsequent structural organization of the participating sulfur atoms precede the chemical reaction.
Thus, the single molecule reduction assay that we have previously developed is able to distinguish the chemistry of a simple SN2 reaction from more elaborate pathways for the reduction of disulfide bonds which are unique to Trx enzymes. It is of interest then to consider the application of these sensitive single molecule techniques to study the evolution of chemical mechanisms in this family of enzymes. The simplest evolutionary hypothesis is that the ancient forms of Trx had capabilities that were only comparable to those of simple reducing agents. Evolutionary pressures then drove the enzymes towards developing unique and more efficient mechanisms of reduction. As a first step towards understanding the evolution of Trx chemistry, we have applied the single molecule assay to examine the chemical mechanisms of reduction of a sample of eight extant Trx enzymes covering four kingdoms of life. From this analysis we report several new advances. First, we demonstrate that the differences in the chemical mechanisms of reduction between E. coli Trx and human TRX (also known as TXN) that we had reported previously, can now be generalized to enzymes of bacterial vs. eukaryotic-origin. We also demonstrate a third mechanism for disulfide bond reduction that corresponds to a single-electron transfer reaction and that is present in all Thioredoxin enzymes. Finally, through a computational analysis of Trx structures, we show that the changes in the catalytic chemistry correlate well with a deepening of the binding grove observed in Trxs of eukaryotic origin. These latter results identify the evolution of the binding groove as an important structural adaptation controlling the disulfide reduction chemistry in the Trx family of enzymes.
RESULTS
Selection of thioredoxins from different species
In order to investigate the variety of catalytic mechanisms developed by Trx we have selected of a set of Trxs belonging to a representative group of species from different kingdoms, i.e., animalia, bacteria, protista and plant (covering two domains of life: bacteria and eukaryotes). Trx is widely distributed in all living organisms from bacteria to mammals. In addition, the existence of a second paralogous Trx gene (TXN2) seems to be common in animals, protists and Gram-negative bacteria 14-18. In protists and animals, Trx1 is located in the cytoplasm whereas Trx2 is present in mitochondria18,19. Interestingly, mitochondrial Trx2 from mammals has been shown to have higher similarity with E. coli Trx1 than with cytosolic Trx1 from mammals16. In the case of plants, a rich variety of Trx genes can be found encoding more than 20 different types of Trxs20 that are classified into six isoforms: Trxf, h, m, x, y and o. The f, m, x and y forms are plastidic Trxs; h forms are mainly cytosolic and o forms are found in mitochondria21. In this study, we have included both human cytosolic and mitochondrial Trxs from animals; poplar Trxh1 (featuring a CPPC active site instead of the canonical CGPC), poplar Trxh3 and pea chloroplastic Trxm from plants; E. coli Trx1 and Trx2 from bacteria, and finally we have chosen Plasmodium falciparum (malaria parasite) Trx1 from protists.
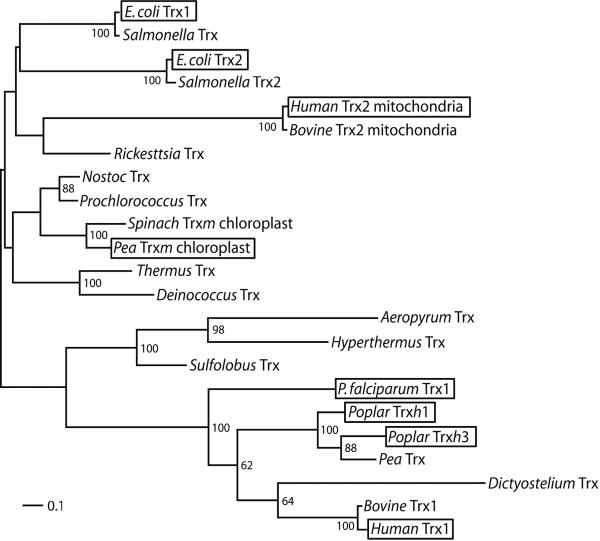
A sequence alignment of the Trxs of interest shows that the residues around the active site are highly conserved (Supplementary Fig. 1). The construction of a phylogenetic tree (Fig.1), incorporating additional Trxs from the three domains of life, classifies E. coli Trx1 and Trx2, human Trx2 and pea Trxm as “bacterial type” Trxs (top branches in Fig.1) and human Trx1, poplar Trxh1 and h3 and P. falciparum Trx1 as “eukaryotic-like” Trxs (botton branches in Fig.1). The construction of a larger tree incorporating over 200 Trx sequences (not shown) corroborates that the sequences used are widely distributed and that they are representative for the entire Trx family.
Figure 1. Phylogeny of Trx homologs from representative species of the three domains of life.
Branch lengths were estimated using maximum likelihood with rate variation modeled according to a gamma distribution. Scale bar represents amino acid replacements per site per unit evolutionary time. Posterior probabilities are shown at nodes of the phylogeny when greater than 50%. The lack of strong node supports deep in the phylogeny results from the ambiguous placement of mitochondrial sequences, possibly due to long-branch attraction effects with non-bacterial sequences. In contrast, there is strong support for the grouping of chloroplast and cyanobacteria (not shown). Boxes highlight the proteins experimentally studied in this work.
Force-dependent chemical kinetics of disulfide reduction
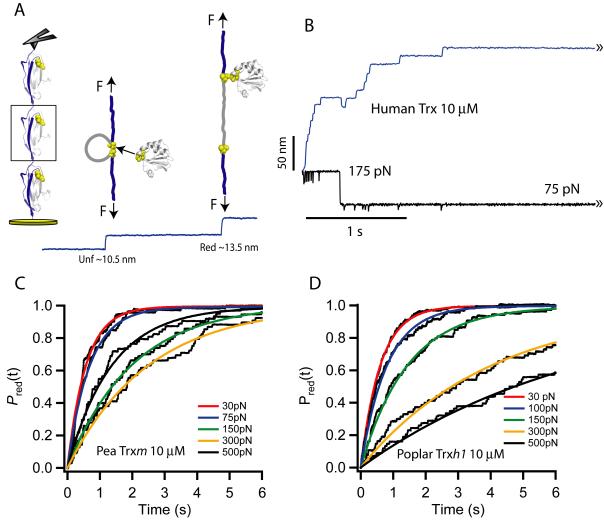
Similar to our previous work, we have used an atomic force microscope in its force-clamp mode to study the chemistry of disulfide reduction by Trx11,22. Briefly, we have used as a substrate a polyprotein composed of eight domains of the 27th module of human cardiac titin in which each module contains an engineered disulfide bond between the 32nd and 75th positions (I27G32C-A75C)8 11. A first pulse of force (175 pN, 0.3 s) is applied to the polyprotein which allows the rapid unfolding of the I27G32C-A75C modules up to the disulfide bond. The individual unfolding can be registered as steps of ~10.5 nm per module. After the first pulse, the disulfide bonds become exposed to the solvent where the Trx molecules are present in the reduced form due to the presence of Trx reductase and NADPH (Trx system)6. A second pulse of force is applied to monitor single disulfide reductions by Trx enzymes that are recorded as a second series of steps of ~13.5 nm per domain (Fig.2A and B). We have accumulated several traces per force (15-50) which have been averaged and fitted with a single exponential with a time constant τ (Fig.2C and D). We thus obtain the reduction rate at a given force (r = 1/τ).
Figure 2. Single molecule force-clamp detection of disulfide bond reduction events catalyzed by thioredoxin enzymes.
(A) Graphic representation of the force-clamp experiment. A first force pulse rapidly unfolds the I27 modules of a polyprotein, exposing the buried disulfide bonds to the solvent. A second force pulse monitors single-disulfide reduction events which are uniquely identified by the extension of the residues trapped behind the disulfide bond. (B) Trace showing the unfolding and consequent disulfide reductions of a (I27G32C-A75C)8 polyprotein. In the example shown the unfolding pulse was set at 175 pN for 0.3 s and the reduction pulse was set to 75 pN for several seconds. (C) Summed and averaged traces of disulfide bond reductions at different forces (second pulse) for pea Trxm (10 μM) and (D) for poplar Trxh1 (10 μM). The smooth curves are single-exponential fits from which we measure the rate of reduction as r = 1/τ, where τ is the time constant measured by the fits at each given force.
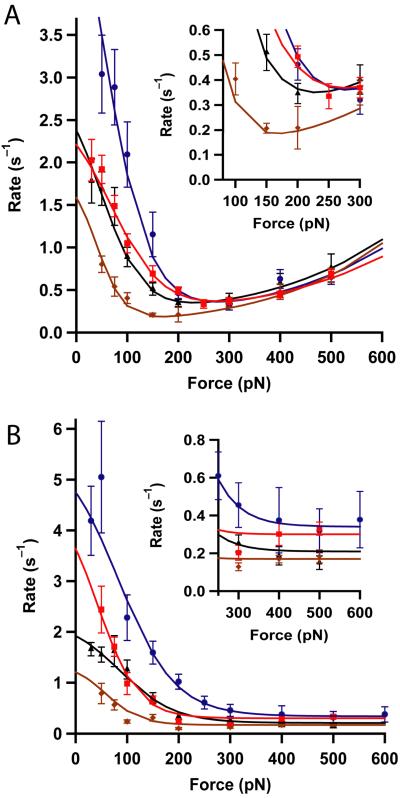
We have applied our single molecule assay to obtain the force dependency of the rate of reduction by the selected Trxs (Fig.3). From these data we can readily distinguish three different types of force-dependencies. First, all tested Trx enzymes showed a negative force dependency in the range 30-200 pN. Second, all Trx enzymes from bacterial origin show that after reaching a minimum rate at around 200 pN, the rate of reduction increases exponentially at higher forces. Third, at forces higher than 200 pN, enzymes from eukaryotic origin show a rate of reduction that becomes force independent. Therefore, our previous observations in E. coli and human Trxs can now be generalized to bacterial and eukaryotic-origin Trxs.
Figure 3. Force-dependency of the rate of disulfide reduction by Trx enzymes from different species.
(A) Bacterial-origin Trxs: human mitochondrial Trx2 (blue), E. coli Trx1 (red), pea chloroplastic Trxm (black), E. coli Trx2 (brown). While the Michaelis-Menten (low force) mechanism differs in magnitude among the Trxs, the simple SN2 like reaction observed at higher forces is very similar in all of them. The inset shows a magnified view of the traces in the region where they reach a minimum. (B) Eukaryotic-origin Trxs: human Trx1 (blue, from ref 11), Plasmodium Trx (red), poplar Trxh1 (black), and poplar Trxh3 (brown). The most noticeable feature is the absence of the SN2 like reaction at high forces in all eukaryotic Trxs. The inset shows an expansion of the minimum rate of reduction attained at high forces. In all experiments the concentration of Trx was 10 μM. The smooth lines are fits of the kinetic model described in the supplementary information. The kinetics parameters obtained are summarized in Table 1. The error bars are given by the standard error of the mean obtained with the bootstrap method.
We have previously proposed that the reduction mechanism observed when the substrate is stretched at low forces (30-200 pN) is similar to a Michaelis-Menten (MM-SN2) reaction in which the formation of an enzyme-substrate complex is determinant11. Upon binding, the substrate disulfide bond needs to rotate in order to achieve the correct geometry necessary for an SN2 reaction to occur, i.e. the three involved sulfur atoms forming a ~180° angle11,22,23. This rotation causes the shortening of the substrate polypeptide along the stretching force axis determined by the negative value of Δx12 in our kinetic model (Table 1, Fig.4 and Supplementary Fig. 2). This mechanism is rapidly inhibited as the force increases generating the negative dependence of the reduction rate with the pulling force in all Trx enzymes (Fig.3). Here we demonstrate that, while the absolute rate of reduction varies from enzyme to enzyme, the general characteristics of this mechanism of reduction are apparent in all of them.
Table 1. Kinetic parameters for Trxs from different species.
The parameters were obtained using the kinetic model described in supplementary information. They are the result of numeric optimization of the global fit using the downhill simplex method. The errors correspond to the standard deviation.
| α0 (μM-1·s-1) | β0 (s-1) | γ0 (μM-1·s-1) | k10 (s-1) | Δx12 (Å) | Δx02 (Å) | λ0(s-1) | |
|---|---|---|---|---|---|---|---|
| E. coli Trx1 | 0.25±0.02 | 24±2 | 0.012±0.002 | 4.7±0.5 | -0.75±0.05 | 0.16±0.01 | 0.08±0.02 |
| E. coli Trx2 | 0.18±0.04 | 23±3 | 0.009±0.002 | 4.5±0.5 | -1.41±0.03 | 0.19±0.01 | 0.07±0.03 |
| Human Trx2 | 0.65±0.15 | 25±2 | 0.019±0.004 | 3.8±0.6 | -0.84±0.05 | 0.19±0.02 | 0.06±0.04 |
| Pea Trxm | 0.28±0.03 | 22±2 | 0.012±0.002 | 5.0±0.6 | -0.93±0.08 | 0.17±0.02 | 0.09±0.04 |
| P. falciparum Trx1 | 0.43±0.05 | 25±2 | - | 4.8±0.5 | -1.01±0.05 | - | 0.34±0.02 |
| Human Trx1 | 0.52±0.05 | 33±2 | - | 3.1±0.9 | -0.71±0.05 | - | 0.35±0.02 |
| Poplar Trxh3 | 0.12±0.03 | 30±3 | - | 4.4±0.4 | -1.16±0.08 | - | 0.17±0.02 |
| Poplar Trxh1 | 0.22±0.02 | 28±3 | - | 5.5±0.6 | -0.68±0.07 | - | 0.17±0.03 |
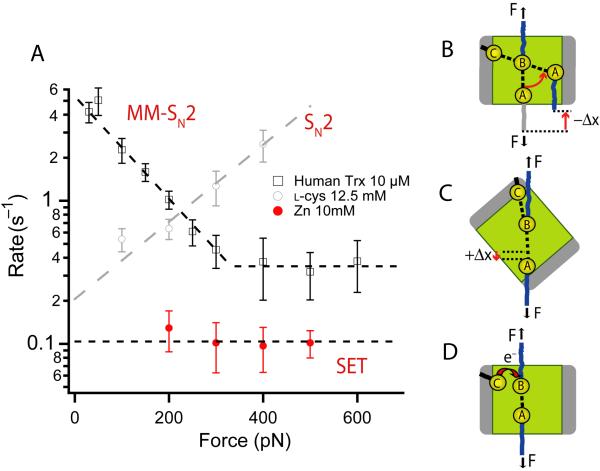
Figure 4. The three chemical mechanisms of disulfide reduction detected by force-clamp spectroscopy.
A) Force dependency of the rate of reduction of disulfide bonds by different reducing agents. Human Trx (black squares, from ref 11) shows the characteristic enzymatic mechanism of reduction, marked by a negative force dependency that reaches a force independent minimum. L-cysteine (gray circles, from ref 12) shows the characteristic SN2 like mechanism marked by an exponential increase in the rate of reduction with the applied force. Finally, metallic Zn (red circles) demonstrates a reduction mechanism that appears force independent. Bacterial thioredoxin enzymes make use of all three mechanisms. B) Schematic representation of the Michaelis-Menten reduction mechanism present in all Trx enzymes. The substrate disulfide bond is shown in the binding groove of the enzyme. Rotation of the substrate disulfide bond against the pulling force is required for the 180° alignment with the catalytic cysteine, and for reduction to occur. C) We speculate that the simple SN2-like mechanism observed at high forces results from aligning the substrate disulfide bond with the catalytic cysteine, without entering the binding groove. This conformation is favored by a shallow binding groove allowing for the simple SN2-type lengthening of the substrate disulfide bond at the transition state, which is the origin of the exponential dependency of the rate of reduction. D) Representation of the single-electron transfer (SET) mechanism ubiquitous to all Trxs. This mechanism can occur irrespective of the orientation of the disulfide bond and is more visible in eukaryotic Trxs at high forces.
According to the parameters obtained from the fitting to a simple MM-SN2 type kinetic model (Table 1), we found that an extrapolation to zero force predicts rate constants ranging from 1.2 × 105 M-1·s-1 for poplar Trxh3 to 6.5 × 105 M-1·s-1 for human Trx2. These values are remarkably similar to those previously obtained using insulin disulfides as substrates and E. coli Trx8. The value of Δx12, remains below 1 Å except for E. coli Trx2 and poplar Trxh3 with values over 1 Å (Table 1). These high values of Δx12 represent a higher rotation angle of the substrate disulfide bond for the SN2 reaction22. This mechanism is unique to Trx enzymes and it seems to be the result of evolutionary pressure toward developing an efficient mechanism of disulfide reduction not possible with simple chemical reagents12.
When forces over 200 pN are applied to the substrate, the MM-SN2 mechanism is blocked and a second force-dependent mechanism of reduction becomes dominant. This is true in all types of Trx enzymes. In enzymes of bacterial origin this high force mechanism (Fig. 3A) appears to be analogous to that of simple chemical compounds such as cysteine, glutathione or dithiotreitol (DTT) which reduce disulfide bonds through a force-dependent SN2 thiol/disulfide exchange reaction with bond elongation at the transition state12,13. We have incorporated this reaction in our kinetic model (k02), obtaining a value for the elongation of the disulfide bond at the transition state of ~0.18 Å (Δx02 in Table 1), similar to the values obtained when using cysteine as a nucleophile (~0.2 Å)12. The absence of the SN2 like mechanism seems to be common to all eukaryotic-origin Trx. Instead, the rate of the reaction becomes force-independent at higher forces (Fig. 3B). This force-independent mechanism gives a constant rate of reduction that ranges from 0.2 to 0.4s-1 (inset in Fig.3B). We speculate that in enzymes of bacterial origin, the minimum value of the reduction rate is also limited by this force-independent “floor” in the rate of reduction which varies in the range 0.2 to 0.4s-1 (inset in Fig.3A). We have incorporated this mechanism into our kinetic model in the form of a constant parameter λ (see kinetic model in supplementary information and Supplementary Fig. 2), which contributes equally to the reduction rate throughout the entire range of forces (Table 1).
An interesting possibility that may explain this force-independent chemical mechanism is a single-electron transfer reaction (SET) via tunneling, a process that has been observed in enzymes containing metallic complexes3,24. In addition, it has been suggested that SET are highly favored when steric hindrance occurs25. To test whether an electron transfer mode of reduction would be force independent, we have investigated the kinetics of disulfide reduction under force by a metal. Some metals participate in oxidation/reduction processes in proteins via electron-transfer reactions governed by the reduction potential26,27. Concretely, we have studied the reduction of disulfide bonds by Zn nanoparticles (diameter <50 nm)28. In sharp contrast to all other reducing agents that we have studied, the rate of reduction of disulfide bonds by Zn is force-independent (Fig. 4A). Due to the experimental difficulty of working at low forces with Zn nanoparticles, we only include experiments at forces over 200 pN. Our results support the idea that the force-independent mechanism is a barrier free electron tunneling reaction that does not require the precise orientation for the SN2 reaction.
Another piece of evidence in support of the SET mechanism can be obtained from the analysis of the concentration dependencies of the MM-SN2 and SET reduction mechanisms. We have obtained data for reduction kinetics by human Trx at different forces and concentrations (50-600 pN and 2-15 μM of Trx) showing that the low force MM-SN2 mechanism (50-200 pN) is clearly dependent on the concentration of the enzyme, whereas the high force SET mechanism (>300 pN) is essentially concentration independent (Supplementary Fig. 3). As expected for first-order MM-SN2 mechanism where substrate binding to the groove is determinant, the rate of reduction shows linear concentration dependence when working below saturating concentrations of Trx enzyme (<15 μM)11. On the other hand, considering that the Trx/NADPH system is in equilibrium due to the presence of TrxR29,30, the redox potential of Trx will remain constant (from the Nernst equation). Therefore, the potential difference between Trx and substrate, and thus the rate of electron transfer31, will be also constant in this Trx concentration range. Hence, the SET mechanism should be essentially independent of the enzyme concentration32,33; this is verified in Supplementary Fig. 3.
Our results suggest that there are three distinct mechanisms of reduction that operate simultaneously in a Trx enzyme. These mechanisms are identified by their force-dependency as illustrated in Figure 4. The most complex mechanism is characterized by a negative force dependency and is unique to enzymatic catalysis by Trx (Fig. 4A, B). This enzymatic mechanism of reduction is characterized by a MM-SN2 reaction between the substrate polypeptide and the binding groove of the enzyme, followed by a rotation of the substrate disulfide bond to gain position for the SN2 reduction mechanism 11,22 (Fig. 4A, B). A much simpler mechanism is that of a regular SN2 reaction, characterized by a rate of reduction that increases exponentially with the applied force. This mechanism is well represented by nucleophiles such as L-cysteine (Fig. 4A), glutathione and DTT11,12. In this mechanism the substrate disulfide bond and the catalytic cysteine of the enzyme orient themselves with the pulling force, without needing a rotation of the substrate disulfide bond (Fig. 4C). We suspected that this mechanism would be possible only if the Trx enzyme had a shallow binding grove that allowed many other orientations of the substrate-enzyme complex. Finally, the third mechanism is the force-independent barrier-free electron tunneling transfer mechanism illustrated by the action of metallic zinc (Fig. 4D). It is inevitable that if the disulfide bond gets close enough to the thiolate anion of the catalytic cysteine, the electron tunneling will occur, albeit at a very low rate.
Thus, comparing the data of Figures 3 and 4, it is clear that the main difference between enzymes of bacterial and eukaryotic origin is the elimination of the high force simple SN2-like mechanism of reduction. We speculate that this drastic change in the catalytic chemistry would be caused by changes in the structure of the enzyme as it evolved. The most salient feature in the structure of Trx enzymes is the binding groove into which the target polypeptide first binds, to be subsequently reduced by the exposed thiol of the catalytic cysteine (Fig. 5A).
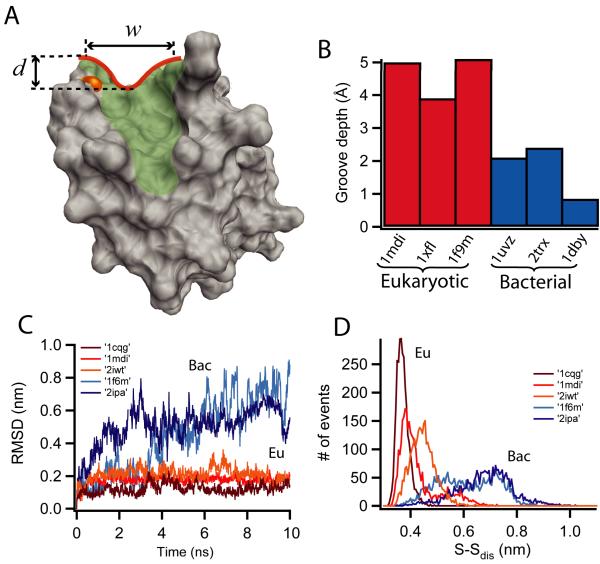
Figure 5. Structural analysis and molecular dynamics simulations of the binding groove in thioredoxin enzymes.
(A) Geometric dissection of the hydrophobic binding groove, shaded in green and outlined in red for human Trx1 (pdb: 3trx). The depth and width of the binding groove in the region surrounding the catalytic cysteine are indicated by arrows and the active cysteine is colored in orange. (B) Groove depth for three eukaryotic-origin Trxs (red): human Trx1 (pdb:1mdi); A. thaliana Trxh1 (pdb:1xfl) and spinach Trxf (pdb:1f9m), and three bacterial-origin Trxs (blue): human Trx2 (pdb:1uvz); E. coli Trx1 (pdb:2trx) and C. reinhardtii Trxm (pdb:1dby). It is clear that the eukaryotic binding grooves are deeper than their bacterial counterparts. (C) Molecular dynamics simulation of the substrate mobility within the binding groove for different eukaryotic (red) and bacterial (blue) Trxs. Three eukaryotic complexes were used: human Trx with the substrate REF-1 (pdb:1cqg); human Trx with NF-κB (pdb:1mdi) and barley Trxh2 with protein BASI (pdb:2iwt). The two bacterial complexes were: E. coli Trx1 with Trx reductase, (pdb:1f6m) and B. subtilis Trx bound to ArsC (pdb:2ipa). The large RMSD of the bacterial Trxs (blue) indicates a high substrate mobility which may facilitate collisions orientated so as favoring SN2 reactions. Eukaryotic Trxs (red) are highly restricted which may explain the different chemical behavior at high forces as compared to bacterial Trxs (Fig.3). (D) Diatomic distance distribution of the S-S bond in the Trx-substrate mixed intermediate, using the same structures as in (C). Again we infer higher mobility of the substrate in bacterial-origin Trxs as indicated by the broader distance distributions of the bacterial complexes (blue) as compared to those of eukaryotic Trxs (red).
Structural analysis and molecular dynamics simulations
In order to study the role of the structure in the chemical behavior of Trxs, we have analyzed the structure of the binding groove of a set of bacterial and eukaryotic-origin Trxs. We have studied three eukaryotic-origin enzymes: human Trx1, A. thaliana Trxh1 and spinach Trxf, and three bacterial-origin enzymes: human mitochondrial Trx2, E. coli Trx1 and C. reinhardtii Trxm. From the X-ray or NMR structures we have defined structural axes which allow us to calculate the depth and width of the binding groove in the region surrounding the catalytic cysteine (see Fig. 5A and supplementary information for details). We found that eukaryotic Trx enzymes have binding grooves that are several Å deeper than those of bacterial origin (Fig.5B). By contrast, the width of the binding groove remained the same (Supplementary Fig. 4). We explored the consequences of a deepening binding groove using molecular dynamics (MD) simulations to probe the mobility of a bound polypeptide (see supplementary information for procedures). For the MD simulations we have considered a set of enzyme structures obtained with mixed disulfide intermediates between the catalytic cysteine and a cysteine in the bound substrate. Such structures capture the general disposition of the substrate in the catalytic site of the Trx enzyme. We have used three eukaryotic complexes: human Trx1 with the substrate REF-1, human Trx1 with NF-κB and barley Trxh2 with protein BASI, and two bacterial complexes: E. coli Trx1 with Trx reductase and B. subtilis Trx with ArsC complex.In order to compare these structures, 13 residues of the substrates are taken into account, with the binding cysteine always set to be the 7th residue. For the MD simulations we have removed the substrate-enzyme disulfide bond to allow substrate mobility. Fig. 5C shows that the shallow binding groove of bacterial Trxs allows the substrate to be mobile. By contrast, the deeper groove found in Trx enzymes of eukaryotic origin tends to freeze the substrate into a much smaller range of conformations. Similarly, the measured distribution of the distances between the reacting sulfur atoms is smaller and more narrowly distributed in the deeper binding groove of Trx enzymes of eukaryotic origin than in those with the shallower grooves found in enzymes of bacterial origin (Fig. 5D).
As an additional test, we have carried out MD simulations in which the substrate has been removed from the pdb file for two thioredoxin complexes; one from eukaryotic origin, barley Trxh2 with protein BASI (2iwt), and the other from bacterial origin, B. subtilis Trx with ArsC complex (2ipa). As shown in Supplementary Fig. 5, there is no significant difference in the dynamics of the groove between the bacterial and the eukaryotic thioredoxins. In fact, the averaged value of the RMSD difference is 0.035±0.028 for 2iwt and 0.023±0.031 for 2ipa (the error is given by the S.D.). These results corroborate that the large differences in the mobility of the substrate that we report (Fig.5C and D) are due to the different binding constraints of the groove.
Finally, we have compared the B-factor distribution of the substrate from the pdb files. The B-factors of protein crystal structures reflect the fluctuation of atoms around their average positions and provide information about protein dynamics. In particular we have compared the B-factor distribution of eukaryotic barley Trxh2 bound to protein BASI (2iwt) with the one from E. coli Trx bound to Trx reductase (1f6m) both from X-ray experiments (Supplementary Fig. 6). Consistent with our simulation results (Fig. 5C), the substrate in 1f6m (bacterial-origin thioredoxin) has larger B-factor than that of eukaryotic thioredoxin 2iwt.
These structural observations suggest that a major feature in the evolution of thioredoxin enzymes has been an increase in the depth of the binding groove, increasing the efficiency of the MM-SN2 mechanism and eliminating the simple SN2 mechanism of catalysis.
DISCUSSION
Over the past 4 billion years the chemistry of living organisms has changed continuously in response to changes in atmospheric conditions and biological phenomena. For example, the large increase in the level of atmospheric oxygen that occurred about ~2.5 billion years ago is thought to have triggered a chemical expansion34,35 that had a large impact on the chemistry of enzymatic reactions, especially those involving redox transformations35,36. However, understanding how enzymes have adapted their chemical mechanisms to evolutionary pressures remains a challenge in molecular biology.
Here we show that single molecule force-clamp spectroscopy can be a valuable tool to examine the evolution of Trx catalysis by studying the chemistry of eight Trx enzymes from four different kingdoms. We show that three different chemical mechanisms for disulfide reduction can be distinguished in Trx enzymes by their sensitivity to a mechanical force applied to their substrate. Common to all Trx enzymes is a highly efficient Michaelis-Menten type mechanism of disulfide bond reduction, characterized by a negative force dependency (Fig. 4A and 4B). Common to all enzymes is also a low rate force-independent mechanism of reduction that, owing to its similarity to metallic zinc, may be due to a barrier free electron tunneling mechanism (Fig. 4A and 4D). Finally, enzymes of bacterial-origin show an additional mechanism of reduction, comparable to that of a simple SN2 reaction, showing a force dependency similar to that of glutathione or cysteine12,13 (Fig. 4A and 4C). This simple SN2 mechanism appears to have been eliminated from Trx enzymes that have a eukaryotic origin, suggesting that the mechanism of disulfide bond reduction by Trx enzymes was altered at an early stage of eukaryotic evolution.
We identify the physical characteristics of the binding groove as important factors in the evolution of Trx catalysis. The appearance of the hydrophobic binding groove allowed Trxs to bind the substrate in a specific fashion, generating a stabilizing interaction that made the enzyme capable of regulating the geometry and orientation of the substrate disulfide bond in the catalytic site of the enzyme. This binding mechanism results in the Michaelis-Menten type kinetics of reduction observed in all Trx. Noteworthy is the fact that as the binding groove deepens in enzymes of eukaryotic origin, the SN2-like mechanism of reduction disappears. These observations are in agreement with the view that the SN2-like mechanism of reduction observed in bacterial Trx enzymes results from less specific enzyme-substrate interactions (Fig. 4C). The emergence of eukaryotes gave rise to vastly more complex biological systems bringing a palette of new functions and targets37. It is tempting to speculate that the deepening of the binding groove in eukaryotic Trx (Fig.5) may have been an important structural adaptation that improved the specificity of substrate-enzyme interactions.
However, evolutionary optimization of Trx activity is clearly a much more complex multi-parameter function involving other structural features and cofactors. Most importantly, thioredoxins work in concert with another enzyme named thioredoxin reductase (TrxR), which converts oxidized Trx to its active dithiol form. There are major differences in the structure and mechanism of TrxR across the evolutionary tree, and it is reasonable to consider that the evolution of the chemical mechanisms in Trx has been tightly associated with the evolution of TrxR. In our experiments we have used generic bacterial and eukaryotic TrxR to keep the thioredoxin enzymes in the reduced state. We anticipate that our assay can be expanded by contrasting the effect of different TrxR enzymes in the observed chemistry of Trx.
From a biological point of view, an interesting hypothesis is that the simple SN2 like mechanism present in bacterial Trxs might be related to their ability to live in extreme environments where elevated mechanical forces might result as a consequence of high pressures or extreme salinity that cause cells to swell or shrink38-40. As we have shown here, under such conditions the enzymatic Michaelis-Menten type mechanism of reduction would become inoperative. In support of this view, Trx has been shown to promote high-pressure resistance in E. coli 41.
Our work generally demonstrates the usefulness of combining single molecule force spectroscopy together with molecular dynamics simulations in probing enzymatic chemistry. We observe significant differences in the chemical mechanisms of extant Trx enzymes. Of great interest would be to track the evolution of these chemical mechanisms using resurrected ancient Trx enzymes. Owing to an extensive sequence database and the development of sophisticated maximum likelihood algorithms for the reconstruction of ancient DNA sequences42, reconstructing the evolution of chemical mechanisms in this class of important enzymes now seems entirely plausible. We anticipate that the enzymatic studies carried out on Trx at the single-molecule level, can serve as a starting point to investigate the chemistry of other enzymes such as C-S lyases43 or proteases44 where the catalyzed rupture of covalent bonds is the fundamental process.
METHODS
Protein expression and purification
Preparation of (I27G32S-A75C)8 polyprotein has been extensively described11,22. The expression and purification of the different Trxs used have also been described elsewhere: P. falciparum45, drosophila Trx146, poplar Trxh47 and Trxh348, pea Trxm49, E. coli Trx150, E. coli Trx217 and human Trx214.
Sequence analysis
We carried out sequence alignment using ClustalW and modified by hand. Tree topology and branch lengths of the tree were estimated using Mr. Bayes (v. 3.5) with rate variation modeled according to a gamma distribution. The following GI numbers were accessed from GenBank: Bacteria E. coli Trx1 (67005950), Salmonella Trx1 (16767191), E. coli Trx2 (16130507), Salmonella Trx2 (16765969), human Trx2 mitochondria (21361403), bovine Trx2 mitochondria (108935910), Rickettsia Trx (15603883), Nostoc Trx (17227548), Proclorococcus Trx (126696505), spinach Trxm chloroplast (2507458), pea Trxm chloroplast (1351239), Thermus Trx (46199687), Deinococcus Trx (15805968), Archaea Aeropyrum Trx (118431868), Hyperthermus Trx (124027987), Sulfolobus Trx (15897303), Eukaryote Plasmodium falciparum Trx (75024181), poplar Trx h1 (19851972), poplar Trx h3 (2398305), pea Trx (27466894), Dictyostelium Trx (165988451), bovine Trx (27806783), human Trx1 (135773).
Single molecule force-clamp experiments
The details of our custom-made atomic force microscope have been described previously51. We used silicon nitride cantilever (Veeco) with a typical spring constant of 20 pN nm-1 which was calibrated using the equipartition theorem. The force-clamp mode provides an extension resolution of ~0.5 nm and a piezoelectric actuator feedback of ~5ms. The buffer used in all the experiments was: 10 mM HEPES, 150 mM NaCl, 1mM EDTA, 2mM NADPH at pH 7.2. Before the beginning of the experiment, Trx reductase, bacterial or eukaryotic depending on the case, was added to a final concentration of 50 nM. The different Trxs were added to the desired concentration. The reaction mixture and the substrate were added and allowed to absorb onto a freshly evaporated gold-coverslip before the experiments. The force-clamp experiment consists of a double-pulse force protocol. The first pulse was set at 175 pN during 0.3-0.4 s. The second pulse can be set at different forces and was held long enough to capture all the possible reduction events. The experiments using metallic Zn were carried out in citrate buffer 100 mM at pH 6. After adding Zn nanoparticles (Sigma) to a concentration of 10 mM, the solution was sonicated to allow resuspension. The pH of the buffer was verified during the time of the experiment and no appreciable changes were observed. In addition, to verify the behavior of the substrate in citrate buffer, several control experiments in the absence of Zn nanoparticles were carried out and no reduction events were detected. Data collection and analysis were carried out using custom written software in IGOR Pro 6.03 (Wavemetrics). The collected traces (15-50 per force) containing the reduction events were summated and averaged. The resulting averaged traces were fitted with single exponential from which the rate constant can be obtained. The analysis of the force-dependent reduction kinetics was carried out using the kinetic model in the supplementary information.
Supplementary Material
Acknowledgments
We thank Dr. Sergi Garcia-Manyes and Dr. Joseph Morrone assistance in writing the manuscript and all the Fernandez laboratory members for helpful discussions. This work was supported by National Institutes of Health grants HL66030 and HL61228 to J. M.F, grant GMO55090 to J.B., grant GM43340 to B.J.B., grant I.C.E. (Columbia University) to B.J.B and J.M.F., grant BIO2006-07332 from the Spanish Ministry of Education and Science and FEDER Funds to J.M.S.-R.
References
- 1.Kraut DA, Carroll KS, Herschlag D. Challenges in enzyme mechanism and energetics. Annu Rev Biochem. 2003;72:517–71. doi: 10.1146/annurev.biochem.72.121801.161617. [DOI] [PubMed] [Google Scholar]
- 2.Henzler-Wildman KA, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–44. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 3.Dai S, Friemann R, Glauser DA, Bourquin F, Manieri W, Schurmann P, Eklund H. Structural snapshots along the reaction pathway of ferredoxinthioredoxin reductase. Nature. 2007;448:92–6. doi: 10.1038/nature05937. [DOI] [PubMed] [Google Scholar]
- 4.Mori T, Vale RD, Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–4. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 5.Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–4. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–71. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 7.Lillig CH, Holmgren A. Thioredoxin and related molecules--from biology to health and disease. Antioxid Redox Signal. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- 8.Holmgren A. Reduction of disulfides by thioredoxin. Exceptional reactivity of insulin and suggested functions of thioredoxin in mechanism of hormone action. J Biol Chem. 1979;254:9113–9. [PubMed] [Google Scholar]
- 9.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254:9627–32. [PubMed] [Google Scholar]
- 10.Holmgren A. Tryptophan fluorescence study of conformational transitions of the oxidized and reduced form of thioredoxin. J Biol Chem. 1972;247:1992–8. [PubMed] [Google Scholar]
- 11.Wiita AP, Perez-Jimenez R, Walther KA, Grater F, Berne BJ, Holmgren A, Sanchez-Ruiz JM, Fernandez JM. Probing the chemistry of thioredoxin catalysis with force. Nature. 2007;450:124–7. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ainavarapu SRK, Wiita AP, Dougan L, Uggerud E, Fernandez JM. Single-molecule force spectroscopy measurements of bond elongation during a bimolecular reaction. Journal of the American Chemical Society. 2008;130:6479–6487. doi: 10.1021/ja800180u. [DOI] [PubMed] [Google Scholar]
- 13.Wiita AP, Ainavarapu SR, Huang HH, Fernandez JM. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proc Natl Acad Sci U S A. 2006;103:7222–7. doi: 10.1073/pnas.0511035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damdimopoulos AE, Miranda-Vizuete A, Pelto-Huikko M, Gustafsson JA, Spyrou G. Human mitochondrial thioredoxin. Involvement in mitochondrial membrane potential and cell death. J Biol Chem. 2002;277:33249–57. doi: 10.1074/jbc.M203036200. [DOI] [PubMed] [Google Scholar]
- 15.Miranda-Vizuete A, Damdimopoulos AE, Gustafsson J, Spyrou G. Cloning, expression, and characterization of a novel Escherichia coli thioredoxin. J Biol Chem. 1997;272:30841–7. doi: 10.1074/jbc.272.49.30841. [DOI] [PubMed] [Google Scholar]
- 16.Spyrou G, Enmark E, Miranda-Vizuete A, Gustafsson J. Cloning and expression of a novel mammalian thioredoxin. J Biol Chem. 1997;272:2936–41. doi: 10.1074/jbc.272.5.2936. [DOI] [PubMed] [Google Scholar]
- 17.Ye J, Cho SH, Fuselier J, Li W, Beckwith J, Rapoport TA. Crystal structure of an unusual thioredoxin protein with a zinc finger domain. J Biol Chem. 2007;282:34945–51. doi: 10.1074/jbc.M704044200. [DOI] [PubMed] [Google Scholar]
- 18.Boucher IW, McMillan PJ, Gabrielsen M, Akerman SE, Brannigan JA, Schnick C, Brzozowski AM, Wilkinson AJ, Muller S. Structural and biochemical characterization of a mitochondrial peroxiredoxin from Plasmodium falciparum. Mol Microbiol. 2006;61:948–59. doi: 10.1111/j.1365-2958.2006.05303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powis G, Montfort WR. Properties and biological activities of thioredoxins. Annu Rev Biophys Biomol Struct. 2001;30:421–55. doi: 10.1146/annurev.biophys.30.1.421. [DOI] [PubMed] [Google Scholar]
- 20.Gelhaye E, Rouhier N, Navrot N, Jacquot JP. The plant thioredoxin system. Cell Mol Life Sci. 2005;62:24–35. doi: 10.1007/s00018-004-4296-4. [DOI] [PubMed] [Google Scholar]
- 21.Meyer Y, Riondet C, Constans L, Abdelgawwad MR, Reichheld JP, Vignols F. Evolution of redoxin genes in the green lineage. Photosynthesis Research. 2006;89:179–192. doi: 10.1007/s11120-006-9095-3. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Jimenez R, Wiita AP, Rodriguez-Larrea D, Kosuri P, Gavira JA, Sanchez-Ruiz JM, Fernandez JM. Force-Clamp Spectroscopy Detects Residue Co-evolution in Enzyme Catalysis. J Biol Chem. 2008;283:27121–9. doi: 10.1074/jbc.M803746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvalho AT, Swart M, van Stralen JN, Fernandes PA, Ramos MJ, Bickelhaupt FM. Mechanism of thioredoxin-catalyzed disulfide reduction. Activation of the buried thiol and role of the variable active-site residues. J Phys Chem B. 2008;112:2511–23. doi: 10.1021/jp7104665. [DOI] [PubMed] [Google Scholar]
- 24.Kappler U, Bailey S. Molecular basis of intramolecular electron transfer in sulfite-oxidizing enzymes is revealed by high resolution structure of a heterodimeric complex of the catalytic molybdopterin subunit and a c-type cytochrome subunit. J Biol Chem. 2005;280:24999–5007. doi: 10.1074/jbc.M503237200. [DOI] [PubMed] [Google Scholar]
- 25.Costentin C, Saveant JM. Competition between S(N)2 and single electron transfer reactions as a function of steric hindrance illustrated by the model system alkylCl + NO- Journal of the American Chemical Society. 2000;122:2329–2338. [Google Scholar]
- 26.Holm RH, Kennepohl P, Solomon EI. Structural and Functional Aspects of Metal Sites in Biology. Chem Rev. 1996;96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 27.McLendon G, Komar-Panicucci S, Hatch S. Applying Marcus's theory to electron transfer in vivo. Electron Transfer-from Isolated Molecules to Biomolecules. 1999;107(Pt 2):591–600. [Google Scholar]
- 28.Erlandsson M, Hallbrink M. Metallic zinc reduction of disulfide bonds between cysteine residues in peptides and proteins. International Journal of Peptide Research and Therapeutics. 2005;11:261–265. [Google Scholar]
- 29.Aslund F, Berndt KD, Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem. 1997;272:30780–6. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Z, Arscott LD, Ballou DP, Williams CH., Jr. The relationship of the redox potentials of thioredoxin and thioredoxin reductase from Drosophila melanogaster to the enzymatic mechanism: reduced thioredoxin is the reductant of glutathione in Drosophila. Biochemistry. 2007;46:7875–85. doi: 10.1021/bi700442r. [DOI] [PubMed] [Google Scholar]
- 31.Yasui S, Itoh K, Tsujimoto M, Ohno A. Irreversibility of single electron transfer occurring from trivalent phosphorus compounds to Iron(III) complexes in the presence of ethanol. Bulletin of the Chemical Society of Japan. 2002;75:1311–1318. [Google Scholar]
- 32.Hazzard JT, Marchesini A, Curir P, Tollin G. Direct measurement by laser flash photolysis of intramolecular electron transfer in the three-electron reduced form of ascorbate oxidase from zucchini. Biochim Biophys Acta. 1994;1208:166–70. doi: 10.1016/0167-4838(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 33.Farver O, Pecht I. Low activation barriers characterize intramolecular electron transfer in ascorbate oxidase. Proc Natl Acad Sci U S A. 1992;89:8283–7. doi: 10.1073/pnas.89.17.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falkowski PG. Evolution. Tracing oxygen's imprint on earth's metabolic evolution. Science. 2006;311:1724–5. doi: 10.1126/science.1125937. [DOI] [PubMed] [Google Scholar]
- 35.Raymond J, Segre D. The effect of oxygen on biochemical networks and the evolution of complex life. Science. 2006;311:1764–7. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 36.Kirschvink JL, Kopp RE. Palaeoproterozoic ice houses and the evolution of oxygen-mediating enzymes: the case for a late origin of photosystem II. Philos Trans R Soc Lond B Biol Sci. 2008;363:2755–65. doi: 10.1098/rstb.2008.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemaire SD, Guillon B, Le Marechal P, Keryer E, Miginiac-Maslow M, Decottignies P. New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2004;101:7475–80. doi: 10.1073/pnas.0402221101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma A, Scott JH, Cody GD, Fogel ML, Hazen RM, Hemley RJ, Huntress WT. Microbial activity at gigapascal pressures. Science. 2002;295:1514–6. doi: 10.1126/science.1068018. [DOI] [PubMed] [Google Scholar]
- 39.La Duc MT, Dekas A, Osman S, Moissl C, Newcombe D, Venkateswaran K. Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean room environments. Appl Environ Microbiol. 2007;73:2600–11. doi: 10.1128/AEM.03007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch AL. Shrinkage of Growing Escherichia-Coli-Cells by Osmotic Challenge. Journal of Bacteriology. 1984;159:919–924. doi: 10.1128/jb.159.3.919-924.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malone AS, Chung YK, Yousef AE. Genes of Escherichia coli O157:H7 that are involved in high-pressure resistance. Appl Environ Microbiol. 2006;72:2661–71. doi: 10.1128/AEM.72.4.2661-2671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008;451:704–7. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- 43.Jones PR, Manabe T, Awazuhara M, Saito K. A new member of plant CS-lyases. A cystine lyase from Arabidopsis thaliana. J Biol Chem. 2003;278:10291–6. doi: 10.1074/jbc.M212207200. [DOI] [PubMed] [Google Scholar]
- 44.Beynon RJ, Bond JS, NetLibrary Inc. Practical approach series. 2nd edn. Oxford University Press; Oxford; New York: 2001. Proteolytic enzymes: a practical approach; p. 340. xviii. [Google Scholar]
- 45.Kanzok SM, Schirmer RH, Turbachova I, Iozef R, Becker K. The thioredoxin system of the malaria parasite Plasmodium falciparum. Glutathione reduction revisited. J Biol Chem. 2000;275:40180–6. doi: 10.1074/jbc.M007633200. [DOI] [PubMed] [Google Scholar]
- 46.Kanzok SM, Fechner A, Bauer H, Ulschmid JK, Muller HM, Botella-Munoz J, Schneuwly S, Schirmer R, Becker K. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science. 2001;291:643–6. doi: 10.1126/science.291.5504.643. [DOI] [PubMed] [Google Scholar]
- 47.Behm M, Jacquot JP. Isolation and characterization of thioredoxin h from poplar xylem. Plant Physiology and Biochemistry. 2000;38:363–369. [Google Scholar]
- 48.Gelhaye E, Rouhier N, Vlamis-Gardikas A, Girardet JM, Sautiere PE, Sayzet M, Martin F, Jacquot JP. Identification and characterization of a third thioredoxin h in poplar. Plant Physiology and Biochemistry. 2003;41:629–635. [Google Scholar]
- 49.Lopez Jaramillo J, Chueca A, Jacquot JP, Hermoso R, Lazaro JJ, Sahrawy M, Lopez Gorge J. High-yield expression of pea thioredoxin m and assessment of its efficiency in chloroplast fructose-1,6-bisphosphatase activation. Plant Physiol. 1997;114:1169–75. doi: 10.1104/pp.114.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez-Jimenez R, Godoy-Ruiz R, Ibarra-Molero B, Sanchez-Ruiz JM. The efficiency of different salts to screen charge interactions in proteins: a Hofmeister effect? Biophys J. 2004;86:2414–29. doi: 10.1016/S0006-3495(04)74298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandez JM, Li HB. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–1678. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.