Abstract
Adenosine-to-inosine (A-to-I) editing of RNA transcripts is an increasingly recognized cellular strategy to modulate the function of proteins involved in neuronal excitability. We have characterized the editing of transcripts encoding the α3 subunit of heteromeric GABAA receptors (Gabra3), in which a genomically encoded isoleucine codon (ATA) is converted to a methionine codon (ATI) in a region encoding the predicted third transmembrane domain of this subunit. Editing at this position (I/M site) was regulated in a spatiotemporal manner with ∼90% of the Gabra3 transcripts edited in most regions of adult mouse brain, but with lower levels of editing in the hippocampus. Editing was low in whole-mouse brain at embryonic day 15 and increased during development, reaching maximal levels by postnatal day 7. GABA-evoked current in transfected cells expressing nonedited α3(I)β3γ2L GABAA receptors activated more rapidly and deactivated much more slowly than edited α3(M)β3γ2L receptors. Furthermore, currents from nonedited α3(I)β3γ2L receptors were strongly outwardly rectifying (corresponding to chloride ion influx), whereas currents from edited α3(M)β3γ2L receptors had a more linear current/voltage relationship. These studies suggest that increased expression of the nonedited α3(I) subunit during brain development, when GABA is depolarizing, may allow the robust excitatory responses that are critical for normal synapse formation. However, the strong chloride ion influx conducted by receptors containing the nonedited α3(I) subunit could act as a shunt to prevent excessive excitation, providing the delicate balance necessary for normal neuronal development.
Keywords: GABAA receptors, ion channel structure–function, binding–gating transduction, kinetics, development, synaptogenesis
Introduction
RNA editing is an increasingly recognized mechanism for expanding sequence diversity within the transcriptome through the site-selective conversion of adenosine to inosine (A-to-I editing) by hydrolytic deamination (Polson et al., 1991). Because inosine preferentially base pairs with cytosine, an inosine within RNA transcripts is read as guanosine during translation, often changing the amino acid coding potential of mRNAs that can dramatically alter the functional properties of the encoded protein products (Gott and Emeson, 2000; Emeson and Singh, 2001). A-to-I editing is catalyzed by a family of enzymes known as adenosine deaminases that act on RNA (ADARs) that can selectively modify adenosine residues within duplex regions of pre-mRNAs (Bass, 2002). These enzymes modulate neuronal activity by altering the amino acid sequence and function of ligand- and voltage-gated ion channels, as well as G-protein-coupled receptors (Sommer et al., 1991; Egebjerg and Heinemann, 1993; Lomeli et al., 1994; Burns et al., 1997; Bhalla et al., 2004). The mouse GABAA receptor α3 subunit transcript (mGabra3) was recently identified as an ADAR substrate, in which a genomically encoded isoleucine (I) is converted to a methionine (M) codon (Ohlson et al., 2007). The altered amino acid was predicted to reside toward the extracellular portion of the TM3 (third transmembrane domain) of the protein, a region that is important for the function and trafficking of GABAA α subunits (Gallagher et al., 2004).
GABAA receptors are pentameric ligand-gated chloride channels, usually composed of 2α, 2β, and either a γ or a δ subunit (Hevers and Luddens, 1998). In the mature brain, activation of GABAA receptors produces a hyperpolarizing influx of chloride ions. However, altered regulation of chloride ion transport during development changes the function of GABAA receptors, creating an excitatory response to GABA that can stimulate action potentials as well as relieve the voltage-dependent block of NMDA receptors. These depolarizing currents are crucial for a number of developmental processes, including proliferation and synaptogenesis (Ben-Ari et al., 2007; Cancedda et al., 2007). Recent studies have indicated that Gabra3 RNA transcripts undergo near complete editing (I/M site) in the adult mouse brain, but not in neonatal animals (Ohlson et al., 2007). Whereas the α1 subunit is the predominant α subtype in the adult brain, the α3 subunit is more highly expressed in the developing CNS (Laurie et al., 1992), raising questions regarding the functional significance of this RNA processing event. The present studies extend the analysis of Gabra3 editing and use voltage-clamp recording with recombinant receptors and rapid GABA application to assess the functional significance of A-to-I conversion within pentameric GABAA receptors.
Materials and Methods
RNA isolation and quantitative analyses of Gabra3 editing/expression.
RNA was isolated from mouse brain of 129S6 (12–14 week) or FVB/J (12 week) male mice, unless otherwise specified. Adult rat whole-brain RNA (Sprague Dawley) was isolated as previously described (Burns et al., 1997), and human whole-brain RNA was purchased from BioChain Institute. As described in supplemental Methods (available at www.jneurosci.org as supplemental material), first-strand cDNA and PCR amplicons were generated corresponding to the predicted I/M site duplex region within Gabra3 transcripts or for other GABAA subunit isoforms (supplemental Table 2, available at www.jneurosci.org as supplemental material).
The extent of Gabra3 editing was quantified using a custom-designed TaqMan SNP (single-nucleotide polymorphism) genotyping assay (supplemental Table 1, available at www.jneurosci.org as supplemental material) (Applied Biosystems). Gabra3 mRNA expression was quantified using a TaqMan MGB gene expression assay for Gabra3 (Mm00433440_m1). Postrun analyses were performed with an ABI Prism 7900HT system (SDS v2.3; Applied Biosystems). All reactions were run in triplicate and quantified as described previously (Feng et al., 2006).
Minigene construction and analysis.
A wild-type Gabra3 minigene (543 bp) was constructed using a nonedited Gabra3 amplicon. HeLa cells were cotransfected using FuGENE 6 (Roche) with 0.5 μg of a Gabra3 minigene along with 4 μg of either an empty pRC-CMV expression vector (Promega) or a FLAG-tagged ADAR1 (p150) or ADAR2b expression vector. Thirty-six hours after transfection, RNA was isolated and purified using Versagene RNA cell and DNase kits (Gentra Systems), and whole-cell extracts were prepared from a parallel sample for Western blotting analysis, as described previously (Schreiber et al., 1989). After reverse transcription (RT)-PCR amplification and sequence analysis of the Gabra3 minigene, editing was quantified directly from electropherogram traces (Kwok et al., 1994); for details, see supplemental Methods (available at www.jneurosci.org as supplemental material).
Expression of recombinant GABAA receptors.
Complementary DNAs encoding the human GABAA receptor α3(I), α3(M), and γ2L subunits were individually subcloned into the mammalian expression vector pcDNA3.1 (Invitrogen); cDNA encoding the human GABAA receptor β3 subunit was subcloned into pCMV6-XL5 (Origene). HEK293T cells were transfected using a calcium phosphate precipitation technique, and subsequently selected as described in supplemental Methods (available at www.jneurosci.org as supplemental material).
For editing analysis, RNA was isolated from HEK293T cells 2 d after transfection using TRI Reagent (Molecular Research Center), and RT-PCR and bulk sequence analysis were performed as described for Gabra3 minigene analysis.
Electrophysiological recording and drug application.
Lifted whole-cell voltage-clamp recordings were performed at room temperature using a rapid drug application device to apply GABA to transiently transfected HEK293T cells (Lagrange et al., 2007). Data was acquired and analyzed as described in supplemental Methods (available at www.jneurosci.org as supplemental material). Uncompensated series resistance error is a possible confounding factor in whole-cell voltage clamp. However, all kinetic differences reported here were confirmed by multiple statistical techniques, as described in the supplemental Methods (available at www.jneurosci.org as supplemental material).
Statistical analysis.
Numerical data were expressed as mean ± SEM. Statistical analysis was performed using GraphPad Prism. Data were compared using a Mann–Whitney test for pairs of data. Changes in editing or expression during development were tested using a generalized Spearman rank-order correlation and in brain region analysis by nonparametric one-way ANOVA. The normalized peak response to 5 or 10 ms of GABA application was compared with the peak current during 4 s GABA applications, using a Wilcoxon signed-rank test to compare with a hypothetical value of 1.0. Statistical significance was taken as p ≤ 0.05.
Results
Gabra3 is highly edited in adult mammalian brain
The mouse Gabra3 transcript was recently reported as a substrate of RNA editing (Ohlson et al., 2007). To assess the editing of Gabra3 transcripts from other mammalian species, we used an RNA folding algorithm (Mfold) (Mathews et al., 1999; Zuker, 2003), which predicted a conserved short duplex structure contained entirely within exon 9 (Fig. 1A). Gabra3 editing was verified directly in RNA isolated from whole human and rat brain through bulk-sequencing analyses of the region surrounding the I/M site. The presence of a mixed A/G peak in electropherogram traces demonstrated the editing of all three species (Fig. 1A). Because inosine has base-pairing properties similar to guanosine, A-to-I editing events appear as A-to-G discrepancies between genomic and cDNA sequences. The presence of a small adenosine peak at the editing site indicated that a portion of the Gabra3 transcripts was not edited (Fig. 1A), despite previous observations that this RNA modification goes to completion in the brain of adult FVB/N mice (Ohlson et al., 2007). To assess whether the extent of Gabra3 mRNA editing varies by mouse strain, we compared the extent of editing in FVB/J animals to those used in our studies (129S6), revealing a significant difference, with mean editing levels of 97.0% and 82.6%, respectively.
Figure 1.
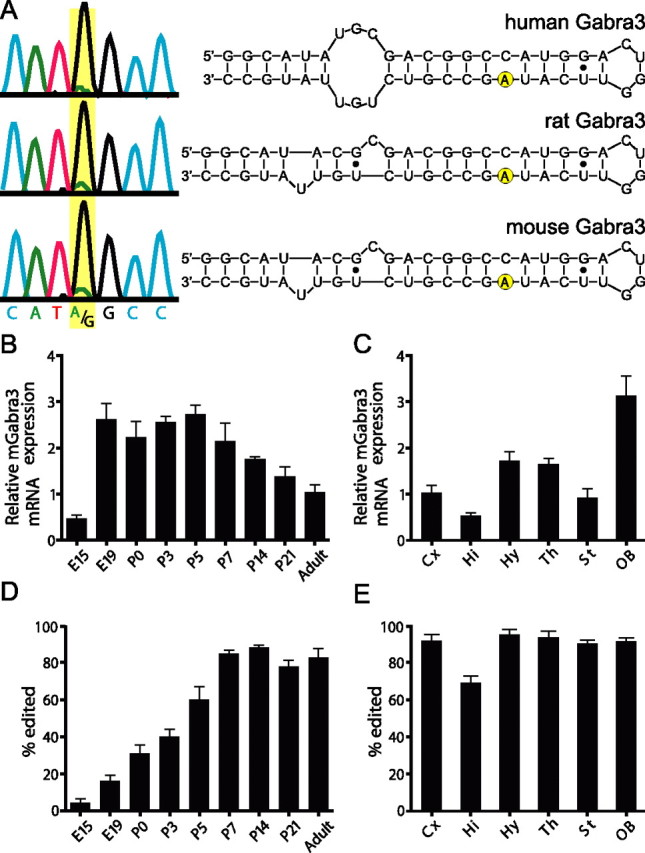
Developmental regulation of GABAA receptor α3 subunit RNA expression and editing (I/M site). A, Bulk-sequence analysis of Gabra3 RT-PCR amplicons generated from whole human, rat, and mouse brains showed A-to-I editing as a mixed A/G peak in each electropherogram trace. Predicted mRNA secondary structures surrounding the I/M editing site are illustrated to the right of each sequencing trace with the editing site indicated in yellow. B, C, Quantitative analyses of Gabra3 mRNA levels showed varied expression during development (p ≤ 0.05; B) and among adult brain regions (p ≤ 0.05; C). D, E, RNA editing analyses (I/M site) in developing mouse brain (D) or adult brain regions (E) showed variation during development (p ≤ 0.01) and decreased editing levels in hippocampus compared with other brain regions (p ≤ 0.05). All data represent a minimum of 100 individual cDNA clones taken from ≥4 different animals. Cx, Cortex; Hi, hippocampus; Hy, hypothalamus; Th, thalamus; St, striatum; OB, olfactory bulb.
Gabra3 represents one of six different genes encoding subtypes of the GABAA receptor α subunit. Although only mouse Gabra2 and Gabra6 genes have an adenosine residue at a position analogous to the Gabra3 I/M site, the presence of an extended duplex in transcripts encoding other GABAA receptor α subunits could allow editing of other adenosine residues within that region. Bulk sequence analyses of cDNAs generated from other α subunits identified no additional editing sites (supplemental Fig. 1, available at www.jneurosci.org as supplemental material), demonstrating that editing of this region is unique to the Gabra3 transcript.
A small exonic RNA duplex is required for I/M site editing
To identify the sequences that direct editing of Gabra3 RNAs, a minigene containing the predicted mouse Gabra3 RNA duplex (Fig. 1A) was introduced into HeLa cells and assessed for editing at the I/M site (supplemental Fig. 2B, available at www.jneurosci.org as supplemental material). We found that a low level of baseline editing was significantly increased by the coexpression of ADAR1 or ADAR2. These results demonstrated not only that both ADAR1 and ADAR2 could efficiently modify the Gabra3 I/M site, but also that the minigene contained the cis-active elements necessary to promote site-selective adenosine deamination.
To further assess whether the predicted duplex was essential for Gabra3 editing, we developed a mutant Gabra3 minigene (M1) containing six mutations to disrupt the proposed structure of the RNA duplex (supplemental Fig. 2A, available at www.jneurosci.org as supplemental material). Coexpression of the mutant minigene with ADAR1 or ADAR2 revealed only background levels of editing (supplemental Fig. 2B, available at www.jneurosci.org as supplemental material), demonstrating that these six nucleotides were essential to maintain normal levels of A-to-I conversion. Western blotting analyses confirmed that the reduced editing levels for the mutant minigene did not result from decreased ADAR expression (data not shown). To determine whether reduced editing resulted from disrupted secondary structure or the mutation of specific sequence determinants, we introduced six compensatory mutations predicted to restore the structure of the mutant Gabra3 duplex (M2). These compensatory sequence alterations successfully restored editing to wild-type levels for both ADARs (supplemental Fig. 2B, available at www.jneurosci.org as supplemental material), indicating that the predicted RNA duplex was essential for I/M site editing in Gabra3 mRNAs.
Gabra3 editing and expression levels are regulated in a spatiotemporal manner
We performed a developmental analysis of Gabra3 mRNA expression using a quantitative RT-PCR approach with RNA isolated from whole-mouse brain. Similar to previous reports in rat (Laurie et al., 1992), Gabra3 expression was found to be low at embryonic day 15 (E15), but increased fivefold by E19. The expression level stayed approximately constant through postnatal day 5 (P5), then began a gradual decline into adulthood (p ≤ 0.0008) (Fig. 1B). A similar analysis of RNA isolated from dissected brain regions in adult mice revealed that Gabra3 mRNA levels varied up to sixfold among the brain regions examined, with the olfactory bulb and hippocampus having the highest and lowest levels of Gabra3 expression, respectively (p ≤ 0.002) (Fig. 1C).
Because Gabra3 editing may represent a cellular strategy for the spatiotem-poral regulation of α3 subunit-containing GABAA receptor function, we quantified the extent of Gabra3 editing during development and in dissected adult brain regions. Results from this analysis demonstrated that I/M site editing was low at E15 (15 ± 2%), then increased gradually through early development, and eventually reached maximal level (80–85%) by P7, which persisted into adulthood (Fig. 1D). Examination of Gabra3 editing patterns in the adult brain indicated that 90–95% of transcripts were edited in most brain regions examined, except for the hippocampus (70 ± 4%) (Fig. 1E).
RNA editing regulates the function of α3 subunit-containing GABAA receptors
To investigate functional consequences of Gabra3 mRNA editing, we used whole-cell voltage-clamp recording and a rapid drug delivery system to apply GABA to lifted HEK293T cells that had been transiently cotransfected with plasmids containing human β3, γ2L, and either nonedited α3(I) or edited α3(M) cDNAs. The β3 and γ2L subunit subtypes were chosen because they are coexpressed in the same brain regions as the α3 subunit, in both mature and developing animals (Laurie et al., 1992; Pirker et al., 2000). To ensure that the nonedited α3(I) subunit was not modified in our heterologous expression system, an expression vector containing a nonedited Gabra3 minigene was transiently transfected into HEK293T cells, and minigene-derived RT-PCR amplicons were sequenced in bulk (as in supplemental Fig. 2, available at www.jneurosci.org as supplemental material). This analysis demonstrated no detectable editing of Gabra3 transcripts, thus validating the use of HEK293T cells to evaluate the consequences of Gabra3 transcript editing.
GABA (1 mm, 4 s) consistently evoked currents from cells expressing either nonedited α3(I)β3γ2L (3910 ± 324 pA; n = 20) or edited α3(M)β3γ2L (2306 ± 323 pA; n = 28) receptors, but the peak current amplitudes were significantly larger from the nonedited receptors (p < 0.01) (supplemental Table 1, available at www.jneurosci.org as supplemental material)
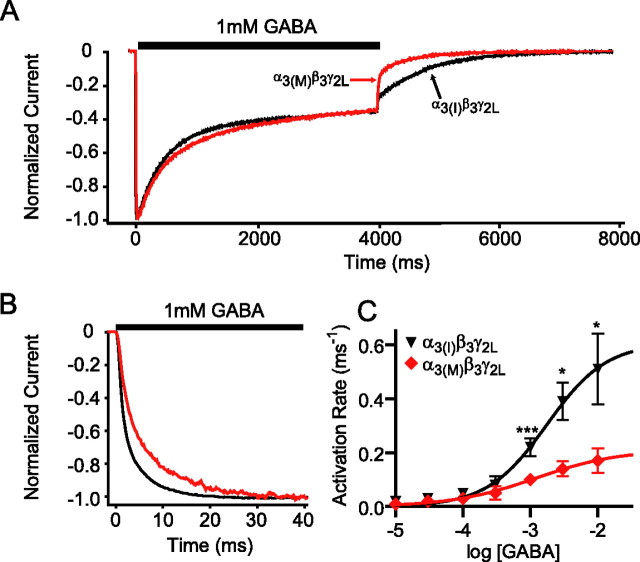
Although all α3β3γ2L currents activated and desensitized very slowly compared with the more widely studied α1 subunit-containing receptors (Barberis et al., 2007; Picton and Fisher, 2007), nonedited α3(I)β3γ2L currents activated more rapidly than edited α3(M)β3γ2L currents (Fig. 2B,C). Furthermore, nonedited α3(I)β3γ2L currents deactivated much more slowly after 4 s of 1 mm GABA (Fig. 2A; supplemental Table 1, available at www.jneurosci.org as supplemental material) [473 ± 45 ms (n = 15) vs 283 ± 48 ms (n = 19) for α3(I)β3γ2L and α3(M)β3γ2L, respectively; p < 0.01]. Similar results were found using briefer, more synaptically relevant (5 or 10 ms) GABA applications (supplemental Table 1, available at www.jneurosci.org as supplemental material). In contrast, both the rate and extent of desensitization of the two differently edited receptors were similar (Fig. 2; supplemental Table 1, available at www.jneurosci.org as supplemental material). As previously described (Barberis et al., 2007; Picton and Fisher, 2007), high GABA concentrations were required to activate α3β3γ2L receptors, with peak current EC50s of 65 and 153 μm, for nonedited α3(I)β3γ2L and edited α3(M)β3γ2L receptors, respectively. However, because the GABA-mediated depolarizations seen during development may last for several seconds (Ben-Ari et al., 2007), we also measured the residual current at the end of a 4 s application of varying concentrations of GABA to better assess the responses to slow GABA fluctuations. We found a maximal response that was ∼40% of the peak current amplitude, with an EC50 of 17–18 μm for both nonedited α3(I)β3γ2L and edited α3(M)β3γ2L GABAA receptors.
Figure 2.
RNA editing slows activation of α3 subunit-containing GABAA receptors. A, Representative GABA-evoked currents from nonedited α3(I)β3γ2L (black) and edited α3(M)β3γ2L (red) receptors were recorded using a rapid-application drug delivery system to expose lifted, whole-cell voltage-clamped HEK 293T cells to GABA (1 mm, 4 s). B, The same representative currents are shown on an expanded time base. C, Nonedited α3(I)β3γ2L receptors (black triangles) activated more quickly than edited α3(M)β3γ2L receptors (red diamonds) at all [GABA] ≥ 1 mm. *p ≤ 0.05, ***p ≤ 0.001, α3(I)β3γ2L versus α3(M)β3γ2L by Mann–Whitney test.
The slow kinetic properties of α3 subunit-containing receptors prompted us to further explore how the editing variants respond differentially to the very brief surges of GABA (1 mm, ≈1 ms) present in the synaptic cleft. The currents from both edited and nonedited α3-containing receptors elicited by brief GABA application (5 ms, 1 mm) were truncated severely compared with the maximal activation seen during more prolonged GABA exposure (Fig. 3A1,A2). However, applying a second brief pulse after a short (≤ 80 ms) interval allowed more complete activation (Fig. 3B1,B2), and repetitive brief GABA applications (5–20 Hz) produced a frequency-dependent summating response (Fig. 3C1,C2).
Figure 3.
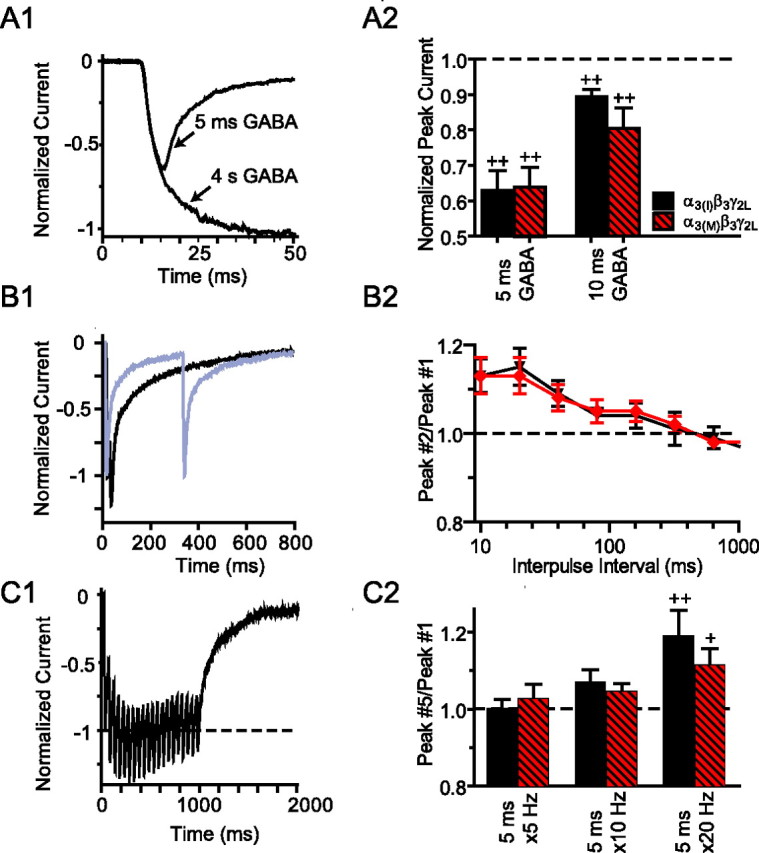
α3β3γ2L receptors produce summating responses during repetitive stimulation. A, Very brief GABA (5 ms, 1 mm) exposure elicits submaximal responses from α3(I)β3γ2L or α3(M)β3γ2L receptors, as shown by representative currents in A1 and summarized in A2. Peak currents are normalized to the peak response to 4 s of 1 mm GABA. B1, Application of a second brief GABA pulse after a short (20 ms, black trace) interval allows more complete activation. When the interval between GABA applications is prolonged (320 ms, gray trace), the first and second responses are equal. B2, Pairs of brief GABA applications produce a consistently greater response from α3 subunit-containing GABAA receptors when the interval between GABA applications is <80 ms. C, α3 subunit-containing GABAA receptors produce a summating response to high-frequency repetitive brief GABA applications, as shown by the representative current shown in C1 and summarized in C2. +p ≤ 0.05, ++p ≤ 0.01 versus a hypothetical value of 1.0 by Wilcoxon signed-rank test.
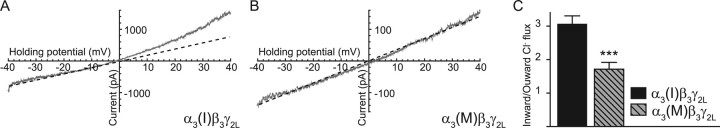
GABAA receptor activation may be either excitatory or inhibitory, depending on the chloride reversal potential. To elucidate the propensity of α3 subunit-containing GABAA receptors to mediate inward (representing chloride efflux) versus outward current, we used ramp current–voltage (I/V) protocols (−50 mV to +50 mV over 1 s) in the absence and presence of a low concentration (3 μm) of GABA. This concentration was chosen to minimize the effects of desensitization and series resistance error that can occur with higher GABA concentrations. The subtracted I/V plots of currents from nonedited α3(I)β3γ2L receptors had a strong outward rectification [chloride ion influx/efflux = 3.1 ± 0.2 (n = 13); p ≤ 0.0006], compared with the more linear currents from edited α3(M)β3γ2L receptors [chloride ion influx/efflux = 1.7 ± 0.2 (n = 14)] (Fig. 4). Similar results were found with hyperpolarizing ramp I/Vs (+50 to −50 mV over 1 s) (data not shown).
Figure 4.
RNA editing alters the voltage-dependent conductance of α3 subunit-containing GABAA receptors. A, B, Representative ramp I/V traces from the nonedited α3(I)β3γ2L or edited α3(M)β3γ2L GABAA receptors. The membrane potential was ramped from −50 mV to +50 mV in external solution and then again in 3 μm GABA. C, Conductance was calculated as the ratio of the chord conductance between +30 and +40 mV to the conductance between −30 and −40 mV. The currents from nonedited α3(I)β3γ2L receptors display a strong outward rectification, corresponding to increased chloride ion influx at depolarized potentials. ***p ≤ 0.001.
Discussion
RNA editing increases genomic diversity by allowing the posttranscriptional modification of RNA transcripts that can encode multiple gene products with altered functional properties (Gott and Emeson, 2000; Emeson and Singh, 2001; Bass, 2002). The GABAA receptor α3 is modified by A-to-I editing in multiple mammalian species, and the level of editing is regulated in a strain-specific manner. Such differences in Gabra3 editing could result in physiological or behavioral variation and may explain strain-specific differences in seizure thresholds (Frankel et al., 2001) and also suggest that human interindividual variability in editing levels may impact neural development, brain function, and/or disease susceptibility.
The I/M site secondary structure is rare in the repertoire of codon-altering editing events because it is contained completely within an exon, whereas most duplexes require the presence of intron sequences and thus editing must occur before splicing within nuclear pre-mRNAs (Gott and Emeson, 2000). Maintenance of the duplex structure in mature Gabra3 transcripts may increase the time-frame during which the substrate RNA may be modified by providing the potential for editing to occur outside the nucleus. Cytoplasmic A-to-I conversion may be relevant to disease because the expression of an ADAR1 isoform (p150) that is present in the cytoplasm is induced in response to viral infection, inflammation, and interferon induction (Yang et al., 2003).
We found that RNA editing of α3 subunit transcripts results in GABAA receptors with smaller amplitudes, slower activation, and faster deactivation. The molecular bases for the altered GABAA receptor properties remain unknown. However, the I/M editing site is immediately adjacent to the extracellular transmembrane 2/3 linker, a region known to be important for channel gating (Ernst et al., 2005). Given this location, it is tempting to suggest that a change in intrinsic channel gating properties, possibly involving reduced open state stability, may underlie these differences. However, the nearly twofold current amplitude difference in cells expressing the edited versus unedited α3 subunit raises the possibility that RNA editing also regulates the number of functional GABAA receptors on the cell surface, thereby adding another level of control over the spatial and temporal distribution of GABAA receptor expression.
Our results suggest that the unusual properties of α3β3γ2L receptors may allow them to serve a unique role in conveying inhibitory information. These receptors are virtually insensitive to the low GABA concentrations (≈1 μm) found outside of the synaptic cleft. Moreover, these slowly activating α3 subunit-containing GABAA receptors will likely respond poorly to the very brief GABA available during an individual synaptic event. However, slow deactivation allows these receptors to produce a summating response during repetitive stimulation of sufficiently high frequency, thereby conveying high-frequency signals but attenuating the response to lower-frequency input. This is in direct contrast to the progressively smaller responses elicited during high-frequency stimulation of the more ubiquitous α1 subunit-containing receptors (Lagrange et al., 2007).
The function of GABAA receptors changes throughout development, and the presence of elevated levels of nonedited α3 subunit-containing GABAA receptors occurs during a period of robust synaptic development (Fig. 1) (Ben-Ari et al., 2007). Increased α3 expression may underlie some of the distinctly slow IPSC kinetics observed during development (Ortinski et al., 2004), although the potential for other subunit combinations, including multiple α subunit subtypes within individual pentamers, makes it difficult to extrapolate our data directly to brain slice physiology. Perhaps more importantly, GABAA receptors have the unusual characteristic of being able to produce hyperpolarization, depolarization, or shunting, depending on the chloride ion reversal potential. The high intracellular concentration of chloride ions in developing neurons produces an excitatory response to GABA that appears to be important for normal embryonic neuronal development (Cancedda et al., 2007), as well as newly born neurons generated in mature animals (Ge et al., 2006; Ben-Ari et al., 2007). Our results predict that nonedited α3(I)-containing receptors are ideally suited to respond to prolonged elevations of GABA, thereby producing robust, long-lived depolarization that may trigger the production of sodium and calcium-mediated action potentials. Once these action potentials depolarize the membrane past the chloride equilibrium potential, α3(I) subunit-containing receptors would convey a strong influx of chloride ions, thereby shunting excess excitation. RNA editing may introduce a functional flexibility that allows nonedited GABAA receptors to play an important role in promoting neuronal growth and connectivity during development, with the edited (M) isoform being preferentially suited to the inhibitory functions of GABAA receptors in more mature animals.
Footnotes
This work was supported by National Institute of Neurological Disorders and Stroke Grants K08 NS045122 (A.H.L.), R01 NS33300 (R.L.M.), and R01 NS33323 (R.B.E.). We thank Emmanuel Botzolakis and Kevin Erreger for helpful discussions and the Vanderbilt University Medical Center Biostatistics Shared Resource for statistical expertise and analyses. E.Y.R. designed and performed experiments characterizing editing and expression; A.H.L. designed and performed the electrophysiology experiments.
References
- Barberis A, Mozrzymas JW, Ortinski PI, Vicini S. Desensitization and binding properties determine distinct α1β2γ2 and α3β2γ2 GABAA receptor-channel kinetic behavior. Eur J Neurosci. 2007;25:2726–2740. doi: 10.1111/j.1460-9568.2007.05530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y, Gaiarsa JL, Tyzio R, Khazipov R. GABA: a pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol Rev. 2007;87:1215–1284. doi: 10.1152/physrev.00017.2006. [DOI] [PubMed] [Google Scholar]
- Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat Struct Mol Biol. 2004;11:950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Cancedda L, Fiumelli H, Chen K, Poo MM. Excitatory GABA action is essential for morphological maturation of cortical neurons in vivo. J Neurosci. 2007;27:5224–5235. doi: 10.1523/JNEUROSCI.5169-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egebjerg J, Heinemann SF. Ca2+ permeability of unedited and edited versions of the kainate selective glutamate receptor GluR6. Proc Natl Acad Sci USA. 1993;90:755–759. doi: 10.1073/pnas.90.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emeson R, Singh M. Adenosine-to-inosine RNA editing: substrates and consequences. In: Bass B, editor. RNA editing. Oxford: Oxford UP; 2001. pp. 109–138. [Google Scholar]
- Ernst M, Bruckner S, Boresch S, Sieghart W. Comparative models of GABAA receptor extracellular and transmembrane domains: important insights in pharmacology and function. Mol Pharmacol. 2005;68:1291–1300. doi: 10.1124/mol.105.015982. [DOI] [PubMed] [Google Scholar]
- Feng Y, Sansam CL, Singh M, Emeson RB. Altered RNA editing in mice lacking ADAR2 autoregulation. Mol Cell Biol. 2006;26:480–488. doi: 10.1128/MCB.26.2.480-488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel WN, Taylor L, Beyer B, Tempel BL, White HS. Electroconvulsive thresholds of inbred mouse strains. Genomics. 2001;74:306–312. doi: 10.1006/geno.2001.6564. [DOI] [PubMed] [Google Scholar]
- Gallagher MJ, Song L, Arain F, Macdonald RL. The juvenile myoclonic epilepsy GABAA receptor α1 subunit mutation A322D produces asymmetrical, subunit position-dependent reduction of heterozygous receptor currents and α1 subunit protein expression. J Neurosci. 2004;24:5570–5578. doi: 10.1523/JNEUROSCI.1301-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Goh EL, Sailor KA, Kitabatake Y, Ming GL, Song H. GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature. 2006;439:589–593. doi: 10.1038/nature04404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- Hevers W, Lüddens H. The diversity of GABAA receptors. Pharmacological and electrophysiological properties of GABAA channel subtypes. Mol Neurobiol. 1998;18:35–86. doi: 10.1007/BF02741459. [DOI] [PubMed] [Google Scholar]
- Kwok PY, Carlson C, Yager TD, Ankener W, Nickerson DA. Comparative analysis of human DNA variations by fluorescence-based sequencing of PCR products. Genomics. 1994;23:138–144. doi: 10.1006/geno.1994.1469. [DOI] [PubMed] [Google Scholar]
- Lagrange AH, Botzolakis EJ, Macdonald RL. Enhanced macroscopic desensitization shapes the response of alpha4 subtype-containing GABAA receptors to synaptic and extrasynaptic GABA. J Physiol (Lond) 2007;578:655–676. doi: 10.1113/jphysiol.2006.122135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie DJ, Wisden W, Seeburg PH. The distribution of thirteen GABAA receptor subunit mRNAs in the rat brain. III. Embryonic and postnatal development. J Neurosci. 1992;12:4151–4172. doi: 10.1523/JNEUROSCI.12-11-04151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomeli H, Mosbacher J, Melcher T, Höger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Ohlson J, Pedersen JS, Haussler D, Ohman M. Editing modifies the GABAA receptor subunit alpha3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinski PI, Lu C, Takagaki K, Fu Z, Vicini S. Expression of distinct alpha subunits of GABAA receptor regulates inhibitory synaptic strength. J Neurophysiol. 2004;92:1718–1727. doi: 10.1152/jn.00243.2004. [DOI] [PubMed] [Google Scholar]
- Picton AJ, Fisher JL. Effect of the alpha subunit subtype on the macroscopic kinetic properties of recombinant GABAA receptors. Brain Res. 2007;1165:40–49. doi: 10.1016/j.brainres.2007.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirker S, Schwarzer C, Wieselthaler A, Sieghart W, Sperk G. GABAA receptors: immunocytochemical distribution of 13 subunits in the adult rat brain. Neuroscience. 2000;101:815–850. doi: 10.1016/s0306-4522(00)00442-5. [DOI] [PubMed] [Google Scholar]
- Polson AG, Crain PF, Pomerantz SC, McCloskey JA, Bass BL. The mechanism of adenosine to inosine conversion by the double-stranded RNA unwinding/modifying activity: a high-performance liquid chromatography-mass spectrometry analysis. Biochemistry. 1991;30:11507–11514. doi: 10.1021/bi00113a004. [DOI] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’ prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Yang JH, Nie Y, Zhao Q, Su Y, Pypaert M, Su H, Rabinovici R. Intracellular localization of differentially regulated RNA-specific adenosine deaminase isoforms in inflammation. J Biol Chem. 2003;278:45833–45842. doi: 10.1074/jbc.M308612200. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]