Abstract
Magnetic resonance (MR) microscopy has become an important tool for small animal cardiac imaging. In relation to competing technologies (microCT and ultrasound), MR is limited by spatial resolution, temporal resolution, and acquisition time. All three of these limitations have been addressed by developing a 4D (3D plus time) radial acquisition sequence. The signal-to-noise has been optimized by minimizing the echo time (300 μs). The temporal resolution and throughput have been improved by center-out trajectories resulting in TR < 2.5 ms. The contrast has been enhanced through the use of a liposomal blood pool agent that reduces T1 of the blood to < 400 ms. We have developed protocols for three specific applications: a) high-throughput with spatial resolution of 87×87×352 μm3 (voxel volume=2.7 nL) and acquisition time of 16 minutes; b) high-temporal resolution with spatial resolution of 87×87×352 μm3 (voxel volume=2.7 nL); temporal resolution at 4.8 ms and acquisition time of 32 minutes; and c) high-resolution isotropic imaging at 87×87×87 μm3 (voxel volume=0.68 nL), and acquisition time of 31 minutes. The 4D image arrays allow direct measure of cardiac functional parameters dependent on chamber volumes, e.g. ejection fraction, end diastolic volume, and end systolic volume.
Keywords: 4D, radial, mouse, heart, liposomal Gd
Introduction
Magnetic resonance microscopy (MRM) has become a standard tool for investigating cardiac structure and function in the mouse. The majority of MR work on mice has been done with 2D acquisition methods with limited resolution along the third axis (1-4). 3D studies have shown promise in the clinical arena as they increase signal-to-noise-ratio (SNR) per unit time and allow for higher in-plane resolution, ultimately allowing more sensitive calculation of cardiac function. But 3D applications in small animal imaging thus far have been limited. While 3D studies have been performed in the rat (5), those studies had limited temporal resolution (systole and diastole). Feintuch et al. have shown 4D (3D spatial+time) data in the mouse at limited spatial resolution (8 nL), limited temporal resolution (12 ms), and with long scan times of 1–2 hours (6). Recent work in the clinical arena has extended methods for cardiac functional assessment with 4D acquisitions that can be divided into two categories: 4D human studies that use contrast (7,8); and 4D studies that use inherent MR contrast (black blood, steady-state imaging techniques) (9,10). In both cases, the increased resolution along the Z-axis provides reduction in volume averaging and increased precision in measurement of end systolic and diastolic volume (ESV, EDV), and ejection fraction (EF). The utility of a routine method for high-throughput assessment of cardiac function in the mouse is clear. But extension of clinical techniques to the mouse is not straightforward. The 25-gram mouse is nearly 3000-times smaller than a human. To view the mouse anatomy at anatomic resolution comparable to that achieved in the clinical domain (voxel volumes of 48 mm3 @ 0.96 sec/frame) (10) will require voxels on the order of 16 nL (<275 μm in every dimension). Because of the higher heart rate in the mouse (450–550 beats per minute), achieving a frame rate similar to humans will require a temporal resolution of 12 ms. We describe a method for functional 4-dimensional cardiac assessment of the mouse using radial encoding of a free induction decay (FID) (11). The method is relatively fast (16–32 min/study), with spatial resolution nearly 8-times that of previous studies. We have incorporated the method into a pipeline that permits rapid acquisition, reconstruction, archive, and analysis. The method has been applied to a cohort of 20 C57BL/6J mice to demonstrate what we believe to be a much more precise measurement of cardiac function than existing MRI methods because it is less susceptible to partial volume effects.
Methods
Magnet Specifications
All MR studies were performed at the Duke Center for In Vivo Microscopy on a 7T, 120 mm bore, Magnex magnet with a GE EXCITE console (EPIC 12.4; GE Medical Systems, Milwaukee, WI). The system employs Resonant Research shielded gradient coils (Billerica, MA) that provide a maximum gradient strength of 770 mT/m and slew rate of 6160 mT/m/ms. The gradients are driven by high-power (12 KW) amplifiers (Copley Controls Model 266). The RF front-end of the GE console has been modified for operation at higher field through the use of a second synthesizer and radiofrequency (RF) mixer providing both up and down conversion in the RF chain via sum and difference signals. The strong gradients with high slew rates, high duty cycle, and high power amplifiers are critical for achieving the very short repetition times over small fields of view required for efficient acquisition.
RF Coil
A 2.5 cm × 3.0 cm transmit/receive surface coil was built to maximize SNR. The coil was embedded in the support cradle as a single unit to speed animal setup. The cradle and coil can accommodate a mouse from 15 grams up to 50 grams. The coil excites a roughly 2.25 cm-long volume of tissue along the long axis of the animal allowing excitation of only the volume of interest and therefore providing a short acquisition time.
Pulse Sequence
The 3D radial encoding sequence developed in the EPIC 12.4 pulse programming environment is shown in Figure 1a. The sequence is prospectively cardiac-gated as seen in Figure 1b allowing arbitrary definition of the number of acquisition windows defining the phases of the cardiac cycle. The current technique employs center-out sampling, where the slope of the gradient is sampled to minimize echo time. The resulting 4D datasets are quite large. In its current implementation, the upper limit of the 4D array is defined by the total number of views permissible in the EPIC software (524,000). For eight phases of the cardiac cycle, with 62,280 views in each of the 3D arrays and 212 points per view, the resulting raw dataset is 407.4 MB. To facilitate post-processing of relevant cardiac functional parameters, subroutines in the EPIC code were included to permit graphic prescription of the acquisition axes parallel to the short and long axes of the mouse heart.
Figure 1.
The 3DRA pulse sequence is presented in (A) for RF and all three readout gradients. The distance between the endpoints of the trajectories on the surface of the bounding sphere (Δk) is uniform. The view-ordering scheme is demonstrated with two view-order randomization factors shown in (B). 3DRA sequence is prospectively gated and the number of phases acquired is determined by the heart rate of the mouse.
The limited signal from smaller voxels is one of the fundamental challenges in MR microscopy. We have increased the SNR by minimizing the effective echo time (TE) and reduced scan time by minimizing repetition time (TR). The lower limits on TE and TR are imposed by the rise time of the gradients, bandwidth of the acquisition, spatial resolution, and duty cycle of the gradient and gradient amplifiers. Figure 1a shows the radial sequence in detail to demonstrate how the echo and repetition time have been minimized with center-out trajectories. The sequence can be used with a slice-selective gradient or with a hard pulse excitation.
There are several ways to determine the k-space sampling pattern by defining the end points of each trajectory on the surface of a uniform sphere (12). We used a precalculated geodesic table. Geodesic spheres have been used routinely in analysis of the earth as they result in nearly perfect isotropic sampling of the sphere. Because this approach is computationally involved, the end points are precalculated and stored in a wave table and then uploaded onto the scanner. For each array size, there are several wave tables to allow for flexibility in sampling. The most basic table is sampled at the Nyquist limit, and each subsequent wave table reduces sampling by √2. For images with anisotropic resolution, the maximum distance (kz) was reduced resulting in an oblate spheroid instead of the isotropic sphere (13). Moving to a spiral technique similar to the technique proposed in (12) would allow direct calculation and possibly more flexibility in the sampling of k-space.
The rapid movement of both the heart (450–550 beats per minute) and lungs (100 breaths per minute) in the mouse causes significant phase errors in the 3D radial acquisition (3DRA). While radial acquisitions are relatively immune to motion compared to Cartesian sequences, accumulation of phase due to motion can cause streaking artifacts, the extent of which depends on the view ordering of the sequence (14). For example, acquisition of a 2D radial image where θ is incremented by , where nviews is the total number of views acquired, will distribute the phase due to local motion over a small angle of the radial dataset, resulting in streaking artifacts. One way to minimize these phase errors is to acquire the radial data such that each successive view rotates by a large angle, which will distribute the phase errors due to motion more uniformly over the volume of k-space. The 3D radial sequence uses a precalculated wave table. In order to minimize phase errors we alter the view order so that each sequential view represents a significant rotation in k-space. The wave table for each phase of the cardiac cycle (represented by ϕ) is identical (i.e. k(ϕ1,nl) = k(ϕm,nl) where n is the total number of views acquired per 3D phase). Moving from k(ϕ1,n1) to k(ϕ1,n2) results in a less than 1% change in end point position in k-space. To minimize phase errors, we use two view randomization factors (Figure 1b): randϕ and randn. For the 10 ms temporal resolution sequence, as seen in Figure 1b, 4 views can be acquired for each phase of the heart cycle for each heartbeat, ultimately requiring n/4 heartbeats to acquire a full 4D dataset. If no view randomization were implemented, this would result in the following sequence order: k(ϕ1,n1),k(ϕ1,n2),k(ϕ1,n3),k(ϕ1,n4),k(ϕ2,n1),k(ϕ2,n2),k(ϕ2,n3),k(ϕ2,n4). Since there is minimal change from k(ϕ1,n1) to k(ϕ1,n2) (less than 1%), and because the trajectories are identical for all phases of the heart cycle, acquiring the data sequentially results in significant phase artifact. The variable randϕ changes the order in which the views are acquired between the first phase and the each subsequent phase of the heart cycle, resulting in k(ϕ1,n1) and k(ϕm,n1*) = k(ϕm,m · randϕ + n1), where ϕ is the cardiac phase and m is the phase of the cardiac cycle. The randn factor changes the order of view acquisition within phases resulting in k(ϕ1,n1) and k(ϕ1,nl*) = k(ϕ1,nl · randn). Both randomization factors together result in the following sequence order: k(ϕm,nl*) = k(ϕm,nl · randn + m · randϕ). The randomization factors, if chosen appropriately, result in changing the end point of sequential trajectories by a factor of 100%, in some cases completely reversing the polarity of the gradient and assisting in spreading phase errors in k-space. In addition to reducing artifacts in the image, randomizing the view ordering also results in more evenly distributed current across the three separate gradient windings, which in turn reduces gradient heating. The reduction in gradient heating (3X) is dramatic when compared to a Cartesian scan in which the full current is applied to the readout (frequency encode) gradient for every view.
Nyquist sampled radial acquisitions require more views than their Cartesian counterparts: a fully sampled 256×256×256 3D radial acquisition (3DRA) sequence requires 205,890 views (15), while a comparable Cartesian sequence requires 65,536 views. Despite this shortcoming, one advantage of radial acquisition is that the sequence is much more resistant than Cartesian to undersampling (16,17). We acquired 62,720 views for every 256×256×256 dataset, an undersampling factor of over three, which resulted in very few reconstruction artifacts. Previous work has suggested that 3DRA can be performed with undersampling by up to 8 with limited artifacts (18).
The resulting 4D radial sequence is extremely flexible. Table 1 summarizes three protocols that collectively test the limits of the acquisition method for high-throughput, high-temporal resolution, and high-spatial resolution. The echo time for all three protocols is 300 microseconds and the TR is between 2.4 ms and 3.1 ms depending on the heart rate of the mouse. The first protocol (high-throughput) can be executed in 16 minutes with minimum TE (300 μs) and TR (2.4 ms). The second protocol (high-temporal resolution) is optimized for the highest temporal resolution (4.8–6.2 ms). The third protocol (3D isotropic) is optimized for the highest volumetric spatial resolution (0.68 nL). All protocols used a bandwidth of ± 125 kHz and a nominal flip angle of 45°.
Table 1.
Three mouse heart imaging protocols that were implemented and tested for the three-dimensional radial acquisition designed and implemented in this paper.
| Protocol | Array | Spatial Res (μm) |
Voxel (nL) |
Temporal Res (ms) |
# of cardiac phases |
Scan Time (minutes) |
CNR blood/Myo |
|---|---|---|---|---|---|---|---|
| Hi throughput | 256×256×64 | 87×87×352 | 2.7 | 9.6-12.4 | 10-12 | 16 | 16.6 |
| Hi temporal res | 256×256×64 | 87×87×352 | 2.7 | 4.8-6.2 | 16-25 | 32 | 14.5 |
| Hi res 3D | 256×256×256 | 87×87×87 | 0.68 | 9.6-12.4 | 8-12 | 31 | 13.9 |
In order to measure the reproducibility of the three protocols, as well as the benefit obtained by moving to higher spatial resolution, a test was implemented. A total of 10 datasets were acquired consecutively on the same mouse on the same day. After the 6th dataset, the cradle and coil were moved by 0.5 mm to test the reproducibility of the high-throughput pulse sequence and estimate the error associated with partial volume effects when working at a lower resolution scan. The first three datasets were acquired with the high-throughput protocol (2.7 nL resolution), followed by three datasets acquired with resolution similar to other published papers (87×87×1000 μm3 - 7.7 nL). After moving the cradle, two more datasets of both the high-throughput protocol and the lower spatial resolution studies were acquired. The data was analyzed in ImageJ <http://rsb.info.nih.gov/ij/>. Precision was determined by the standard deviation of the analyzed datasets.
Reconstruction and Data Processing
Fourier space is sampled non-uniformly because of two separate effects. The most obvious is the fact that one is sampling with radial trajectories. Data is also sampled non-uniformly along any given trajectory. Since data in a given view is sampled uniformly in time, data acquired during the rising part of the gradient is thus not sampled uniformly along that radial trajectory. We employed a non-uniform fast Fourier transform (NUFFT) algorithm that addresses both problems at the same time. NUFFT is a gridding reconstruction that uses a least squares optimized kernel for interpolation that provides a balance between computation time and accuracy of interpolation potentially allowing fast and accurate results (19-21). Solving the least squares solution compensates for varying sampling density due to trajectory design and varying sampling density due to slope sampling. Reconstruction time for each 3D dataset took around 3 minutes, and for one typical high-throughput image reconstruction, the total reconstruction time was 25 minutes. Because the trajectories started in the center and progressed out, despite the use of slope-sampling, no eddy current corrections were needed as the artifacts are minimal.
Ventricular volumes were calculated using ImageJ. 3D short integer stacks were imported into ImageJ where the images were cropped to include only slices through the left ventricle. The left ventricle was excised using threshold segmentation and the total number of voxels of the left ventricle were calculated and multiplied by voxel volume to obtain volumetric measurements.
Volume rendering was performed using Vitrea (Vital Images, Inc., Minneapolis, MN) with segmentation provided by the cardiac function package. All the data shown in this manuscript is available to the interested reader at CIVMSpace, <http://www.civm.duhs.duke.edu/mrm200701/index.html>, a web portal for data-sharing and collaboration at the Center for In Vivo Microscopy.
Contrast Agent
The liposomal blood pool agent used for this work has been described in detail elsewhere (22). Briefly, a lipid mixture consisting of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (Genzyme, MA), cholesterol (Sigma, St. Louis, MO) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(poly(ethylene glycol))-2000] (Genzyme, Cambridge, MA) in the ratio 55:40:5 was dissolved in ethanol at 60°C. The mixture was then hydrated with 0.5 mol/L gadodiamide solution (Omniscan®) and stirred at 60°C. The resulting solution was sequentially extruded on Lipex thermoline extruder (Northern Lipids, Vancouver, BC, Canada) with 3 passes through 200 nm Nuclepore membrane (Whatman, Newton, MA), 4 passes through 100 nm membranes and 10 passes through 50 nm membrane. The external phase was then cleaned using a MicroKros module (Spectrum Labs, CA) of 400-kDa cut-off.
The size distribution of the liposomes in the final formulation was determined by dynamic light scattering (DLS) using a ZetaPlus Analyzer (Brookhaven Instruments, Incorporated, Chapel House, UK). The resultant size of liposomes as determined using DLS was 72 nm with a polydispersity index of 0.12. The concentration of gadolinium (Gd) in the liposomal-Gd formulation was quantified using inductively coupled plasma optical emission spectroscopy (ICPOES; Model Optima 4300D, Perkin Elmer, Norwalk, CT) operating at a wavelength of 336.223 nm. The concentration of gadolinium was 35.4 mM.
Animal Handling and Preparation
All animal studies were approved by the Duke University Institutional Animal Care and Use Committee. A total of 20 C57BL/6J mice were used in this study. Animals were induced with isoflurane and given a tail vein injection of liposomal Gd contrast agent at a dose of 0.2 mmol/kg resulting in an average injection of 0.12 mL. Animals were free-breathing and were maintained under anesthesia using an isoflurane nose cone. The ECG and breathing rate of the animals were monitored using an SA Instruments Inc. (Edison, NJ, USA) system. Heart rate was maintained between 450 and 550 beats per minute by adjusting the mixture of isoflurane and O2. A rectal temperature probe was used to monitor core body temperature in the magnet and adjusted to maintain stable body temperature (37 ± 0.5°C) throughout the course of the studies.
Results
The importance of view order randomization is illustrated with two datasets; one acquired without view order randomization (Figure 2a) and one acquired with view order randomization (Figure 2b). Both images were from the same mouse and are displayed for the same slice and same cardiac phase, while keeping all other acquisition parameters identical. Total acquisition time for both datasets was 16 minutes. To minimize changes that might have occurred over time, the images were acquired sequentially. The SNR of the myocardium in the highlighted region in Figure 2b is double that of Figure 2a, while the SNR of the blood in Figure 2b is 1.5-times that of the SNR of blood pool in Figure 2a. The improvement in SNR of the myocardium results in a nearly 30% increase in contrast-to-noise-ratio (CNR), which is critical for the semi-automated ventricular volume analysis. Note the phase artifact in Figure 2a along the septum consisting of a hypointense band in the right ventricle and a hyperintense band along the septal wall of the left ventrical. The similar banding artifacts are not present in Figure 2b. Note the of detail around the papillary muscle and the virtual disappearance of the outer wall of the left ventricle in Figure 2a. The variation in signal from the blood pool is 25% over the entire blood volume in Figure 2a, whereas the variation of the blood pool in Figure 2b is < 7 %. By calculating ventricular volumes for these datasets, the difference from one dataset to the next was over 20% and the segmentation of the dataset without view randomization took twice as long (2 minutes). The net effect of view randomization is an increase in contrast-to-noise and reduction in signal variation across the blood pool—both of which contribute to improving the consistency of myocardial volume calculations.
Figure 2.
Demonstration of the signal variations that occur when view randomization in 3DRA is not implemented (A) compared to images acquired with two randomization factors (B). Note the artifacts along the septum in (A) (arrow) and the reduced SNR relative to (B).
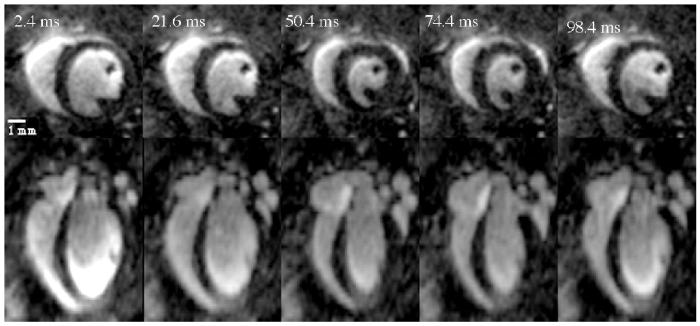
The images for the high-throughput protocol detailed in Table 1 are presented in Figure 3. The top row of Figure 3 shows the short axis view of five phases (out of a total 10) of the cardiac cycle for a representative mid-ventricle slice. The bottom row presents the same cardiac phases for a long axis view of the same dataset. The volumetric images had a temporal resolution of 9.6 ms as 4 views (TR=2.4 ms) were acquired per R-to-R interval for each phase of the heart cycle. Total acquisition time of the sequence was 16 minutes. The high-throughput protocol, with resolution of 87×87×352 μm3, had high SNR (myocardium ∼ 6.4; blood ∼ 23) and high CNR between blood and myocardium (16.6) with consistent blood pool signal that varied by < 11% over the entire blood volume. Contrast between myocardium and blood allowed visualization of wall motion, thickening of the septum, and movement of papillary muscles in the mouse heart. The consistency of the blood pool signal allowed for easy threshold segmentation.
Figure 3.
The high-throughput protocol produces images with a resolution of 87×87×352 μm3. Shown here are five representative phases of the heart cycle (from a total of 10 acquired) for a short axis slice (top row) and long axis slice (bottom row). The higher resolution (87×87 μm2) in the short axis plane is evident allowing one to follow changes in septal wall thickness and the papillary muscles throughout the cardiac cycle.
The high-throughput protocol was used to characterize the cardiac function in 10 C57BL/6J mice. The results are summarized in Table 2. Threshold segmentation allowed the determination of ejection fraction (EF), end diastolic volume (EDV), end systolic volume (ESV), and stroke volume (SV). The average weight of the 10 mice was 25.7 ± 3.4 g and the EDV calculated for the 6 mice was 59.08 ± 8.60 μL, ESV was 22.59 ± 4.09 μL, SV was 36.90 ± 4.70 μL, and EF was 62.18 ± 2.9%. The calculated values are comparable to the numbers presented elsewhere (23-25).
Table 2.
All three protocols (high-throughput, high-temporal resolution, and high-spatial resolution) were analyzed to determine end diastolic volume (EDV), end systolic volume (ESV), stroke volume (SV), and ejection fraction (EF). In addition, a study was performed on one mouse 10 times with two different resolutions to determine reproducibility of the high-throughput studies as compared to a lower-resolution study.
| Protocol | N | Repetitions | Voxel size (nL) |
Weight (g) |
EDV (μL) |
ESV (μL) |
SV (μL) |
EF |
|---|---|---|---|---|---|---|---|---|
| High-throughput | 10 | 1 | 2.7 | 25.7± 3.4 |
59.08± 8.60 |
22.59± 4.09 |
36.90± 4.70 |
0.62± .03 |
| High-temporal resolution |
5 | 1 | 2.7 | 26.1± 1.8 |
65.39± 10.06 |
26.25± 2.20 |
39.14± 8.15 |
0.60± .03 |
| High-spatial resolution |
5 | 1 | 0.68 | 25.6± 2.5 |
62.39± 9.07 |
23.28± 2.98 |
39.11± 6.68 |
0.63± .03 |
| High-throughput reproducibility |
1 | 5 | 2.7 | 25.4 | 71.06± 1.45 |
29.80± 0.66 |
41.25± 1.68 |
0.58± .01 |
| Lower spatial resolution reproducibility |
1 | 5 | 7.7 | 25.4 | 75.64± 3.74 |
30.55± 1.29 |
45.09± 3.89 |
0.60± .02 |
Images collected using the high-temporal resolution protocol detailed in Table 1 can be seen in Figure 4. Five phases of the 21-phase dataset are displayed at the same slice for each representative phase. The top row of Figure 4 shows the short axis images and the bottom row shows the long axis images. The temporal resolution of the dataset was 4.6 ms, with two views per phase for each R-to-R interval (TR=2.4 ms). Images were reconstructed at 87×87×352 μm3 resolution with a total acquisition time of 32 minutes. The SNR was lower than the high-throughput protocol (blood ∼ 19.3 myocardium ∼ 5.2), as was the CNR (14.1). The variation of the blood pool signal in each short axis slice (14%) is higher than the high-throughput. Greater variation was seen in the long axis view (28%). The high-temporal resolution protocol was applied to 5 mice and EDV, ESV, EF, and SF were calculated and listed in Table 2.
Figure 4.
High-temporal resolution dataset presents 5 phases of the heart cycle out of a total 21 phases acquired. Short axis images for 3D dataset are visible on the top row and the bottom row presents the same dataset in a long axis view.
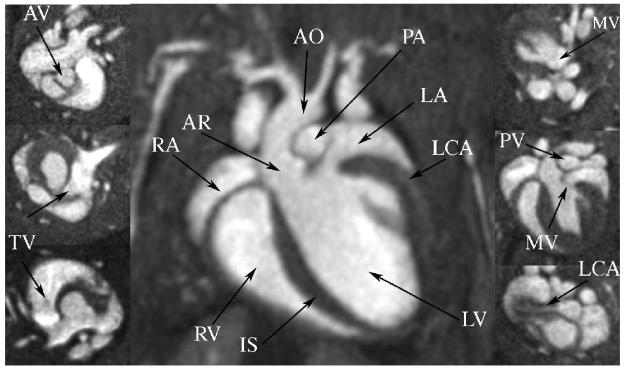
Figures 5, 6, and 7 show the results for the high-spatial resolution protocol described in Table 1. Five animals were imaged using the high-spatial resolution protocol. The top row of Figure 5 shows selected short axis images for a mid-ventricle region for five representative phases of the heart cycle. The bottom row of Figure 5 presents the images along the long axis view. Images were reconstructed at 87×87×87 μm3 for a resulting voxel volume of 0.65 nL. The temporal resolution was 12.4 ms and total acquisition time was 31 minutes. The SNR of blood was 17.7, while the SNR of myocardium was 3.8, resulting in a CNR of 13.9. The blood pool signal varied over the 3D volume by 15%, which supported threshold segmentation of the left ventricle, as is demonstrated in the volume-rendered image of the blood seen in Figure 6. The figure allows visualization of the vasculature surrounding the heart including carotid arteries, aorta, pulmonary artery, and pulmonary vein, as well as some of the microvasculature of the lung and surrounding rib cage. The isotropic high-resolution images allow visualization of the mouse heart structures including all four heart valves of the mouse heart, along with the coronary arteries (Figure 7 arrows). The high-spatial resolution protocol was applied to five mice and the calculated values for EDV, ESV, SV, and EF are presented in Table 2.
Figure 5.
Top row shows short axis view of 87 μm isotropic mouse heart images for 5 representative phases of the cardiac cycle. Bottom row shows the long axis view for the same 5 phases. All 8 phases of the heart cycle were acquired in 31 minutes.
Figure 6.
3D visualization of the high-resolution dataset, depicted in diastole (A) and systole (B). Excellent contrast-to-noise between myocardium and blood allows for visualization of veins and arteries.
Figure 7.
With isotropic 87 μm images, visualization of left coronary artery (LCA) and all 4 valves was possible. Aortic valve (AV) was visualized most clearly as the three individual leaflets could be seen followed by the tricuspid valve (TV), mitral valve (MV), and pulmonary valve (PV). Center image is a 4-chamber view of the heart showing right ventricle (RV) and atria (RA); left ventricle (LV) and atria (LA); the interventricular septum (IS); the aorta (AO); and the aortic root (AR).
The mouse heart is particularly susceptible to changes in anesthesia and temperature requiring careful monitoring of vital signs throughout the course of the studies. To test the baseline variability of the mouse's cardiac function, we acquired 5 datasets on the same mouse over a period of ∼ 3 hours. The results shown in Table 2 demonstrate a 2% baseline variability on EDV and ESV, resulting in an EF variability of 1%. The same experiment was repeated at lower spatial resolution (87×87×1000 μm3), comparable to the spatial resolution of other published literature. The variability in EDV and ESV was 4.9% and 4.2%, respectively. This resulted in a 2% precision in EF estimation. The results of a t-test performed on the end diastolic volume with the high-throughput protocol and the lower spatial resolution study were statistically different with a p<0.037. The reproducibility of the high-throughput protocol was more than twice as good as that of the lower spatial resolution study (2.0% verses 4.9% standard deviation). Shifting the cradle by 0.5 mm did not impact the reproducibility of the high-throughput protocol, as the range of the five analyzed datasets were all within 2% of one another. However, for the lower spatial resolution study, shifting the table by 0.5 mm resulted in a 7.9% increase in EDV, more than three-times the variability obtained in the high-throughput study. This suggests that the high-throughput protocol is reproducible and that over the course of the 3 hours that the study was being performed, the animal was stable and the functional measurements (EF) were consistent to 1%.
Discussion
Relative to ultrasound, MR has suffered from limited temporal resolution and long acquisition times. In order to address these shortcomings, we have implemented a radial pulse sequence to increase SNR, increase temporal resolution, and allow for rapid data acquisition. Radial acquisition (RA) was responsible for the creation of the first MR images (11), but until recently has fallen out of favor because non-Cartesian acquisitions have several disadvantages. Non-Cartesian datasets (such as radial acquisitions) require longer reconstruction time and suffer from lower SNR than their Cartesian counterparts (26). Radial acquisitions, particularly the center-out trajectories implemented here, require more than double the number of views for Nyquist sampling than a standard Cartesian sequence. Despite these shortcomings, advantages over standard Cartesian imaging: motion resistance; resistance to undersampling; and short echo and repetition times. The motion resistance of radial acquisitions is a result of several features: inherent first order and second order moment nulled gradients (center-out trajectories only) and oversampling of the center of Fourier space. Radial acquisitions vastly oversample the center of Fourier space, as each view samples the center, which allows variation due to motion (or view-to-view variation) to be minimized by spreading the motion effects in the radial direction (16). The resistance of RA to undersampling is also due to the oversampling of the center as any artifacts resulting from undersampling will be spread in the radial dimension leaving the center of Fourier space fully sampled (27). In addition, radial acquisitions are well suited to self-gating techniques (28), as phase information due to the motion of the heart and lungs is collected as the center of k-space is sampled for every view. The technique implemented here could be extended to include self-gating techniques, which may result in further improvement in acquisition time. In this implementation, the short echo times achievable with RA were used to maximize the signal of the very small 0.68 nL isotropic voxels (29).
While MR microscopy has become a standard tool for mouse heart imaging, there is room for improvement in spatial resolution, temporal resolution, and acquisition time. The high-throughput images, acquired in 16 minutes, will allow us to improve the precision of functional measurements relative to previous MR work. The majority of literature reports slice-selective techniques with z resolution of 1mm (1-4). Moving to a 3D acquisition has allowed us to improve the resolution in z by a factor of 3. To date, only one 4D method has been implemented on a mouse, acquiring 200-micron isotropic images (8 nL) in 1-2 hours (6). The high-throughput protocol outlined in this paper produces 87×87×352 μm3 (2.7 nL) images with 10–12 phases of the heart cycle in 15 minutes. This represents an improvement in spatial resolution of 3X with a 4X reduction in acquisition time.
The short repetition times possible with RA create the ability to obtain flexible temporal resolution. Temporal resolution is limited by TR and most literature on MR mouse heart imaging reports TRs of 7.7-14 ms (2,30,31), resulting in a maximum temporal resolution of 7.7 ms. The fastest repetition time reported for traditional Cartesian sequences of the mouse heart is 4.3 ms (1,32), whereas with the radial acquisition technique proposed here, it is possible to obtain temporal resolutions as low as 2.4 ms.
Higher spatial resolution scans have been performed at single time points in the cardiac cycle with resolutions of 78×78×300 μm3 (1.8 nL) (33), 100×100×200 μm3 (2 nL) (34), and 49×98×300 μm3 (1.4 nL) (35). These data were acquired with a variation of slice-selective acquisitions and thin (6.4mm) 3D slabs, so full coverage of the heart would require 51 to 70 minutes for a single phase of the heart cycle. The high-resolution protocol with isotropic resolution of 87 μm (0.68 nL) is an improvement in spatial resolution of 2-3 times, including 8 phases of the heart cycle at 34 minutes, i.e. up to 50% shorter acquisition time. The protocol allows routine visualization of the coronary arteries and valves, a feat which has only been demonstrated in two other publications (30,34). Sosnovik et al. have acquired CINE studies with 1 mm slices (22.5 nL) in which the left coronary is seen throughout the cycle (30). Acquisition time was 8–10 minutes per slice.
Thus coverage of the entire heart will take at least 60 minutes. The images in Figures 5-7 were acquired in half the time with 34-times higher spatial resolution. The CNR between the blood and myocardium is very high (13.9) and the high-spatial resolution allows for visualization of all four heart valves and the left coronary artery. Furthermore, the higher spatial resolution, with 0.68-2.7 nL voxels, reduces the partial volume effects resulting in a more reproducible study.
Conclusion
A 4D radial acquisition was developed to increase the SNR resulting in high-resolution cine mouse heart images. Three separate protocols were designed to increase throughput, increase temporal resolution, and increase spatial resolution. The high-throughput images, acquired in 16 minutes, with high-spatial resolution of 87×87×352 μm3 allows us to significantly improve the sensitivity and reproducibility of functional measurements relative to previous MR work. The datasets in Figures 5-7 (87×87×87 μm3) are the highest resolution CINE MR images of the whole live mouse heart to date. The high-spatial resolution will allow calculations of the ventricular volumes to be less susceptible to partial volume effects and visualization of abnormal morphologic phenotypes, e.g. valve defects, aortic stenosis, or other abnormalities of the supra-aortic vessels. The methods represent a significant step forward in our ability to characterize both structure and function in the mouse heart.
Acknowledgements
This work was performed at the Duke Center for In Vivo Microscopy, an NCRR/NCI Biomedical Technology Resource Center (P41 RR005959, U24 CA092656). We would like to thank Gary Cofer for his help with coil design, Sally Gewalt, and Dr. Jiayu Song for help with reconstruction, Dr. Laurence Hedlund for advice with animal support and handling, Jeff Brandenburg for his help setting up the website materials, and Sally Zimney for editorial assistance in preparing the manuscript.
References
- 1.Dawson D, Lygate CA, Saunders J, Schneider JE, Ye X, Hulbert K, Noble JA, Neubauer S. Quantitative 3-dimensional echocardiography for accurate and rapid cardiac phenotype characterization in mice. Circulation. 2004;110(12):1632–1637. doi: 10.1161/01.CIR.0000142049.14227.AD. [DOI] [PubMed] [Google Scholar]
- 2.Zhou R, Pickup S, Glickson JD, Scott CH, Ferrari VA. Assessment of global and regional myocardial function in the mouse using cine and tagged MRI. Magn Reson Med. 2003;49(4):760–764. doi: 10.1002/mrm.10423. [DOI] [PubMed] [Google Scholar]
- 3.Cassidy PJ, Schneider JE, Grieve SM, Lygate C, Neubauer S, Clarke K. Assessment of motion gating strategies for mouse magnetic resonance at high magnetic fields. J Magn Reson Imaging. 2004;19(2):229–237. doi: 10.1002/jmri.10454. [DOI] [PubMed] [Google Scholar]
- 4.Berr S, Roy RJ, French BA, Yang Z, Gilson W, Kramer CM, Epstein F. Black Blood Gradient Echo Cine Magnetic Resonance Imaging of the Mouse Heart. Magn Reson Med. 2005;53:1074–1079. doi: 10.1002/mrm.20487. [DOI] [PubMed] [Google Scholar]
- 5.Brau ACS, Hedlund LW, Johnson GA. Cine magnetic resonance microscopy of the rat heart using cardiorespiratory-synchronous projection reconstruction imaging. J Magn Reson Imaging. 2004;20:31–38. doi: 10.1002/jmri.20089. [DOI] [PubMed] [Google Scholar]
- 6.Feintuch A, Zhu Y, Bishop J, Davidson L, Dazai J, Bruneau B, Henkelman R. 4D cardiac MRI in the mouse. NMR Biomed. 2007;20:360–365. doi: 10.1002/nbm.1164. [DOI] [PubMed] [Google Scholar]
- 7.Barger AV, Grist TM, Block WF, Mistretta CA. Single breath-hold 3D contrast-enhanced method for assessment of cardiac function. Magn Reson Med. 2000;44:821–824. doi: 10.1002/1522-2594(200012)44:6<821::aid-mrm1>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 8.Amano Y, Herfkens RJ, Shifrin RY, Alley MT, Pelc NJ. Three-dimensional cardiac cine magnetic resonance imaging with ultrasmall superparamagnetic iron oxide blood pool agent (NC100150) J Magn Reson Imaging. 2000;11:81–86. doi: 10.1002/(sici)1522-2586(200002)11:2<81::aid-jmri2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 9.Peters DC, Ennis DB, Rohatgi P, Syed MA, McVeigh ER, Arai AE. 3D breath-held cardiac function with projection reconstruction in steady state free precession validated using 2D cine MRI. J Magn Reson Imaging. 2004;20:411–416. doi: 10.1002/jmri.20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhettmann DW, Saranathan M, Wu KC, Azevedo CF, Bluemke DA, Foo TKF. High Temporal Resolution Breathheld 3D FIESTA CINE imaging: Validation of ventricular function in patients with chronic myocardial infarction. J Magn Reson. 2007;25:1141–1146. doi: 10.1002/jmri.20923. [DOI] [PubMed] [Google Scholar]
- 11.Lauterbur PC. Image formation by induced local interactions - examples employing nuclear magnetic resonance. Nature. 1973;242:190–191. [PubMed] [Google Scholar]
- 12.Wong ST, Roos MS. A strategy for sampling on a sphere applied to 3D selective RF pulse design. Magn Reson Med. 1994;32(6):778–784. doi: 10.1002/mrm.1910320614. [DOI] [PubMed] [Google Scholar]
- 13.Chen BT, Yordanov AT, Johnson GA. Ventilation-synchronous MR microscopy of pulmonary structure and ventilation in mice. Magn Reson Med. 2005;53(1):69–75. doi: 10.1002/mrm.20307. [DOI] [PubMed] [Google Scholar]
- 14.Theilmann RJ, Gmitro AF, Altbach MI, Trouard TP. View-ordering in radial fast spin-echo imaging. Magn Reson Med. 2004;51(4):768–774. doi: 10.1002/mrm.20031. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein MA, KIng KF, Zhou XJ. Handbook of MRI Pulse Sequences. Elsevier Academic Press; San Diego: 2004. [Google Scholar]
- 16.Glover G, Pauly J. Projection reconstruction techniques for reduction of motion effects in MRI. Magn Reson Med. 1992;28:275–289. doi: 10.1002/mrm.1910280209. [DOI] [PubMed] [Google Scholar]
- 17.Peters DC, Korosec FR, Grist TM, Block WF, Holden JE, Vigen KK, Mistretta CA. Undersampled projection reconstruction applied to MR angiography. Magn Reson Med. 2000;43:91–101. doi: 10.1002/(sici)1522-2594(200001)43:1<91::aid-mrm11>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 18.Barger AV, Block WF, Toropov Y, Grist TM, Mistretta CA. Time-Resolved contrast-enhanced imaging with isotropic resolution and broad coverage using undersampled 3D projection trajectory. Magn Reson Med. 2002;48:297–305. doi: 10.1002/mrm.10212. [DOI] [PubMed] [Google Scholar]
- 19.Song J, Liu QH. Improving non-cartesian MRI reconstruction through discontinuity subtraction. Int J Bio Imag. 2006:1–9. doi: 10.1155/IJBI/2006/87092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sha L, Guo H, Song AW. An improved gridding method for spiral MRI using nonuniform fast Fourier transform. J Magn Reson. 2003;162:250–258. doi: 10.1016/s1090-7807(03)00107-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu QH, Nguyen N. An accurate algorithm for nonuniform fast Fourier transform (NUFFT's) IEEE Microwave and Guided Wave Letters. 1998;8(1):18–20. [Google Scholar]
- 22.Ghaghada K, Bockhorst K, Mukundan J,S, Annapragada A, Narayana P. High resolution vascular imaging of the rat spine using liposomal blood pool MR agent. Am J Neuroradiol. 2007;28(1):48–53. [PMC free article] [PubMed] [Google Scholar]
- 23.Heijman E, de Graaf W, Niessen P, Nauerth A, van Eys G, de Graaf L, Nicolay K, Strijkers GJ. Comparison between prospective and retrospective triggering for mouse cardiac MRI. NMR Biomed. 2007;20(4):439–447. doi: 10.1002/nbm.1110. [DOI] [PubMed] [Google Scholar]
- 24.Badea C, Fubara B, Hedlund L, Johnson G. 4D micro-CT of the mouse heart. Molecular Imaging. 2005;4(2):110–116. doi: 10.1162/15353500200504187. [DOI] [PubMed] [Google Scholar]
- 25.Nahrendorf M, Badea C, Hedlund L, Figueiredo J, Sosnovik D, Johnson GA, Weissleder R. High resolution imaging of murine myocardial infarction with delayed enhancement and cine micro-CT. Am J Physiol Heart Circ Physiol. 2007;292:3172–3178. doi: 10.1152/ajpheart.01307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauzon ML, Rutt BK. Effects of polar sampling in k-space. Magn Reson Med. 1996;36(6):940–949. doi: 10.1002/mrm.1910360617. [DOI] [PubMed] [Google Scholar]
- 27.Lethmate R, Wajer FTAW, Cremillieux Y, Van Ormondt D, Graveron-Demilly D. Dynamic MR-imaging with radial scanning, a post-acquisition keyhold approach. EURASIP Journal on Applied Signal Processing. 2003;5:405–412. [Google Scholar]
- 28.Hiba B, Richard N, Croisille P. Cardiac and respiratory double self-gated cine MRI in the mouse at 7T. Magn Reson Med. 2006;2006(55):506–513. doi: 10.1002/mrm.20815. [DOI] [PubMed] [Google Scholar]
- 29.Gewalt SL, Glover GH, Hedlund LW, Cofer GP, MacFall JR, Johnson GA. MR microscopy of the rat lung using projection reconstruction. Magn Reson Med. 1993;29(1):99–106. doi: 10.1002/mrm.1910290117. [DOI] [PubMed] [Google Scholar]
- 30.Sosnovik DE, Dai G, Nahrendorf M, Rosen BR, Seethamraju R. Cardiac MRI in mice at 9.4 tesla with a transmit-receive surface coil and a cardiac-tailored intensity-correction algorithm. J Magn Reson Imaging. 2007;26:279–287. doi: 10.1002/jmri.20966. [DOI] [PubMed] [Google Scholar]
- 31.Ross AJ, Yang Z, Berr S, Gilson W, Petersen WC, Oshinski JN, French BA. Serial MRI evaluation of cardiac structure and function in mice after reperfused myocardial infarction. Magn Reson Med. 2002;47(6):1158–1168. doi: 10.1002/mrm.10166. [DOI] [PubMed] [Google Scholar]
- 32.Wiesmann F, Ruff J, Engelhardt S, Hein L, Dienesch C, Leupold A, Illinger R, Frydrychowicz A, Hiller KH, Rommel E, Haase A, Lohse MJ, Neubauer S. Dobutamine-stress magnetic resonance microimaging in mice: acute changes of cardiac geometry and function in normal and failing murine hearts. Circ Res. 2001;88(6):563–569. doi: 10.1161/01.res.88.6.563. [DOI] [PubMed] [Google Scholar]
- 33.Itskovich VV, Choudhury RP, Aguinaldo JGS, Fallon JT, Omerhodzic S, Fisher EA, Fayad ZA. Characterization of aortic root atherosclerosis in ApoE knockout mice: High resolution in vivo and ex vivo MRM with histological correlation. Magn Reson Med. 2003;49(2):381–385. doi: 10.1002/mrm.10360. [DOI] [PubMed] [Google Scholar]
- 34.Ruff J, Wiesmann F, Lanz T, Haase A. Magnetic resonance imaging of coronary arteries and heart valves in a living mouse: techniques and preliminary results. J Magn Reson. 2000;146(2):290–296. doi: 10.1006/jmre.2000.2147. [DOI] [PubMed] [Google Scholar]
- 35.Wiesmann F, Szimtenings M, Frydrychowicz A, Illinger R, Hunecke A, Rommel E, Neubauer S, Haase A. High-resolution MRI with cardiac and respiratory gating allows for accurate in vivo atherosclerotic plaque visualization in the murine aortic arch. Magn Reson Med. 2003;50(1):69–74. doi: 10.1002/mrm.10500. [DOI] [PubMed] [Google Scholar]