Abstract
Macrophage migration inhibitory factor (MIF) is a secreted protein expressed in numerous cell types that counters the anti-inflammatory effects of glucocorticoids and has been implicated in sepsis, cancer and certain auto-immune diseases. Interestingly, the structure of MIF contains a catalytic site resembling the tautomerase/isomerase sites of microbial enzymes. While bona fide physiological substrates remain unknown, model substrates have been identified. Selected compounds that bind in the tautomerase active site also inhibit biological functions of MIF. It had previously been shown that the acetaminophen metabolite, N-acetyl-p-benzoquinone imine (NAPQI), covalently binds to the active site of MIF. In this study, kinetic data indicate that NAPQI inhibits MIF both covalently and non-covalently. The structure of MIF co-crystallized with NAPQI reveals that the NAPQI has undergone a chemical alteration forming an acetaminophen dimer (bi-APAP), and binds non-covalently to MIF at the mouth of the active site. We also find that the commonly used protease inhibitor, phenylmethylsulfonyl fluoride (PMSF), forms a covalent complex with MIF and inhibits the tautomerase activity. Crystallographic analysis reveals the formation of a stable, novel covalent bond for PMSF between the catalytic nitrogen of the N-terminal proline and the sulfur of PMSF with complete, well-defined electron density in all three active sites of the MIF homotrimer. Conclusions are drawn from the structures of these two MIF-inhibitor complexes regarding the design of novel compounds that may provide more potent reversible and irreversible inhibition of MIF.
Macrophage migration inhibitory factor (MIF1) is a pro-inflammatory cytokine that counteracts the anti-inflammatory activities of glucocorticoids (1). MIF is also a potential target for developing treatments for many inflammatory diseases, including septic shock (2, 3) and various auto-immune diseases (4–6) and is implicated in a variety of cancers (7, 8) and in cardiovascular disease (9).
MIF is unusual among cytokines because it both signals through a cell surface receptor and catalyzes chemical reactions through an enzymatic active site. The receptor for MIF has been identified as CD74 (10), which forms a complex with CD44 (11) leading to activation of Src-family kinases. Two separate catalytic activities and active sites have been described: an oxido-reductase requiring Cys 56 and Cys 59 (12), and a tautomerase reaction requiring the N-terminal proline (13). Although a physiological substrate remains unknown, the tautomerase activities have been better characterized. Non-physiological substrates that have been found include D-dopachrome and L-dopachrome methyl ester (13). Two potential physiological substrates for the tautomerase activity also have been described, phenylpyruvic acid and 4-hydroxyphenylpyruvic acid (HPP) (14) but whether these are bona fide substrates is yet unknown. Although a cause-effect relationship has not been established between these enzymatic activities and the cytokine activity of MIF, selected molecules that inhibit MIF tautomerase activity also diminish the pro-inflammatory action of MIF in vitro and in vivo (15, 16). The activity of the inhibitors depends on the homotrimeric form of MIF because the tautomerase active site is at a subunit-subunit interface (17).
Senter et al. previously reported on the inhibition of MIF tautomerase activity by N-acetyl-para-benzoquinone imine (NAPQI), a metabolite of the commonly used analgesic/antipyretic drug, acetaminophen (18). Using mass spectrometry, a portion of MIF was found to be covalently derivatized by NAPQI. At NAPQI concentrations that yielded 96% inhibition, only approximately one-third of MIF was covalently complexed to NAPQI. This indicated that most of the NAPQI formed a non-covalent complex with MIF. We have performed kinetic and structural experiments to elucidate further the nature of the interactions between NAPQI and MIF. In the meanwhile, we also discovered that the serine protease inhibitor PMSF covalently modifies MIF, forming an unexpected nitrogen-sulfur bond, and also report the kinetics of PMSF inhibition of MIF and the structure of PMSF-bound MIF.
Experimental Procedures
Protein Purification
MIF was purified in a similar manner to that previously described (19). MIF was expressed in E. coli BL21(DE3). Harvested cells were frozen, thawed and lysed in 20 mM NaCl, 20 mM Tris (pH 7.5) using a French Pressure Cell. The soluble fractions of the lysate were filtered and the protein was purified from the filtrate by ion-exchange chromatography through DEAE and SP columns connected in series. Fractions were collected from the flow-through, and those containing pure MIF were pooled. Gel filtration was performed afterwards when deemed necessary, and the protein was concentrated.
Preparation of PMS-MIF Complex and Mass-Spectrometry
One microliter of 200 mM PMSF in isopropanol was added to 300 μl 0.43 mM MIF. This solution was allowed to incubate overnight at room temperature. Mass spectrometry showed that not all of the MIF was derivatized (see Supporting Information). Therefore, to increase the fraction of covalently derivatized MIF, 2 μl of 200 mM PMSF was added to the already prepared PMS-MIF complex. Mass spectrometry was used to demonstrate that all of the MIF was then derivatized. The complex was then concentrated and used for crystallization.
Enzyme Kinetics
All measurements were performed using the Tecan Infinite M200 spectrophotometer with a Corning semi-transparent 96-well plate. MIF-PMSF incubation reactions were prepared by mixing 14.9 μl of 0.21 mM MIF (in 20 mM NaCl, 20 mM Tris, pH 7.5) with 0.7 μl of either isopropanol or 0.20 M PMSF in isopropanol, yielding 0.20 mM MIF, 19 mM NaCl, 19 mM Tris and 4.8% isopropanol with or without 9.5 mM PMSF. Reactions were allowed to incubate at room temperature. At various times of incubation, a 0.8 μl aliquot was removed and added to 719 μl of 442 mM borate, pH 6.2. This was mixed and 144 μl were immediately added to an assay well, followed by the addition of 6.0 μl of 25 mM HPP (in 0.5 M ammonium acetate). The well contents were mixed and absorbance measurements were recorded at 306 nm at one-second intervals.
For the MIF-NAPQI incubation reactions, equal volumes of 0.21 mM MIF and either DMSO or 10 mM NAPQI in DMSO were mixed and incubated at room temperature. (Incubation concentrations were 0.10 mM MIF, 10 mM NaCl, 10 mM Tris and 50% DMSO, with or without 5.0 mM NAPQI). At various incubation times, 0.6 μl was added to 287 μl of 443 mM borate, pH 6.2, and the solution was mixed (the final DMSO concentration was 0.1%). The assay was then performed with 144 μl of this solution and 6.0 μl of 25 mM HPP, as in the MIF-PMSF assay. Curve fitting for both the PMSF and NAPQI experiments was performed using Graph Pad Prism 4 software.
Crystallization and X-Ray Data Collection
Crystallization was performed using the hanging-drop vapor diffusion method. For native MIF crystallization, 2 μl of 19 mg/ml MIF (1.6 mM) was mixed with 2 μl of reservoir containing 50% saturated ammonium sulfate, 0.1 M Tris (pH 7.5), and 3% isopropanol. Crystals grew within one day. One crystal was briefly soaked in cryo-protectant consisting of 25% glycerol, 50% saturated ammonium sulfate, 0.1 M Tris (pH 7.5), 3% isopropanol. Diffraction data were collected at beamline X25 of the National Synchrotron Light Source at Brookhaven National Laboratory.
The PMS-MIF complex was crystallized in a similar manner. Two microliters of 0.9 mM PMS-MIF was mixed with 2 μl of reservoir containing 50% saturated ammonium sulfate, 0.1 M Tris (pH 7.5), and 4% isopropanol. A crystal was mounted on a Rigaku x-ray generator with an R-Axis IV++ detector after a brief soak in cryo-protectant consisting of 25% glycerol, 50% saturated ammonium sulfate, 0.1 M Tris (pH 7.5), 4% isopropanol.
For MIF-NAPQI co-crystallization, a 0.1 M NAPQI stock in DMSO was used to prepare a solution containing 1.16 mM MIF, 5 mM NAPQI in 5% DMSO. The complex was crystallized in the above-described manner using 2 M (NH4)2SO4, 0.1 M Tris (pH 7.5), 3% isopropanol for the reservoir. The crystal was soaked in a cryo-protectant consisting of 2.25 M NaCl, 2 M (NH4)2SO4, 50 mM Tris (pH 7.5) and mounted on the x-ray unit.
For all data sets, one-degree oscillation frames were processed with HKL2000 (20). Data collection statistics are listed in Table 1.
Table 1.
Crystallographic Statisticsa
| PMS-MIF | bi-APAP-MIF | Native MIFb | |
|---|---|---|---|
| Integration and Scaling | |||
| Space group | P212121 | P21 | P212121 |
| Unit cell | a = 67.40 Å, b = 67.51 Å, c = 88.83 Å, α = β = γ =90° | a = 69.97 Å, b = 60.90 Å, c = 77.81 Å, α = γ = 90°, β = 93.28° | a = 68.23 Å, b = 68.26 Å, c = 86.51 Å, α = β = γ =90° |
| Resolution, A (highest shell) | 1.55 (1.61–1.55) | 1.95 (2.02–1.95) | 1.25 (1.29–1.25) |
| Unique Reflections | 58,940 (5,478) | 47,844 (4,772) | 110,755 (10,714) |
| Completeness, % | 99.1 (93.2) | 99.7 (100) | 99.0 (96.9) |
| Redundancy | 4.7 (2.8) | 3.6 (3.6) | 4.6 (4.4) |
| Avg. I/Avg. σ | 26.52 (2.37) | 20.5 (2.41) | 17.5 (2.27) |
| Rmerge | 0.069 (0.316) | 0.070 (0.443) | 0.101 (0.498) |
| Refinementc | |||
| R (working) | 19.5% | 20.92% | 17.0% |
| R-free | 20.9% | 24.73% | 18.6% |
| RMS deviations from idealityd | |||
| Bond lengths (Å) | 0.012 | 0.011 | 0.008 |
| Bond angles (°) | 1.3 | 1.5 | 1.1 |
| Number of atoms per asymmetric unit | 3151 | 5580 | 3165 |
| Average B-factors, Å2 | |||
| Overall | 16.4 | 32.1 | 16.1 |
| Protein (number of residues) | 13.1 (114 × 3 chains) | 31.6 (114 ×6) | 13.5 (114 ×3) |
| Water | 32.0 (464) | 37.1 (349) | 29.9 (466) |
| Chloride | NA (0) | 43.6 (15) | NA (0) |
| Sulfate | 45.1 (5) | NA (0) | 22.9 (6) |
| Isopropanol | 29.5 (2) | NA (0) | 22.5 (3) |
| Glycerol | 42.4 (3) | NA (0) | 22.7 (4) |
| Ligands | 16.2 (3 PMS) | 46.0 (1 bi-APAP) | NA (0) |
NA: not applicable
with glycerol from the cryo-protectant bound in the active sites
Data cutoff for refinement of all structures was F > 0.
compared to values from Engh and Huber (46) as computed by the RCSB Protein Data Bank
Structure Determination
Protein coordinates from PDB entry 1CA7 (MIF bound to HPP) were used in molecular replacement with AMoRe (21, 22) to generate an initial model for the PMS-MIF complex. Likewise, protein coordinates from the PMS-MIF complex were used for the search model for the high-resolution native MIF data using AMoRe. For the acetaminophen dimer (bi-APAP)-MIF complex – from the MIF-NAPQI co-crystallization – protein coordinates from our new high-resolution native MIF structure was used as a search model, using Phaser (23) for molecular replacement. Crystallographic refinement was performed using CNS (24). ShelxL (25) was also used for the native MIF structure, and Refmac5 (22, 26) was employed for both the native and PMS-MIF structures. For the PMS-MIF structure, TLS segments were found using the TLSMD server at the University of Washington (27, 28), and refinement of TLS parameters for the PMS-MIF complex was performed using Refmac5. Manual adjustments to the models were made using O (29), COOT (30) and MIFit (31). Crystallographic statistics are listed in Table 1.
Superposition of Protein Structures
Superpositions of protein structures were performed using LSQKAB (32) in the CCP4 package (22). When the PMS-MIF structure was overlaid on the native structure using resides 2–114, it was noticed that residues 32, 51 and 114 had large deviations in all subunits relative to the nearby residues. Pro 1 was not considered in the overlay in order not to bias the analysis of the relative position and orientation of Pro 1 amongst the structures. Therefore, the positions of α-carbons of amino acids 2–31, 37–50 and 52–113 were used for all overlays.
Results
Kinetic Analysis of MIF Covalent Modification
Acetaminophen metabolites are lethal hepatotoxins that act by depleting intracellular glutathione (33). The major metabolite of acetaminophen, NAPQI is formed from acetaminophen via the action of cytochrome P450 (34–36) and it is normally detoxified via reduction by glutathione to form a glutathione-acetaminophen adduct (37). NAPQI was reported to form an irreversible adduct with MIF (18). Kinetic analysis of MIF HPP tautomerase activity at various concentrations of NAPQI does reveal concentration dependent NAPQI-mediated inhibition of MIF. Michaelis-Menten kinetics is not observed, as expected, for an irreversible inhibitor (data not shown). We also identified PMSF as a covalent inhibitor (Supplementary Figure 1) and examined its kinetic properties. In contrast to NAPQI, very little inhibition of enzymatic activity was detected when 20 nM MIF and 1 mM PMSF were mixed immediately prior to performing the assay (data not shown). Therefore, we concluded that the kinetics of MIF modification by PMSF must be slow.
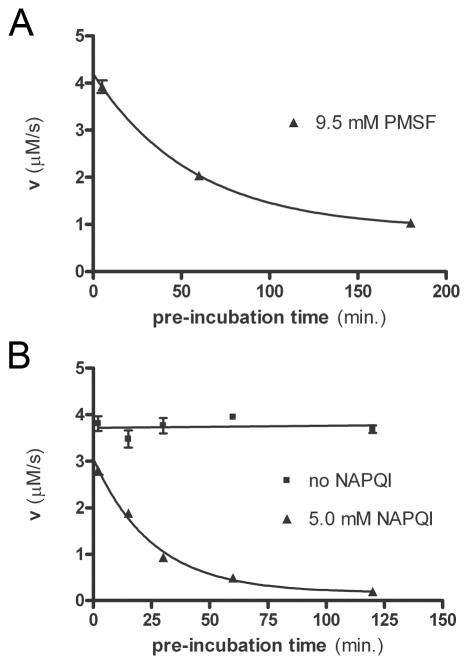
In order to determine the time frame of MIF modification by these two compounds, we performed incubation assays in which we mixed MIF with either NAPQI (dissolved in DMSO), DMSO alone, PMSF (dissolved in isopropanol), or isopropanol alone. After various periods of incubation times, a portion of the sample was diluted into HPP tautomerase assay buffer, and kinetic assays were immediately performed. Figure 1 shows that PMSF and NAPQI both inhibit MIF in a time-dependent fashion, consistent with a mechanism involving covalent modification. The activity of MIF upon incubation with either of these fits a single-exponential decay curve. We define the “half-life” of activity as being the equivalent of the half-life of a species undergoing exponential decay; that is, the amount of pre-incubation time it takes for the enzyme’s activity to decrease by 50%. The formation of covalent adducts using 9.5 mM PMSF or 5.0 mM NAPQI (48-fold molar excess of enzyme) is slow, with half-lives of 39 minutes and 18 minutes for PMSF- and NAPQI-mediated inhibition, respectively. The remaining 21% activity for MIF after a long incubation time with PMSF is likely due to the instability of PMSF in aqueous solutions (38). This instability leads to hydrolysis of PMSF in aqueous solution, which inactivates it.
Figure 1.
Kinetic Analysis Reveals Time-Dependent Inhibition of MIF by both PMSF and NAPQI. HPP tautomerase activity of MIF was measured after various times of incubation with (A) PMSF or (B) NAPQI. Inhibitor concentrations shown are those during the incubations, and were in 48-fold molar excess of MIF. After incubation with inhibitor, the MIF solution was diluted to 0.2 μM MIF in tautomerase assay buffer and activity was measured. Data shown are from triplicate experiments. Mean ± SEM are plotted for all data. For many points, the error bars are too small to be seen beyond the data point symbol.
MIF activity was extrapolated to zero minutes of incubation. Interestingly, MIF displays only 82% of full activity at ‘zero minutes’ of incubation with NAPQI (Figure 1). This suggests that NAPQI is able to inhibit MIF activity even without covalently modifying the protein. This implies two modes of inhibition: both covalent and non-covalent. Initially, a non-covalent complex forms, which is of sufficient affinity to inhibit HPP tautomerase activity. Chemical reactivity between the enzyme and NAPQI is gradual such that adduct formation occurs very slowly. However, once it occurs, the adduct is irreversible. This is consistent with findings from Senter et al. who found that MIF activity is inhibited by 96% under conditions in which they estimate that only 1/3 of the protein has been covalently modified (18). In contrast, extrapolation of the PMSF-treated MIF data to zero minutes confirms that there is no immediate inhibition. This is interpreted as PMSF binding to MIF quite poorly; however, once it is bound, a covalent complex is quickly and irreversibly formed.
Crystallographic Structures of MIF-Inhibitor Complexes
(A) Structure of MIF Co-Crystallized with NAPQI
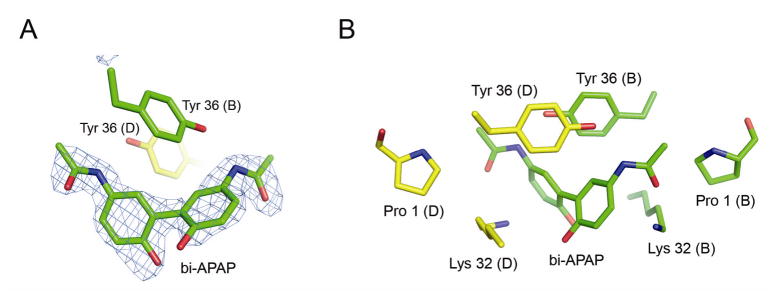
Upon determining the structure of MIF co-crystallized with NAPQI, we found that the bound ligand was in fact a dimer of acetaminophen (Figure 2A). The acetaminophen dimer (bi-APAP) has been found to be catalyzed from acetaminophen by horseradish peroxidase in the presence of H2O2 (39). It has been reported by Potter and Hinson that NAPQI is able to react non-enzymatically with acetaminophen to form acetaminophen dimers and longer acetaminophen polymers (37). They also found that when NAPQI is incubated in the absence of acetaminophen, low levels of acetaminophen dimer formation occur. However, it is possible that this was due to very small amounts of acetaminophen present in the NAPQI stock. Nonetheless, they did not find NAPQI-NAPQI or NAPQI-acetaminophen dimers. We therefore modeled the ligand found in our structure as an acetaminophen dimer (bi-APAP).
Figure 2.
Acetaminophen dimer (bi-APAP) bound to MIF. A. Fo-Fc omit map calculated by omitting bi-APAP from the electron density calculation, contoured at 3σ. The ligand is found between two MIF trimers related by crystallographic symmetry. Subunit B of one trimer is colored with carbon atoms in green and subunit D of a symmetry-related trimer is colored with carbon atoms in yellow. Aromatic hydrophobic interactions with a Tyr 36 residue of each molecule are shown. B. Bi-APAP bridges one of the active sites of one trimer with an active site of the other. This figure, as well as figures 3A, 4 and 5, was prepared using Pymol (49).
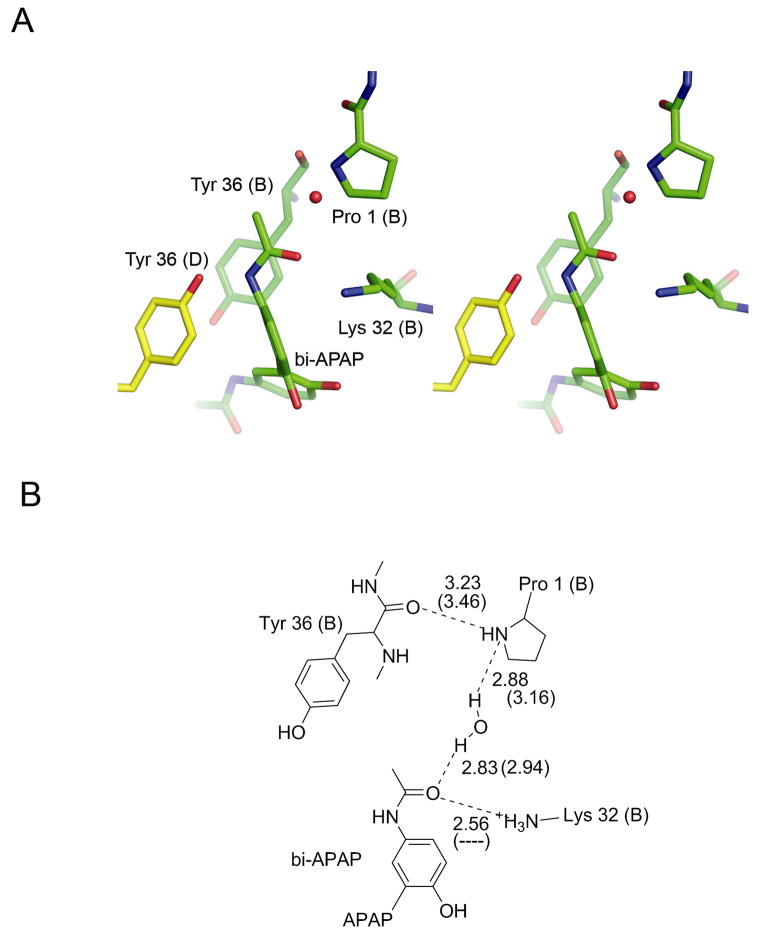
The acetaminophen dimer was bound non-covalently at the mouth of the active site, instead of in the active site cleft as seen with other ligands. The co-crystals grew with a different morphology and space group than we have previously observed with MIF in the same crystallization conditions. This new crystal form contains two MIF trimers in the asymmetric unit. After solving the structure, we found that the different crystal packing was due to the dimerized ligand bridging the active sites of symmetry-related molecules (Supporting Information). Binding of bi-APAP to MIF is facilitated by hydrophobic interactions with Tyr 36 of two symmetry-related copies of the protein that pack with bi-APAP, each tyrosine abutting a different ring of bi-APAP (Figure 2). Each methyl amide group of bi-APAP binds in the mouth of an MIF active site (Figure 2B and Figure 3). Figure 3 shows the detail of these interactions. Although the positioning of bi-APAP in each active site is similar to the other, the interactions are not quite the same. The water-mediated hydrogen bond to the Pro 1 nitrogen is shorter, and presumably stronger, in active site B. Moreover, a hydrogen bond from Lys 32 in active site B is lost in active site D, in which the amino group of Lys 32 is in close proximity to the phenyl ring of bi-APAP, where it can interact with the bi-APAP phenyl ring π-cloud (Figure 3). Therefore, the interactions are stronger with active site B than with active site D.
Figure 3.
Interactions of bi-APAP with active site residues. A. Three-dimensional stereo view of interactions in active site B. The interactions in active site D are similar, but not quite as strong. B. Schematic representation of active site hydrogen bonding. Hydrogen bonds are labeled with the gdistances between non-hydrogen atoms as found in active site B with the distances in active site D in parentheses. Distances are in Ångstroms. Lys 32 in subunit D is not positioned to have a good angle for hydrogen bonding; this lack of a hydrogen bond is noted by a dashed line.
(B) PMS-MIF Covalent Complex
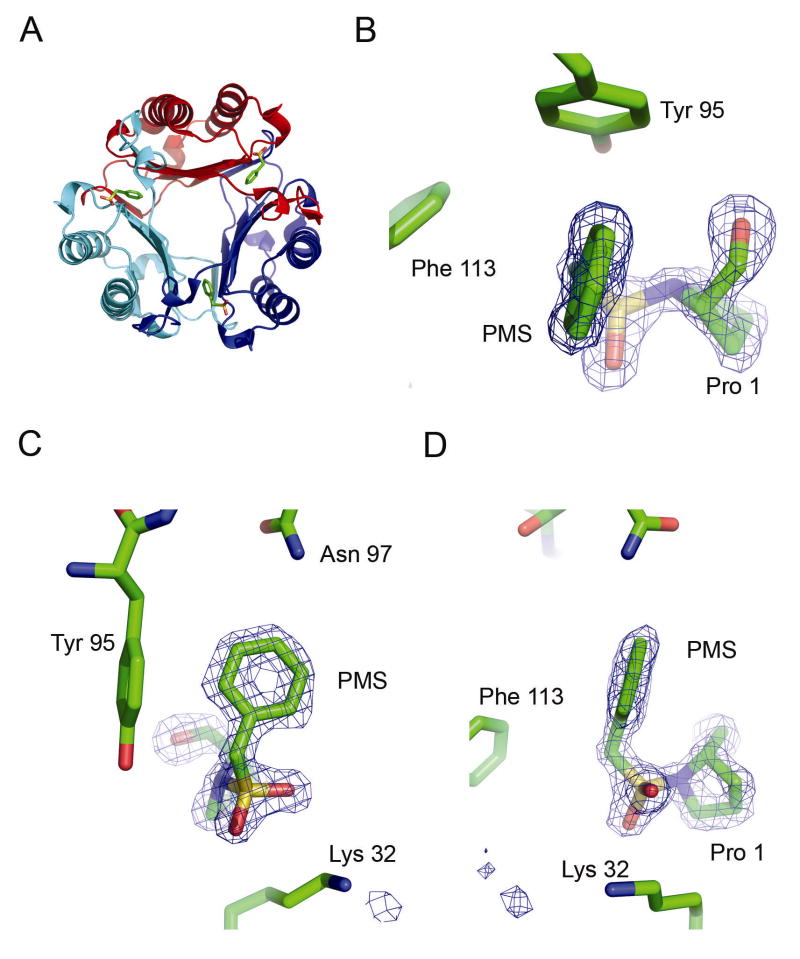
The structure of the PMS-MIF complex was determined crystallographically. The overall tertiary structure is the same as unliganded MIF and MIF bound with many other tautomerase active site inhibitors (Figure 4A). Examination of the 1.55 Å PMSF-treated MIF structure reveals the phenylmethylsulfonyl group bound to MIF (Figure 4B–D), verifying that the fluoride is the leaving group, as seen with serine proteases and as suggested by mass spectrometric results (Supporting Information and Supplementary Figure 1). The PMS group binds covalently to Pro 1, as indicated by the electron density (Figure 4B, D). The phenyl ring is positioned in a stable orientation as revealed by well-defined density (Figure 4C) and by the very low ligand B-factors which are only 1.2 times that of the average protein B-factors and even less than the average B-factor for all atoms in the structure (Table 1). It is also evident from the electron density that the covalent bond formed is between the sulfur of PMS and the nitrogen of Pro 1. This nitrogen has tetrahedral geometry (Figure 4B) suggesting sp3 hybridization.
Figure 4.
Crystallographic Structure of PMS-MIF Complex at 1.55Å. A. Global structure of the PMS-MIF complex. B, C, D. Active site with 4.5σ Fo-Fc electron density calculated omitting Pro 1 and the PMS moiety. The fit to the electron density is displayed in active site A and is similar in the other two active sites. B. View showing the tetrahedral character of the Pro 1 nitrogen. C. View showing phenyl ring of PMS in active site. D. Same as (C), but rotated 90°, showing the covalent bond between Pro 1 and the PMS moiety.
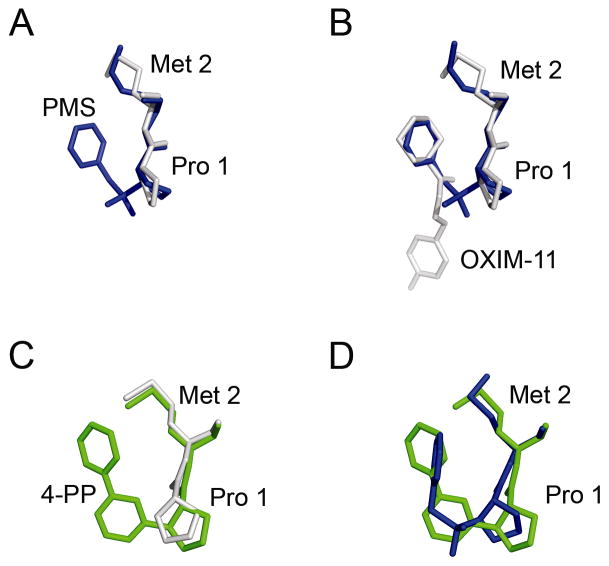
There is a slight protein conformational change in the active site relative to native MIF. We had determined the structure of native MIF to higher resolution than previously published (Supplementary Figure 3). We used this 1.25 Å structure, which only contains glycerol from the cryo-protectant in the active sites, for a superposition with the PMS-MIF structure. We also compared the covalently bound PMS-MIF structure to MIF bound non-covalently to a known inhibitor, OXIM-11 (3). The orientation of Pro 1 changes, as revealed by the overlay of the PMS-MIF complex onto unliganded MIF or onto MIF bound non-covalently to a ligand (Figure 5A–B). This is due to a change in torsion angles between Pro1 and Met 2 (Table 2) and enables an optimum bonding angle for the tetrahedral nitrogen.
Figure 5.
Flexibility of PMS allows binding to MIF with only minimal protein conformational change. A, B. Overlays of the PMS-MIF structure onto (A) unliganded MIF and (B) MIF bound to OXIM-11 (PDB entry 2OOH) (3). In both panels, the PMS-MIF structure is in blue. C, D. Overlay of (C) unliganded MIF (white) and (D) PMS-MIF (blue) onto MIF bound by the less flexible covalently bound ligand 4-PP (PDB entry 3B9S, green) (40).
Table 2.
N-Terminal Backbone Dihedral Angles of native MIF (with active site glycerol), MIF bound to OXIM-11 (PDB entry 2OOH) (3), PMS-MIF and 4-PP-MIF (PDB entry 3B9S) (40)a,b
| Native MIF | OXIM-11-MIF | PMS-MIF | 4-PP-MIF | |
|---|---|---|---|---|
| Pro 1 ψ | 153.2 ± 1.4 | 151.7 ± 0.85 | 168.5 ± 0.61 | 146.6 ± 12.4 |
| Met 2 ϕ | −134.8 ± 1.3 | −136.7 ± 0.97 | −142.5 ± 0.76 | −122.9 ± 6.8 |
| Met 2 ψ | 127.0 ± 3.5 | 127.2 ± 3.2 | 139.7 ± 2.2 | 122.1 ± 0.62 |
| Phe 3 ϕ | −121.5 ± 2.3 | −121.2 ± 1.0 | −123.9 ± 1.9 | −119.6 ± 2.7 |
| Phe 3 ψ | 126.0 ± 0.78 | 127.5 ± 0.50 | 131.8 ± 1.4 | 124.3 ± 0.95 |
The inhibitor 4-IPP also modifies MIF covalently (40). Kinetically, 4-IPP is a much faster inhibitor than PMSF. However, the electron density for the inhibitor molecules in the three active sites is less well-defined than in the PMS-MIF structure. An overlay of the 4-IPP structure onto the native MIF structure reveals that MIF undergoes a much larger conformational change upon binding 4-IPP than upon binding PMSF (Figure 5C). This is due to a change in torsion angles of Met 2 (Table 2). Such an adaptation of the protein allows the phenyl ring of the 4-phenylpyrimidine (4-PP) attached to MIF to occupy the same general location as the phenyl ring of PMS (Figure 5D). The steric imposition 4-PP places upon MIF may explain the poorer electron density. The B-factors of Pro 1 and Met 2 are substantially increased in the complex with 4-PP relative to that with PMS and relative to the native protein (Supporting Information and Supplementary Figure 4). However, the 4-phenylpyrimidine group fills more of the hydrophobic active site than the phenyl ring of PMSF. This is the likely explanation for faster kinetics: 4-IPP likely has a stronger affinity for the active site than does PMSF. However, once PMSF binds, it is able to form an adduct with the protein efficiently, with relatively little perturbation to the protein structure.
Discussion
Our studies compare two MIF tautomerase inhibitors, PMSF and NAPQI. Both molecules demonstrate slow covalent modification of MIF. The slow inhibition obtained with PMSF can be explained by slow formation of a non-covalent Michaelis complex followed by efficient covalent bond formation with the active site proline once PMSF is properly bound in the active site. In contrast, NAPQI inhibition involves rapid MIF-binding – possibly due to bi-APAP that is generated – followed by slow covalent bond formation.
Bi-APAP and its Interaction with MIF
Acetaminophen, which is widely used as an over-the-counter analgesic, is also well known to cause hepatotoxicity and renal toxicity when taken in overly large doses (41). The toxicity is due to the cytP450-mediated production of NAPQI from acetaminophen. Glutathione reacts with NAPQI, forming an acetaminophen-glutathione adduct, thereby detoxifying the NAPQI. With normal acetaminophen dosage, NAPQI is produced in a quantity that is small enough such that cellular concentrations of glutathione are sufficient for detoxification. However, as acetaminophen dosage increases, enough NAPQI is produced to deplete cellular glutathione. Excess NAPQI is able to react non-specifically with cellular proteins, and this process is involved in the mechanism of toxicity (42, 43).
NAPQI is also able to react with acetaminophen to produce bi-APAP and longer oligomers of acetaminophen (37). The reaction of NAPQI with acetaminophen, as well as with cellular proteins, occurs much more slowly than the reaction with glutathione (37) and therefore should not be relevant at a therapeutic dosage. However, at higher acetaminophen doses in mice, bi-APAP levels were found to be correlated to the acetaminophen dose and also to liver and kidney toxicity (36). Bi-APAP is therefore one of a number of acetaminophen-derived species that may be responsible for its toxicity (36). The observed crystallographic interactions between MIF and bi-APAP after co-crystallization of MIF with NAPQI may present a previously unrecognized role for MIF in acetaminophen metabolism that remains to be fully characterized.
Implications for Structure-Based Drug Design
The interaction between bi-APAP and MIF engages the opening of the MIF active site. This is in contrast to all other MIF inhibitors that have been studied structurally in complex with MIF. These generally bind deep in the active site cleft. A MIF inhibitor can be designed containing properties of both bi-APAP and of active-site cleft binding molecules to form a higher affinity inhibitor. This inhibitor conceptually would consist of three chemical groups: (1) an aromatic (or cyclohexyl) group that binds deep in the hydrophobic active site that could also have a polar group attached – such as a hydroxyl – to interact with Asn 97, (2) a polar linker to engage the Tyr 95 hydroxyl group, the Pro 1 imine, and/or the Lys 32 side chain amine, and (3) a phenyl ring similar to bi-APAP that interacts with Tyr 36 (except that it would be within the same trimer instead of a symmetry-related MIF trimer). The potential benefit of attaching a phenyl ring to interact with Tyr 36 to an active-site binding moiety in order to improve inhibition is in agreement with the hypothesis of Orita et al. (44). It should be noted that the MIF inhibitor OXIM-11 binds with its hydroxyphenyl ring at the mouth of the active site (3). However, instead of extending towards Tyr 36, the ring extends towards the other side of the active site opening and is disordered. Therefore it is necessary to design the polar linker to position the outer phenyl ring such that it extends towards Tyr 36.
In contrast to the insights obtained from bi-APAP, PMSF provides instruction for designing covalently-modifying MIF inhibitors. This is the first instance, to our knowledge, in which PMSF has been observed to react with a proline to form an N–S bond. It should be noted that for PMSF to form a covalent bond with proline, it must be the N-terminal residue. Moreover, reactivity would be unlikely if the pKa of the Pro 1 imino group were not 4 units lower than that of a proline imino group in a dipeptide (45). This causes the predominant species at physiological pH to be that of the singly-protonated, neutral imine. The Pro 1 nitrogen, therefore, has a lone electron pair that acts as a nucleophile. The proton that is initially attached to the Pro 1 nitrogen must leave during the course of the reaction according to the mass-spectrometry results (Supplementary Figure 1). In the final complex, the Pro 1 nitrogen contains a lone pair of electrons and has tetrahedral geometry.
The reaction between PMSF and MIF affects the orientation of the catalytic residue, Pro 1 (Figure 5). The methylene group intervening between the phenyl ring and the sulfonyl moiety of PMSF is important for orienting the phenyl ring in a preferred conformation in the hydrophobic cavity. If the phenyl ring were directly bonded to the sulfonyl group, the ring would be restrained to extend in a direction pointing away from Pro 1. This would place it away from the center of the hydrophobic cavity where it would interfere sterically with Tyr 95 and/or Phe 113. This can be contrasted to the covalently-modifying MIF inhibitor 4-IPP, which neither has a flexible linker between the leaving group and the pyrimidine ring nor one between the two aromatic rings. Therefore, when it forms an adduct with MIF, the protein must undergo a large conformational change in order to place the hydrophobic group in the proper place in the active site cavity. Inherent flexibility in the inhibitor therefore optimizes adduct formation. Therefore, each inhibitor has a feature that provides benefit. 4-IPP has a better binding moiety, which provides faster kinetics, whereas PMSF has less rigidity, which causes less adaptation needed by the protein.
The complex between MIF and PMSF demonstrates how flexibility in the protein and ligand work together to allow covalent bond formation and at the same time position the ligand aromatic ring in the same favorable position as seen with non-covalent inhibitors. Using this scaffold it may be possible to design inhibitors that couple reactive groups (such as sulfonyl fluoride) connected by an intervening methylene group to aromatic moieties that have high affinity for the MIF active site. The methylene provides the flexibility needed to place the reactive group near the Pro 1 nitrogen, and the aromatic group squarely in the cavity. The hydrophobic cavity-binding aromatic group can be a multiple ring moiety, such as 4-PP, or a fused ring group. The hydrophobic cavity-binding moiety should provide increased specificity and faster kinetics with respect to modifying the enzyme due to a greater affinity for MIF than a phenyl ring. Therefore, a molecule similar to PMSF with the phenyl ring substituted with a group that binds better in the active site should yield a more potent inhibitor.
Supplementary Material
Acknowledgments
We thank Drs. Young-joo Kim, Anne Roberts and Cristina Furdui for their assistance with mass-spectrometric studies, and Dr. Peter Senter (Seattle Genetics) for helpful discussions. We also thank the staff at the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory for the use of, and assistance at, their facility.
Footnotes
Supported by grants from the NIH and the Alliance for Lupus Research (EJL, RB, LL).
Coordinates and structure factors of PMS-bound MIF, MIF bound to an acetaminophen dimer, and 1.25 Å native MIF have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB entries 3CE4, 3DJI and 3DJH, respectively).
Abbreviations: MIF, macrophage migration inhibitory factor; PMSF, phenylmethylsulfonyl fluoride; PMS, phenylmethylsulfonyl; NAPQI, N-acetyl-para-benzoquinone imine; APAP, acetaminophen; bi-APAP, acetaminophen dimer; 4-IPP, 4-iodo-6-phenylpyrimidine; 4-PP, 4-phenylpyrimidine; Tris, tris(hydroxymethyl)aminomethane; SD, standard deviation; SEM, standard error of the mean; cytP450, cytochrome P450.
Supporting Information Available
Supporting information is provided regarding mass spectrometric results and methods, details regarding the novel crystal form of the bi-APAP-MIF complex, our higher resolution native MIF structure, and B-factor comparison of the N-terminal β-strands of native MIF and MIF inhibited by PMSF or 4-IPP treatment. Text and figures are presented. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Calandra T, Bernhagen J, Metz CN, Spiegel LA, Bacher M, Donnelly T, Cerami A, Bucala R. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377:68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 2.Bernhagen J, Calandra T, Mitchell RA, Martin SB, Tracey KJ, Voelter W, Manogue KR, Cerami A, Bucala R. MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature. 1993;365:756–9. doi: 10.1038/365756a0. [DOI] [PubMed] [Google Scholar]
- 3.Crichlow GV, Cheng KF, Dabideen D, Ochani M, Aljabari B, Pavlov VA, Miller EJ, Lolis E, Al-Abed Y. Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor. J Biol Chem. 2007;282:23089–95. doi: 10.1074/jbc.M701825200. Epub 2007 May 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cvetkovic I, Al-Abed Y, Miljkovic D, Maksimovic-Ivanic D, Roth J, Bacher M, Lan HY, Nicoletti F, Stosic-Grujicic S. Critical role of macrophage migration inhibitory factor activity in experimental autoimmune diabetes. Endocrinology. 2005;146:2942–51. doi: 10.1210/en.2004-1393. Epub 2005 Mar 24. [DOI] [PubMed] [Google Scholar]
- 5.Denkinger CM, Denkinger M, Kort JJ, Metz C, Forsthuber TG. In vivo blockade of macrophage migration inhibitory factor ameliorates acute experimental autoimmune encephalomyelitis by impairing the homing of encephalitogenic T cells to the central nervous system. J Immunol. 2003;170:1274–82. doi: 10.4049/jimmunol.170.3.1274. [DOI] [PubMed] [Google Scholar]
- 6.Mikulowska A, Metz CN, Bucala R, Holmdahl R. Macrophage migration inhibitory factor is involved in the pathogenesis of collagen type II-induced arthritis in mice. J Immunol. 1997;158:5514–7. [PubMed] [Google Scholar]
- 7.Takahashi N, Nishihira J, Sato Y, Kondo M, Ogawa H, Ohshima T, Une Y, Todo S. Involvement of macrophage migration inhibitory factor (MIF) in the mechanism of tumor cell growth. Mol Med. 1998;4:707–14. [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell RA. Mechanisms and effectors of MIF-dependent promotion of tumourigenesis. Cell Signal. 2004;16:13–9. doi: 10.1016/j.cellsig.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Zernecke A, Bernhagen J, Weber C. Macrophage migration inhibitory factor in cardiovascular disease. Circulation. 2008;117:1594–602. doi: 10.1161/CIRCULATIONAHA.107.729125. [DOI] [PubMed] [Google Scholar]
- 10.Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197:1467–76. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi X, Leng L, Wang T, Wang W, Du X, Li J, McDonald C, Chen Z, Murphy JW, Lolis E, Noble P, Knudson W, Bucala R. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleemann R, Mischke R, Kapurniotu A, Brunner H, Bernhagen J. Specific reduction of insulin disulfides by macrophage migration inhibitory factor (MIF) with glutathione and dihydrolipoamide: potential role in cellular redox processes. FEBS Lett. 1998;430:191–6. doi: 10.1016/s0014-5793(98)00654-1. [DOI] [PubMed] [Google Scholar]
- 13.Rosengren E, Bucala R, Aman P, Jacobsson L, Odh G, Metz CN, Rorsman H. The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol Med. 1996;2:143–9. [PMC free article] [PubMed] [Google Scholar]
- 14.Rosengren E, Aman P, Thelin S, Hansson C, Ahlfors S, Bjork P, Jacobsson L, Rorsman H. The macrophage migration inhibitory factor MIF is a phenylpyruvate tautomerase. FEBS Lett. 1997;417:85–8. doi: 10.1016/s0014-5793(97)01261-1. [DOI] [PubMed] [Google Scholar]
- 15.Dios A, Mitchell RA, Aljabari B, Lubetsky J, O’Connor K, Liao H, Senter PD, Manogue KR, Lolis E, Metz C, Bucala R, Callaway DJ, Al-Abed Y. Inhibition of MIF bioactivity by rational design of pharmacological inhibitors of MIF tautomerase activity. J Med Chem. 2002;45:2410–6. doi: 10.1021/jm010534q. [DOI] [PubMed] [Google Scholar]
- 16.Lubetsky JB, Dios A, Han J, Aljabari B, Ruzsicska B, Mitchell R, Lolis E, Al-Abed Y. The tautomerase active site of macrophage migration inhibitory factor is a potential target for discovery of novel anti-inflammatory agents. J Biol Chem. 2002;277:24976–82. doi: 10.1074/jbc.M203220200. Epub 2002 May 7. [DOI] [PubMed] [Google Scholar]
- 17.Lubetsky JB, Swope M, Dealwis C, Blake P, Lolis E. Pro-1 of macrophage migration inhibitory factor functions as a catalytic base in the phenylpyruvate tautomerase activity. Biochemistry. 1999;38:7346–54. doi: 10.1021/bi990306m. [DOI] [PubMed] [Google Scholar]
- 18.Senter PD, Al-Abed Y, Metz CN, Benigni F, Mitchell RA, Chesney J, Han J, Gartner CG, Nelson SD, Todaro GJ, Bucala R. Inhibition of macrophage migration inhibitory factor (MIF) tautomerase and biological activities by acetaminophen metabolites. Proc Natl Acad Sci U S A. 2002;99:144–9. doi: 10.1073/pnas.011569399. Epub 2002 Jan 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun HW, Swope M, Cinquina C, Bedarkar S, Bernhagen J, Bucala R, Lolis E. The subunit structure of human macrophage migration inhibitory factor: evidence for a trimer. Protein Eng. 1996;9:631–5. doi: 10.1093/protein/9.8.631. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr, Sweet RM, editors. Methods in Ezymology. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 21.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallographica Section A. 1994;50:157–163. [Google Scholar]
- 22.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallographica Section D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 23.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–8. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 24.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Sheldrick G. A short history of SHELX. Acta Crystallographica Section A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallographica Section D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 28.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. Journal of Applied Crystallography. 2006;39:109–111. [Google Scholar]
- 29.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A 47(Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.McRee DE. Differential evolution for protein crystallographic optimizations. Acta Crystallogr D Biol Crystallogr. 2004;60:2276–9. doi: 10.1107/S0907444904025491. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallographica Section A. 1976;32:922–923. [Google Scholar]
- 33.Davis M, Ideo G, Harrison NG, Williams R. Hepatic glutathione depletion and impaired bromosulphthalein clearance early after paracetamol overdose in man and the rat. Clin Sci Mol Med. 1975;49:495–502. doi: 10.1042/cs0490495. [DOI] [PubMed] [Google Scholar]
- 34.Dahlin DC, Miwa GT, Lu AY, Nelson SD. N-acetyl-p-benzoquinone imine: a cytochrome P-450-mediated oxidation product of acetaminophen. Proc Natl Acad Sci U S A. 1984;81:1327–31. doi: 10.1073/pnas.81.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart SG, Cartun RW, Wyand DS, Khairallah EA, Cohen SD. Immunohistochemical localization of acetaminophen in target tissues of the CD-1 mouse: correspondence of covalent binding with toxicity. Fundam Appl Toxicol. 1995;24:260–74. doi: 10.1006/faat.1995.1029. [DOI] [PubMed] [Google Scholar]
- 36.Chen C, Krausz KW, Idle JR, Gonzalez FJ. Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. J Biol Chem. 2008;283:4543–59. doi: 10.1074/jbc.M706299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potter DW, Hinson JA. Reactions of N-acetyl-p-benzoquinone imine with reduced glutathione, acetaminophen, and NADPH. Mol Pharmacol. 1986;30:33–41. [PubMed] [Google Scholar]
- 38.James GT. Inactivation of the protease inhibitor phenylmethylsulfonyl fluoride in buffers. Anal Biochem. 1978;86:574–9. doi: 10.1016/0003-2697(78)90784-4. [DOI] [PubMed] [Google Scholar]
- 39.Potter DW, Miller DW, Hinson JA. Horseradish peroxidase-catalyzed oxidation of acetaminophen to intermediates that form polymers or conjugate with glutathione. Mol Pharmacol. 1986;29:155–62. [PubMed] [Google Scholar]
- 40.Winner M, Meier J, Zierow S, Rendon BE, Crichlow GV, Riggs R, Bucala R, Leng L, Smith N, Lolis E, Trent JO, Mitchell RA. A novel, macrophage migration inhibitory factor suicide substrate inhibits motility and growth of lung cancer cells. Cancer Res. 2008;68:7253–7. doi: 10.1158/0008-5472.CAN-07-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamlyn AN, Douglas AP, James O. The spectrum of paracetamol (acetaminophen) overdose: clinical and epidemiological studies. Postgrad Med J. 1978;54:400–4. doi: 10.1136/pgmj.54.632.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pumford NR, Hinson JA, Potter DW, Rowland KL, Benson RW, Roberts DW. Immunochemical quantitation of 3-(cystein-S-yl)acetaminophen adducts in serum and liver proteins of acetaminophen-treated mice. J Pharmacol Exp Ther. 1989;248:190–6. [PubMed] [Google Scholar]
- 43.Pumford NR, Roberts DW, Benson RW, Hinson JA. Immunochemical quantitation of 3-(cystein-S-yl)acetaminophen protein adducts in subcellular liver fractions following a hepatotoxic dose of acetaminophen. Biochem Pharmacol. 1990;40:573–9. doi: 10.1016/0006-2952(90)90558-3. [DOI] [PubMed] [Google Scholar]
- 44.Orita M, Yamamoto S, Katayama N, Aoki M, Takayama K, Yamagiwa Y, Seki N, Suzuki H, Kurihara H, Sakashita H, Takeuchi M, Fujita S, Yamada T, Tanaka A. Coumarin and chromen-4-one analogues as tautomerase inhibitors of macrophage migration inhibitory factor: discovery and X-ray crystallography. J Med Chem. 2001;44:540–7. doi: 10.1021/jm000386o. [DOI] [PubMed] [Google Scholar]
- 45.Swope M, Sun HW, Blake PR, Lolis E. Direct link between cytokine activity and a catalytic site for macrophage migration inhibitory factor. Embo J. 1998;17:3534–41. doi: 10.1093/emboj/17.13.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engh RA, Huber R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallographica Section A. 1991;47:392–400. [Google Scholar]
- 47.Diemand AV, Scheib H. iMolTalk: an interactive, internet-based protein structure analysis server. Nucleic Acids Res. 2004;32:W512–6. doi: 10.1093/nar/gkh403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diemand AV, Scheib H. MolTalk--a programming library for protein structures and structure analysis. BMC Bioinformatics. 2004;5:39. doi: 10.1186/1471-2105-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLano WL. DeLano Scientific; Palo Alto, CA, USA: 2002. http://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.