Abstract
We genotyped 160 P. falciparum infections from Malawi for pfmdr-1 copy number changes and SNPs associated with in vivo tolerance and poor in vitro sensitivity to the component drugs of Coartem. We also measured in vitro susceptibility of 49 of these isolates to a variety of drugs in clinical use or with a potential for use in Africa. All 160 infections carried a single copy of pfmdr-1 but 34% exhibited sequence variation at 4 of the 5 polymorphic sites in pfmdr-1. Isolates carrying 86-Asn and 184-Tyr pfmdr-1 alleles were significantly less sensitive (p<0.001) to mefloquine, lumefantrine, artemether and dihydroartemisinin compared with those bearing 86-Tyr and 184-Phe polymorphisms. This study provides baseline measures prior to policy change: continued surveillance for changes in baseline drug susceptibility, pfmdr-1 copy number and SNPs, and other putative Coartem resistance loci will be necessary to provide an early warning of emerging Coartem resistance in this setting.
Keywords: Plasmodium falciparum, antimalarial resistance, multi-drug resistance gene (pfmdr-1), copy number, single nucleotide polymorphisms (SNPs), artemisinin combination therapy (ACT), Coartem, Malawi
Introduction
In P. falciparum, both copy number variation and single nucleotide polymorphisms (SNPs) of the multi-drug resistance gene (pfmdr-1) contribute to variability in parasite response to a variety of antimalarial drugs. This includes parasite susceptibility to component drugs of the two artemisinin-based combination therapies (ACTs), which are increasingly being deployed throughout the Sub-Saharan African region and South East Asia to replace the failing monotherapies. The two ACTs are a combination of lumefantrine with artemether (Coartem), which most countries in Sub-Saharan Africa have adopted as their first-line treatment for malaria, and a combination of mefloquine with artesunate, which is widely used in South East Asia. In South East Asia, amplification of the pfmdr-1 gene has been strongly associated with elevated mefloquine, halofantrine, quinine, lumefantrine and artemisinin IC50s (Wilson et al., 1993; Price et al., 1999; Pickard et al., 2003; Price et al., 2004; Price et al., 2006). A causal link between pfmdr-1 copy number and resistance to each of these drugs has since been demonstrated using parasites genetically manipulated to express single-copy pfmdr-1 (Sidhu et al., 2006). The knockdown parasites were significantly more sensitive to mefloquine, halofantrine, quinine, lumefantrine and artemisinin compared to the parental clone that carries two functional copies of pfmdr-1 (Sidhu et al., 2006). This finding unequivocally underscores the importance of pfmdr-1 copy number as a determinant of in vitro resistance to these drugs. A similar role for pfmdr-1 amplification in conferring in vivo resistance to ACTs and their component drugs has been proposed and investigated. A survey conducted in Gabon, West Africa, revealed amplification of the pfmdr-1 gene in > 5% of the isolates collected in 1995; this amplification was associated with low-grade in vivo resistance to mefloquine (Uhlemann et al., 2005). However, in samples collected in 2002, no parasites with multiple pfmdr-1 copies were detected. Recently, a clinical trial on the Thai-Burmese border identified pfmdr-1 amplification as the key determinant of MQ treatment failure and a major risk factor for treatment failure with mefloquine/artesunate combination (Price et al., 2004). This finding suggests that pfmdr-1 copy number could be used as a molecular marker of treatment failure for the two treatment regimens in that setting. In another study from Thailand, increase in pfmdr-1 copy number was associated with recrudescence after the administration of a 4-dose regimen of Coartem (Price et al., 2006). A similar study in Zanzibar found that infections carrying the 86-Asn pfmdr-1 polymorphism were significantly more tolerant to Coartem than infections harbouring the mutant, 86-Tyr, allele (Sisowath et al., 2005). Its follow-up study could not establish the role for pfmdr-1 copy number in Coartem resistance as parasites in all breakthrough infections carried single-copy pfmdr-1 (Sisowath et al., 2007). Nonetheless, it appears that the 86-Asn pfmdr-1 polymorphism confers parasite resistance to Coartem if it exists in a genetic background containing multiple copies of the pfmdr-1 gene. Because of its demonstrable role in conferring in vitro resistance to most component drugs of ACTs and its potential role in the development of in vivo resistance to widely used ACTs, surveillance for pfmdr-1 copy number in malaria-endemic countries is of utmost importance.
In this study, we genotyped 160 P. falciparum infections from Malawi for pfmdr-1 copy number changes and SNPs with the view to providing baseline prevalence data on polymorphisms that could potentially undermine the clinical utility of Coartem. We also measured in vitro sensitivity of a subset of these infections to a variety of antimalarial drugs with a potential for clinical use and determined relationships between pfmdr-1 genotype and drug response.
Materials and Methods
Sample collection and DNA extraction
Ninety-two P. falciparum isolates, in the form of whole blood, were collected from children under five years of age presenting to the Queen Elizabeth Central Hospital, Blantyre, Malawi, with uncomplicated falciparum malaria in the year 2007. We sought the consent of parents or guardians in order to draw whole blood samples (0.5mL) from these children. We then extracted parasite DNA from whole blood using the QIAmp DNA Mini Kit (Qiagen Ltd, UK). We also took a subset of these infections (49) and measured in vitro sensitivity to a range of antimalarial drugs. This study was part of an antimalarial drug resistance study approved by the College of Medicine Research and Ethics Committee, University of Malawi. The other 68 P. falciparum isolates were also in the form of whole blood, and were collected from moderately severe malaria patients in 1998 and 1999 as part of the study investigating the efficacy and tolerability of rectal artesunate in children. For this batch of samples, we extracted parasite DNA using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, USA).
Determination of pfmdr-1 copy number
We used a real-time polymerase chain reaction (PCR) assay to determine copy number of pfmdr-1 relative to that of a single copy gene, Seryl-T synthetase, using the comparative cycle threshold (ΔΔCT) method (Price et al., 2004). We then measured copy number of the pfmdr-1 gene relative to that of a standard calibrator parasite, 3D7, which has a single copy of pfmdr-1. In addition, we ran DNA from a Thai isolate, M778, which has 2 copies of pfmdr-1, as a control. All reactions were conducted in quadruplicate in 384-well plates on an ABI 7900HT real-time PCR machine. We repeated copy number measurements for all samples that had high CT values (>30) and those which had wide 95% confidence intervals for the measured copy number. We were able to reduce high CT values to acceptable levels (<30) and obtain tight confidence intervals by concentrating the DNA two to five fold. In the final analysis of copy number, the number of pfmdr-1 copies carried by each isolate was rounded to the nearest integer.
Detection of single nucleotide polymorphisms in pfmdr-1
We genotyped five point mutations in pfmdr-1 using the primer extension method as previously described (Anderson et al., 2005). Briefly, genomic DNA from each isolate was amplified using two sets of primers that span across regions containing all the 5 polymorphic loci in pfmdr-1. The PCR products were digested with shrimp alkaline phosphatase (Amersham) and Exonuclease I (Amersham) to remove unincorporated nucleotides and excess primers. Digested products were then amplified in a second PCR using ABI PRISM SNaPshot™ Multiplex Kit (Applied Biosystems) in the presence of fluorescent-labelled dideoxy nucleotides (ddNTPs) and primers that are one base short of the target SNP. This PCR results in single-base extension of primers at their 3’ ends and generation of products that are fluorescently labelled with a particular dye depending on the base present. The products of these reactions were analysed on an ABI 3100 capillary sequencer and scored using GENOTYPER software. The following are the point mutations that we genotyped: N86Y (the substitution of tyrosine for asparagine), Y184F (the substitution of phenylalanine for tyrosine), S1034C (the substitution of cysteine for serine), N1042D (the substitution of aspartic acid for asparagine) and D1246Y (the substitution of tyrosine for aspartic acid). For all the reactions, we ran P. falciparum DNA from strains 3D7, W2 and 7G8 in parallel with DNA from patient isolates as positive controls.
Estimating the prevalence of pfmdr-1 amplification and SNPs
We determined both the prevalence and binomial exact 95% confidence intervals for the prevalence of isolates with amplified pfmdr-1 using STATA version 8.1 (Stata Corporation, USA). We also used the same program to estimate both the prevalence and 95% confidence intervals for the prevalence of isolates carrying a particular set of SNPs in pfmdr-1.
In vitro drug sensitivity assays
We measured in vitro sensitivity of 49 isolates collected in 2007 to the following drugs: chloroquine, amodiaquine, desethylamodiaquine, quinine, mefloquine, artemether, dihydroartemisinin and lumefantrine. Antimalarial test compounds were obtained from the following sources: chloroquine diphosphate, amodiaquine hydrochloride, desethylamodiaquine and quinine hydrochloride (Sigma, UK), mefloquine hydrochloride (Hoffman-La Roche, Basel, Switzerland), artemether, dihydroartemisinin and lumefantrine (Novartis Pharma AG, Basel, Switzerland). Stock solutions of amodiaquine, quinine, mefloquine, artemether, and dihydroartemisinin were prepared in a 70% ethanol/water mixture while that of lumefantrine was prepared in a 1:1:1 mixture of linoleic acid, triton-x and ethanol (all from Sigma, UK). Those of chloroquine and desethylamodiaquine were prepared in sterile distilled water. All stock solutions were sterilised by passing them through a 20-μm filter and stored at −20°C until required. We used the Sybr green I assay (Johnson et al., 2007) to assess in vitro antimalarial sensitivity of patient isolates. Whenever patient isolates were assayed for drug susceptibility, a laboratory strain with known drug sensitivity status (3D7) was included in the tests as a control. Parasites were plated in the ring stage at 1% hematocrit and 1% parasitemia in 100μL of an antimalarial drug at an appropriate, defined concentration. Plates were placed in a sealed jar and flushed with a gas mixture of 4% O2, 3% CO2, and 93% N2, and incubated at 37°C for 72 hours. When incubation was complete, plates were subjected to three 20-minute freeze–thaw cycles to resuspend the culture. Thereafter, 100μL of Sybr green I solution at 0.2μL of 10000x Sybr green I (Sigma, UK) in 1ml of Lysis buffer were added to each well of a new, duplicate flat-bottomed 96-well microtitre plate. The culture in each of the wells of the original plate was resuspended by mixing with a multichannel pipette. Thereafter, 100μL of the culture was taken from each well and added to the corresponding well of the detection plate. The detection plate was read at excitation and emission wavelengths of 485nm and 538nm respectively with the aid of a Labsystems Fluoroskan II fluorescence plate reader (Global Medical Instrumentation, MN, USA) after an hour of incubation in the dark.
Analysis of drug sensitivity data
Fluorescence counts at various drug concentrations were expressed as a percentage of the control (100%) and plotted against corresponding drug concentrations using Grafit Software (Erithacus Software Ltd, Surrey, England) to generate log dose–response curves from which IC50 values were obtained. IC50s were log-transformed and expressed as geometric mean IC50 and their respective 95% confidence intervals were calculated. The Mann-Whitney U test was used to test whether there were significant differences in median IC50 values between isolates that carry the pfmdr-1 genotype 86-Tyr/184-Phe both in pure and mixed state, and those that carry the pfmdr-1 genotype, 86-Asn/184-Tyr. For all statistical tests, the level of significance (p) was set at 5%.
Results and Discussion
(a) Variation in pfmdr-1 SNPs and copy number
We observed sequence variation at codons 86, 184, 1034 and 1246 of the pfmdr-1 gene (Table 1) in 34% of the isolates but no evidence of pfmdr-1 amplification in all the 160 isolates. Our estimates of pfmdr-1 copy number for these isolates ranged from 0.6 to 1.3 (Figure 1). When we rounded copy numbers to the nearest integer, all isolates had a single copy of pfmdr-1. Failure to detect parasites bearing multiple copies of pfmdr-1 does not necessarily mean that such parasites are absent: 95% confidence intervals for our estimated prevalence of parasites with multiple copies of pfmdr-1 ranged from 0 to 3.4%. This observation is in sharp contrast to the situation in Thailand, where frequencies of >30% have been reported for parasites with multiple pfmdr-1 copies (Price et al., 1999; Price et al., 2006; Nair et al., 2007). This may be explained by differences in patterns of drug use between the two epidemiological settings. In Thailand, the parasite population has experienced selection pressure from nearly all the available antimalarial drugs including mefloquine and artemisinins. For this reason, parasites with multiple copies of pfmdr-1 may have been selected by these drugs. In contrast, Malawi has not used most of these drugs because it has been unable to afford them. Quinine could have potentially selected parasites with multiple copies of pfmdr-1. However, this drug is only used for treating severe disease and first-line treatment failures. Therefore, its total amount of drug pressure relative to the total parasite population is limited and may not have permitted the selection of multiple-copy variants of pfmdr-1. The virtual absence of pfmdr-1 amplification in Malawian isolates offers a glimmer of hope for the therapeutic lifespan of Coartem, a combination therapy that replaced SP as the first-line antimalarial in January 2008. However, this picture is marred by the high prevalence of the pfmdr-1 86-Asn allele, which is associated with in vivo Coartem tolerance (Sisowath et al., 2005; Dokomajilar et al., 2006) and poor in vitro sensitivity to the component drugs of Coartem. This study found that isolates harbouring pfmdr-1 polymorphisms 86-Asn and 184-Tyr were significantly less sensitive to mefloquine, lumefantrine, dihydroartemisinin and artemether compared to those carrying 86-Tyr and 184-Phe polymorphisms (p<0.001). The 86-Asn pfmdr-1 allele was estimated to be at a prevalence of 66% in this study (Table 1). This polymorphism is thought to be the first in a series of mutation steps leading to the selection of Coartem resistance (Hastings and Ward., 2005). We postulate that pfmdr-1 amplification in isolates carrying this allele is the next mutational event that culminates into high-level resistance and Coartem treatment failures. Nonetheless, it is also possible that subsequent mutations may occur at other putative resistance loci within the genome such as in the PfATPase6 gene (Jambou et al., 2005). Although this study did not detect any pfmdr-1 amplification, continued surveillance for pfmdr-1 copy number changes will still be essential because these mutations may arise on several independent occasions in natural populations (Triglia et al., 1991; Nair et al., 2007), and at a much higher rate compared to point mutations (Imwong, M., personal communication). Moreover, parasites with amplified pfmdr-1 may be introduced into the local parasite population by importation from areas where they are prevalent. An examination of the evolutionary origins of chloroquine and SP resistance revealed that drug-resistant parasites from South East Africa share a common lineage with those from South East Asia (Wootton et al., 2002; Roper et al., 2004). This finding suggests that gene flow from founder foci in South East Asia rather than de novo selection of drug resistance mutations at a country level has been the major driving force leading to the dissemination of CQ and SP resistance in Africa. Therefore, in addition to selection of pfmdr-1 multiple-copy variants de novo, we must be wary of the threat posed by their dispersal and migration from South East Asia and other areas where they are prevalent.
Table 1.
Pfmdr-1 SNP profile of 160 P. falciparum isolates from Malawi
|
pfmdr-1 codon |
||||||
|---|---|---|---|---|---|---|
| 86 | 184 | 1034 | 1042 | 1246 | n | Prevalence (%) |
| N | Y | S | N | D | 106 | 66.3 (58.4 — 73.5) |
| Y | F | S | N | D | 20 | 12.5 (7.8 — 18.8) |
| N/Y | Y | S | N | D | 9 | 5.6 (2.6 — 10.4) |
| N/Y | Y/F | S | N | D | 6 | 3.8 (1.4 — 8.0) |
| N | Y/F | S | N | D | 4 | 2.5 (0.7 — 6.3) |
| N/Y | F | S | N | D | 3 | 1.9 (0.4 — 5.4) |
| Y | Y/F | S | N | D | 2 | 1.3 (0.2 — 4.4) |
| N | F | S | N | D | 2 | 1.3 (0.2 — 4.4) |
| Y | Y | S | N | D | 1 | 0.6 (0 — 3.4) |
| N | Y | S | N | Y | 1 | 0.6 (0 — 3.4) |
| N | Y | C | N | D | 1 | 0.6 (0 — 3.4) |
| N | Y | C | N | Y | 1 | 0.6 (0 — 3.4) |
| N/Y | Y/F | S | N | Y | 1 | 0.6 (0 — 3.4) |
| N | Y | S/C | N | D/Y | 1 | 0.6 (0 — 3.4) |
| N/Y | Y/F | S/C | N | D/Y | 1 | 0.6 (0 — 3.4) |
| N | Y | S | N | D/Y | 1 | 0.6 (0 — 3.4) |
Letters are abbreviations for different amino acids in the pfmdr-1 gene product. n = number of isolates with the listed set of alleles. Prevalence in parenthesis represents binomial exact 95% confidence intervals for the measured prevalence. Mutant alleles are indicated in bold; if both the wild type and mutant alleles were found in the same isolate, the genotype carried by the isolate was considered ‘mixed,’ for example, N/Y means the isolate carried both the wild type asparagine (N) residue and the mutant tyrosine (Y) residue at codon 86.
Figure 1.
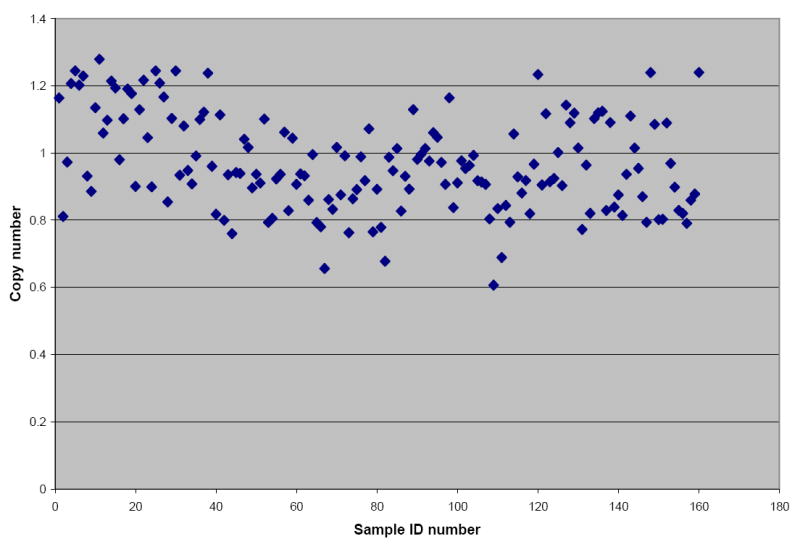
A scatter-gram depicting pfmdr-1 copy number estimates for 160 P. falciparum isolates from Malawi. No isolate with pfmdr-1 copy number ≥ 2 was detected.
(b) In vitro susceptibility of 49 patient isolates to diverse antimalarials
Our in vitro drug susceptibility data indicate that Malawian isolates are sensitive to various quinoline antimalarials, aryl-amino-alcohol dugs and artemisinin compounds (Table 2). These findings are quite consistent with previous observations from the same area (Nkhoma et al., 2007) and support the use of aryl-amino-alcohol and quinoline-based ACT in this setting. Isolates bearing pfmdr-1 polymorphisms 86-Tyr and 184-Phe were significantly more sensitive to mefloquine, lumefantrine, dihydroartemisinin and artemether but less sensitive to chloroquine and amodiaquine compared to those carrying 86-Asn and 184-Tyr polymorphisms (p<0.001). This observation reinforces previous findings that isolates bearing the 86-Tyr pfmdr-1 polymorphism tend to exhibit hypersensitivity to aryl-amino-alcohol drugs and artemisinin compounds (Duraisingh et al., 2000).
Table 2.
In vitro susceptibility of patient isolates to diverse antimalarial compounds
| Drug | n | Geometric mean IC50 (nM) | Tr* (nM) |
|---|---|---|---|
| Chloroquine | 49 | 24.8 (22.8 — 26.4) | 100 |
| Amodiaquine | 49 | 19.5 (18.3 — 20.8) | 80 |
| Desethylamodiaquine | 48 | 64.3 (60.6 — 68.3) | 60 |
| Quinine | 48 | 277.4 (260.8 — 295.2) | 450 |
| Mefloquine | 49 | 22.3 (20.1 — 24.6) | 30 |
| Dihydroartemisinin | 49 | 5.8 (5.1 — 6.6) | NE |
| Artemether | 48 | 10.4 (9.4 — 11.5) | NE |
| Lumefantrine | 48 | 71.5 (64.0 — 79.9) | NE |
Drug sensitivity data are quoted as geometric mean 50% inhibitory concentrations and their associated 95% confidence intervals (in parenthesis).
n = number of isolates successfully assayed for in vitro drug susceptibility
Tr* stands for threshold resistance and represents a previously defined cut-off concentration that distinguishes drug-susceptible parasites from the resistant ones; NE = not established.
IC50s for the reference clone, 3D7: chloroquine = 22 ± 3 nM; amodiaquine = 15 ± 2 nM; desethylamodiaquine = 31 ± 9 nM; quinine = 127 ± 12 nM; mefloquine = 21 ± 6 nM; dihydroartemisinin = 7 ± 2 nM; artemether = 13 ± 4 nM; lumefantrine = 79 ± 8 nM
Conclusion
We have shown that most isolates from Malawi carry 86-Asn and 184-Tyr pfmdr-1 alleles associated with in vivo tolerance and in vitro resistance to the component drugs of Coartem but lack pfmdr-1 duplications that can also undermine the clinical utility of Coartem. We have also shown that Malawian isolates exhibit in vitro sensitivity to a range of quinoline antimalarials, aryl-amino-alcohol dugs and artemisinin compounds, a finding that supports the use of aryl-amino-alcohol and quinoline-based ACTs in this setting. Surveillance for changes in baseline drug susceptibility, prevalence of pfmdr-1 alleles and copy number variants, and changes in other putative resistance loci will be necessary to track the evolution of resistance to Coartem and other ACTs in this setting.
Acknowledgments
We thank all parents and guardians of children that participated in this study through the donation of venous blood samples. We also thank Mrs Enid Mfungwe and Mr Maxwell Kanjala for technical assistance. This work was supported by the Gates Malaria Partnership Re-entry Grant to Dr Standwell Nkhoma and an NIH Grant to Dr Timothy Anderson (Grant Number R01 AI075145-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson TJ, Nair S, Qiu WG, Singlam S, Brockman A, Paiphun L, Nosten F. Are transporter genes other than the chloroquine resistance locus (pfcrt) and multi drug resistance gene (pfmdr) associated with antimalarial drug resistance? Antimicrob Agents Chemother. 2005;272:1153–1161. doi: 10.1128/AAC.49.6.2180-2188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. Selection of Plasmodium falciparum pfmdr1 Alleles following Therapy with Artemether-Lumefantrine in an Area of Uganda where Malaria Is Highly Endemic. Antimicrob Agents Chemother. 2006;50:1893–1895. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol Biochem Parasitol. 2000;108:13–23. doi: 10.1016/s0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- Hastings IM, Ward SA. Coartem (Artemether-Lumefantrine) in Africa: The Beginning of the End? J Infect Dis. 2005;192:1303–1304. doi: 10.1086/432554. [DOI] [PubMed] [Google Scholar]
- Jambou R, Legrand E, Niang M, Khim N, Lim P, Volney B, Ekala MT, Bouchier C, Esterre P, Fandeur T, Mercereau-Puijalon OS. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005;366:1960–1963. doi: 10.1016/S0140-6736(05)67787-2. [DOI] [PubMed] [Google Scholar]
- Johnson JD, Dennull RA, Gerena L, Lopez-Sanchez M, Roncal NE, Waters NC. Assessment and continued validation of the malaria Sybr Green-I based fluorescence assay for use in malaria drug screening. Antimicrob Agents Chemother. 2007;51:1926–1933. doi: 10.1128/AAC.01607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Nash D, Sudimack D, Jaidee A, Barends M, Uhlemann AC, Krishna S, Nosten F, Anderson TJ. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol Biol Evol. 2007;24:562–573. doi: 10.1093/molbev/msl185. [DOI] [PubMed] [Google Scholar]
- Nkhoma S, Molyneux M, Ward S. In vitro antimalarial susceptibility profile and pfcrt/pfmdr-1 genotypes of Plasmodium falciparum field isolates from Malawi. Am J Trop Med Hyg. 2007;76(6):1107–1112. [PubMed] [Google Scholar]
- Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Zalewski C, Kawamoto F, Miller RS, Meshnick SR. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47:2418–2423. doi: 10.1128/AAC.47.8.2418-2423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Cassar C, Brockman A, Duraisingh M, van Vugt M, White NJ, Nosten F, Krishna S. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–2949. doi: 10.1128/aac.43.12.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Uhlemann AC, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Uhlemann AC, van Vugt M, Brockman A, Hutagalung R, Nair S, Nash D, Singhasivanon P, Anderson TJ, Krishna S, White NJ, Nosten F. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis. 2006;42:1570–1577. doi: 10.1086/503423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 Copy Number in Plasmodium falciparum Malaria Heightens Susceptibility to Mefloquine, Lumefantrine, Halofantrine, Quinine and Artemisinin. J Infect Dis. 2006;194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, Gil JP. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- Sisowath C, Ferreira PE, Bustamante LY, Dahlstrom S, Martensson A, Bjorkman A, Krishna S, Gil JP. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop Med Int Health. 2007;12:736–742. doi: 10.1111/j.1365-3156.2007.01843.x. [DOI] [PubMed] [Google Scholar]
- Triglia T, Foote SJ, Kemp DJ, Cowman AF. Amplification of the multidrug resistance gene pfmdr1 in Plasmodium falciparum has arisen as multiple independent events. Mol Cell Biol. 1991;11:5244–5250. doi: 10.1128/mcb.11.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlemann AC, Ramharter M, Lell B, Kremsner PG, Krishna S. Amplification of Plasmodium falciparum multidrug resistance gene 1 in isolates from Gabon. J Infect Dis. 2005;192:1830–1835. doi: 10.1086/497337. [DOI] [PubMed] [Google Scholar]
- Wilson CM, Volkman SK, Thaithong S, Martin RK, Kyle DE, Milhous WK, Wirth DF. Amplification of pfmdr1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol Biochem Parasitol. 1993;57:15–160. doi: 10.1016/0166-6851(93)90252-s. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]


