Summary
The antagonistic pleiotropy theory of aging proposes that aging takes place because natural selection favors genes that confer benefit early on life at the cost of deterioration later in life. This theory predicts that genes that impact development would play a key role in shaping adult lifespan. To better understand the link between development and adult lifespan, we examined the genes previously known to be essential for development. From a pool of 57 genes that cause developmental arrest after inhibition using RNA interference, we have identified 24 genes that extend lifespan in Caenorhabditis elegans when inactivated during adulthood. Many of these genes are involved in regulation of mRNA translation and mitochondrial functions. Genetic epistasis experiments indicate that the mechanisms of lifespan extension by inactivating the identified genes may be different from those of the insulin/insulin-like growth factor 1 (IGF-1) and dietary restriction pathways. Inhibition of many of these genes also results in increased stress resistance and decreased fecundity, suggesting that they may mediate the trade-offs between somatic maintenance and reproduction. We have isolated novel lifespan-extension genes, which may help understand the intrinsic link between organism development and adult lifespan. Key words: developmental arrest; antagonistic pleiotropy; aging; mRNA translation; mitochondria; C. elegans.
Introduction
The use of Caenorhabditis elegans has led to a number of insights into the molecular mechanisms of aging. Mutations in the daf-2 insulin/insulin-like growth factor 1 (IGF-1) receptor gene double adult lifespan (Kenyon et al., 1993; Kimura et al., 1997). DAF-2 functions through a kinase cascade to inhibit the DAF-16/forkhead box O (FOXO) transcription factor, which regulates the expression of many downstream genes (Lin et al., 1997; Ogg et al., 1997; Lee et al., 2003a; Murphy et al., 2003; Oh et al., 2006). Steroid hormone signaling from the reproductive system affects lifespan through DAF-16 (Arantes-Oliveira et al., 2003). Genes such as sir-2.1, jnk-1, smk-1 and cst-1 have been shown to regulate lifespan by modulating DAF-16 activities (Tissenbaum & Guarente, 2001; Oh et al., 2005; Berdichevsky & Guarente, 2006; Lehtinen et al., 2006; Wolff et al., 2006). Dietary restriction extends C. elegans lifespan, although the molecular mechanisms remain to be defined. Down-regulation of the TOR (target of rapamycin) pathway (Jia et al., 2004) and inhibition of mitochondrial respiration and adenosine triphosphate (ATP) synthesis also extend worm lifespan by DAF-16 independent mechanisms (Dillin et al., 2002b; Lee et al., 2003b).
To identify new genes affecting C. elegans lifespan, two groups have performed genome-wide RNA interference (RNAi) screens independently (Hamilton et al., 2005; Hansen et al., 2005). Eighty-nine and 23 genes were identified by each group, respectively. Surprisingly, only three genes were common in the two screens, suggesting the screens have not been saturated. In those screens, animals were treated with RNAi from early larval stages. Genes that play essential roles during development were likely excluded from the screens as knocking down those genes may cause larval arrest or embryonic lethality. However, evolutionary theories of aging suggest that genes that play essential roles during development are likely to be important for aging.
The underlying principle of evolutionary theories of aging is that the force of natural selection declines with age (Medawar, 1952). Two possible hypotheses have been proposed to explain the existence of aging based on this principle. The mutation accumulation theory suggests that deleterious mutations that manifest their effects late in life would accumulate in a population and lead to senescence as they are not removed due to lack of selective pressure (Medawar, 1952). The antagonistic pleiotropy theory suggests the existence of pleiotropic genes that endow benefits early in life at the cost of deleterious effects later to explain the evolution of senescence (Williams, 1957). A prediction of this theory is that genes that are essential for early life development play important roles later on in adulthood and aging. daf-2 represents a good example of an antagonistic pleiotropic gene. Null mutations of daf-2 cause embryonic lethality or L1 larval arrest, whereas weak alleles of daf-2 extend adult lifespan (Kenyon et al., 1993; Kimura et al., 1997; Gems et al., 1998). Another example is the microRNA (miRNA) gene lin-4, which regulates developmental timing and adult lifespan through the downstream transcription factor LIN-14 (Boehm & Slack, 2005).
In this study, we found 24 out of 57 genes that play essential roles during development also regulate adult lifespan. Our findings are consistent with the evolutionary theory of aging, and may help understand the role of developmental processes in longevity.
Results
A previous C. elegans genome-wide RNAi screen performed by Kamath et al. (2003) identified 57 genes that lead to larval arrest phenotypes consequent on RNAi inactivation (Supplementary Table S1 and Fig. S1). We treated animals with these RNAi clones only during adulthood, and measured the adult lifespan. Surprisingly, 24 out of 57 RNAi treatments caused lifespan extension. One gene, atp-3, has been previously reported to extend lifespan (Dillin et al., 2002b; Lee et al., 2003b; Hamilton et al., 2005; Curran & Ruvkun, 2007). Thus, we have identified 23 new genes that affect adult lifespan in C. elegans (Table 1, Fig. 1 and Supplementary Fig. S1). We classified these genes into multiple functional groups. BLAST (basic local alignment search tool) search indicates that most of the identified genes have strong orthologs in other organisms (Supplementary Table S2), suggesting their roles in aging may be conserved.
Table 1.
Lifespan extension in different genetic backgrounds
| Gene | Brief description | Predicted functional group | N2 extension† | daf-2 extension† | daf-16; daf-2 extension† | eat-2 extension† |
|---|---|---|---|---|---|---|
| Y39G10AR.8 | eIF-2γ | Translation initiation | 24%** | 41%** | 9%** | 32%** |
| D2085.3 | eIF-2Bε | Translation initiation | 18%** | 45%** | 12%** | 23%** |
| D2013.7 | eIF-3f | Translation initiation | 28%** | 31%** | 13%** | 41%** |
| C09D4.5 | rpl-19 | Ribosome subunit | 11%** | 50%** | 17%** | 24%** |
| W01B11.3 | Ribosome biogenesis protein Nop58p/Nop5p | Ribosome biogenesis | 15%** | 51%** | 12%** | 35%** |
| F59C6.4 | 3′–5′ exoribonuclease subunit involved in rRNA processing | Ribosome biogenesis | 12%** | 34%** | 14%** | 28%** |
| Y48G1A.4 | Nucleolar protein involved in 40S ribosome biogenesis | Ribosome biogenesis | 21%** | 39%** | 10%* | 19%** |
| C47D12.6 | Threonyl-tRNA synthetase | tRNA synthesis | 12%** | 19%* | 8%* | 11%* |
| F22D6.3 | Asparaginyl-tRNA synthetase | tRNA synthesis | 11%** | 30%** | 9%** | 28%** |
| C48E7.2 | RNA polymerase III subunit | Transcription | 26%** | 29%** | 14%** | 32%** |
| F14B4.3 | RNA polymerase I subunit | Transcription | 15%** | 31%** | 9%* | 27%** |
| R53.4 | Mitochondrial F1F0-ATP synthase subunit f | ATP synthesis | 20%** | 57%** | 9%* | 43%** |
| F59C6.5 | NADH:ubiquinone oxidoreductase subunit NDUFB10/PDSW | ATP synthesis | 16%** | 49%** | 10%* | 23%** |
| ZK973.10 | NADH:ubiquinone oxidoreductase subunit NDUFS4 | ATP synthesis | 17%** | 31%** | 11%** | 30%** |
| C01F1.2 | Cytochrome C oxidase assembly protein | ATP synthesis | 23%** | 24%** | 15%** | 17%** |
| K01C8.6 | Mitochondrial rpl-10 | Mitochondrial translation | 17%** | 38%** | 13%** | 26%** |
| F59A3.3 | Mitochondrial rpl-24 | Mitochondrial translation | 14%** | 28%** | 12%** | 28%** |
| B0511.8 | Mitochondrial rps-30 | Mitochondrial translation | 21%** | 44%** | 17%** | 25%** |
| Y47G6A.10 | AAA+-type ATPase | Mitochondrial translation | 24%** | 43%** | 13%** | 25%** |
| F26E4.4 | Cell death regulator Aven | Mitochondrial | 17%** | 27%** | 9%** | 20%** |
| C56E6.1 | ABC superfamily transporter | Signaling | 16%** | 38%** | 14%** | 27%** |
| K11B4.1 | Uncharacterized conserved protein | Unkown | 16%** | 29%** | 11%** | 28%** |
| B0491.5 | Uncharacterized protein | Unkown | 16%** | 43%** | 9%** | 35%** |
RNAi inactivation of the identified genes during adulthood cause lifespan extension in the wild-type N2, daf-2(e1370), daf-16(mu86); daf-2(e1370) and eat-2(ad465) animals. All lifespan experiments were performed at 20 °C. Lifespan of animals treated with different RNAi were compared with those treated with the control RNAi (empty vector) using the log-rank method.
Mean lifespan extension compared to animals treated with the control RNAi.
P < 0.001;
P < 0.01.
P values were calculated using the log-rank method (Graphic Prism 4).
Fig. 1.
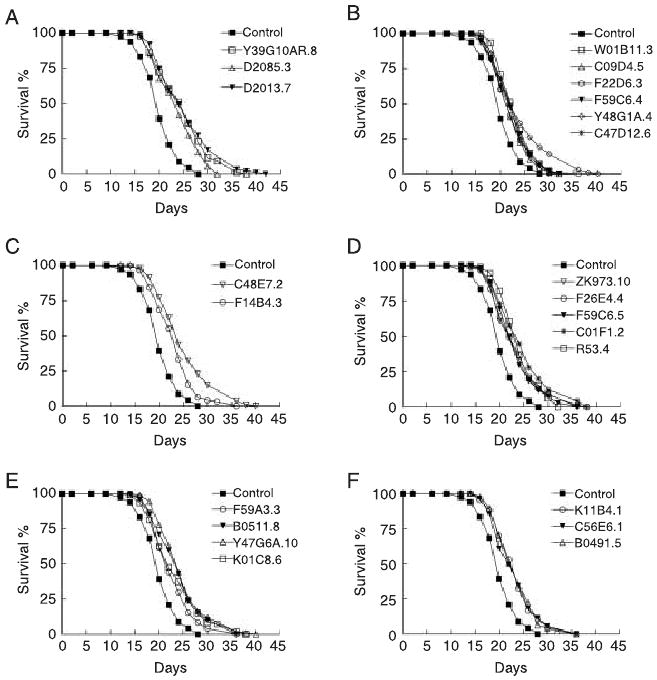
RNAi inactivation of 23 genes extends N2 lifespan. Lifespan of animals treated with RNAi against genes involved in translation initiation (A); ribosome and tRNA biogenesis (B); transcription (C); mitochondrial respiration and adenosine triphosphate (ATP) synthesis (D); mitochondrial translation (E); and signaling and unknown functions (F).
Among the 24 genes identified, most of them are involved in regulation of mRNA translation (42%, 10/24) and mitochondrial functions (38%, 9/24). Previous studies have shown that RNAi inactivation of various components of the eukaryotic initiation factor 4F (eIF4F) complex, which is composed of eIF4E (a 5′-cap binding protein), eIF4A (an ATP-dependent helicase) and eIF4G (a scaffold protein), decreases cap-dependent translation and extends lifespan in C. elegans (Henderson et al., 2006; Curran & Ruvkun, 2007; Hansen et al., 2007; Pan et al., 2007; Syntichaki et al., 2007). In this study, we identified eIFs that have not been previously linked with determining adult lifespan. Genes Y39G10AR.8, D2085.3 and D2013.7 encode eIF-2γ, eIF-2Bε and eIF-3f, respectively. RNAi inactivation of these genes results in most robust lifespan extension in wild-type animals (Table 1 and Supplementary Table S3). The initiation of mRNA translation in eukaryotic cells requires coordinated activities of multiple eIFs. A ternary complex composed of eIF2, guanosine triphosphate (GTP) and methionine transfer RNA (Met-tRNA) associates with the 40S ribosome to form a 43S pre-initiation complex (Hershey & Merrick, 2000). eIF2, composed of eIF2α, β, γ, delivers Met-tRNA to the 40S ribosome. The eIF2γ subunit contains a consensus GTP-binding domain. The 43S pre-initiation complex binds to an mRNA near the 5′-cap facilitated by eIF4 factors, and it scans the mRNA for the AUG start codon. Once the start codon is found, GTP hydrolysis makes the guanosine diphosphate (GDP)-binding eIF2 detach from the ribosome. The guanine nucleotide exchange factor (GEF) eIF2B exchanges GTP for GDP on eIF2 and allows eIF2 to participate in subsequent rounds of translation initiation (Hinnebusch, 2000). The eIF2B∈ subunit has GEF activity and nteracts directly with the eIF2γ subunit (Alone & Dever, 2006). The binding of the ternary complex to the 40S ribosome is promoted by eIF3 (Majumdar et al., 2003). eIF3 consists of an active core and subunits that serve to modulate eIF3 activity. eIF3f, one of these subunits, is a member of the Mov34 family, which is involved in translation initiation, regulation of the proteasome and transcription (Aravind & Ponting, 1998). eIF3f is down-regulated in several human tumors. It is a negative regulator of translation and cell growth, and it is also involved in apoptosis. eIF3f might play important roles in the regulation of eIF3 activity for translation of specific mRNAs (Shi et al., 2006).
We also identified other components of the translation machinery, including genes encoding a ribosomal subunit (C09D4.5), and genes involved in ribosomal biogenesis (W01B11.3, F59C6.4 and Y48G1A.4) and tRNA synthesis (C47D12.6 and F22D6.3). It has been shown by Hansen et al. (2007) that RNAi inactivation of C09D4.5 extends lifespan. Two genes (F14B4.3 and C48E7.2) encoding subunits of RNA polymerase I and III were also identified. As the major transcriptional products of RNA polymerase I and III are precursors of rRNA and tRNA, respectively, these genes are also likely to play essential roles in regulation of mRNA translation. Taken together, various genes involved in post-transcriptional regulation of gene expression are important regulators of C. elegans lifespan.
Among the identified genes that regulate mitochondrial function, four genes are involved in mitochondrial protein synthesis. Three of these genes encode mitochondrial ribosomal proteins and the fourth gene Y47G6A.10 (spg-7) encodes an AAA-type ATPase. Mutations in the human homolog of spg-7 cause hereditary spastic paraplegia. The yeast homolog of spg-7 encodes a mitochondrial inner membrane protein that controls mitochondrial ribosome assembly (Nolden et al., 2005). We also identified other genes that are involved in mitochondrial respiration and ATP synthesis from our screen (Table 1), which is consistent with previous studies showing that inactivation of the genes involved in these processes extends lifespan (Feng et al., 2001; Dillin et al., 2002a; Lee et al., 2003b).
We set out to characterize the genetic mechanisms of lifespan extension using epistasis experiments. Mutations in the daf-2 insulin-like/IGF-1 receptor double lifespan (Kenyon et al., 1993; Kimura et al., 1997). We treated the daf-2(e1370) animals with RNAi during adulthood. Surprisingly, all of the RNAi treatments can further extend daf-2 lifespan (Table 1, Fig. 2 and Supplementary Table S3). However, it is possible that the maximal lifespan extension due to perturbing the insulin-like/IGF pathway may have not been achieved in this long-lived mutant. For example, daf-2 RNAi has been shown to further extend the lifespan of a daf-2 mutant (Arantes-Oliveira et al., 2003). Mutations in DAF-16/FOXO transcription factor functions can fully suppress lifespan extension by daf-2 mutations (Lin et al., 1997; Ogg et al., 1997). Therefore, we examined lifespan phenotypes in the daf-2; daf-16 background to test whether the additive lifespan extensions in the daf-2 background were independent of the insulin-like/IGF signaling pathway. We treated the daf-2; daf-16 double mutant with RNAi against the genes identified from the targeted screen. Lifespan extension was observed in every RNAi treatment (Table 1, Fig. 3 and Supplementary Table S3). Therefore, the genes that we identified are likely to affect lifespan through mechanisms different from the insulin-like/IGF signaling.
Fig. 2.
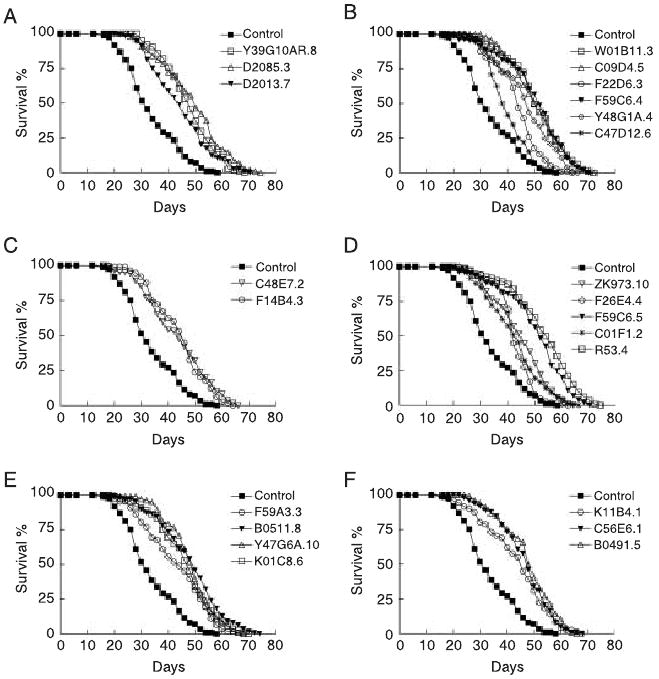
RNAi inactivation of 23 genes further extends daf-2(e1370) lifespan. Lifespan of animals treated with RNAi against genes involved in translation initiation (A); ribosome and tRNA biogenesis (B); transcription (C); mitochondrial respiration and adenosine triphosphate (ATP) synthesis (D); mitochondrial translation (E); and signaling and unknown functions (F).
Fig. 3.
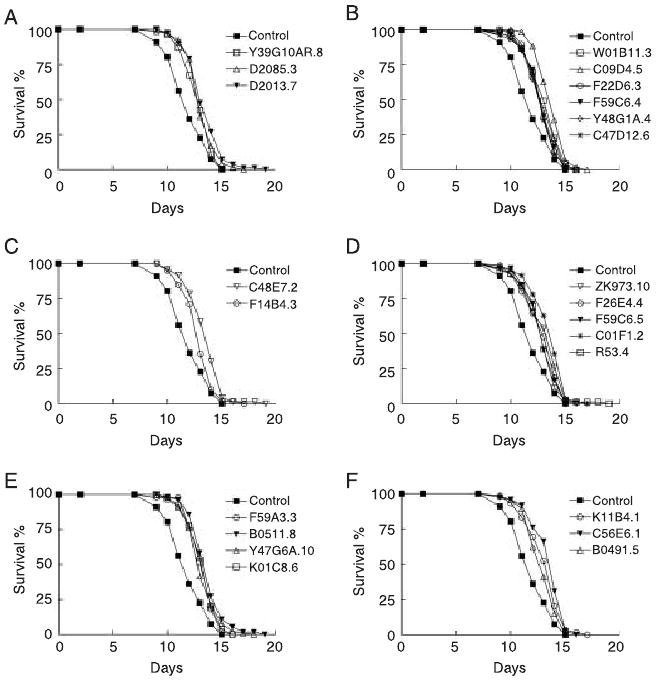
RNAi inactivation of 23 genes extends daf-16(mu86); daf-2(e1370) lifespan. Lifespan of animals treated with RNAi against genes involved in translation initiation (A); ribosome and tRNA biogenesis (B); transcription (C); mitochondrial respiration and adenosine triphosphate (ATP) synthesis (D); mitochondrial translation (E); and signaling and unknown functions (F).
The eat-2 gene encodes a subunit of nicotinic acetylcholine receptor. Mutations in eat-2 result in slow pharyngeal pumping and extended lifespan (Lakowski & Hekimi, 1998). eat-2 mutants have been suggested to be genetic mimics of dietary restriction in previous studies (Lakowski & Hekimi, 1998). We treated the eat-2(ad465) mutant with RNAi. Inactivation of all these genes by RNAi during adulthood further extends eat-2 lifespan (Table 1, Fig. 4 and Supplementary Table S3), suggesting genetic mechanisms of lifespan extension distinct from those regulated by dietary restriction.
Fig. 4.
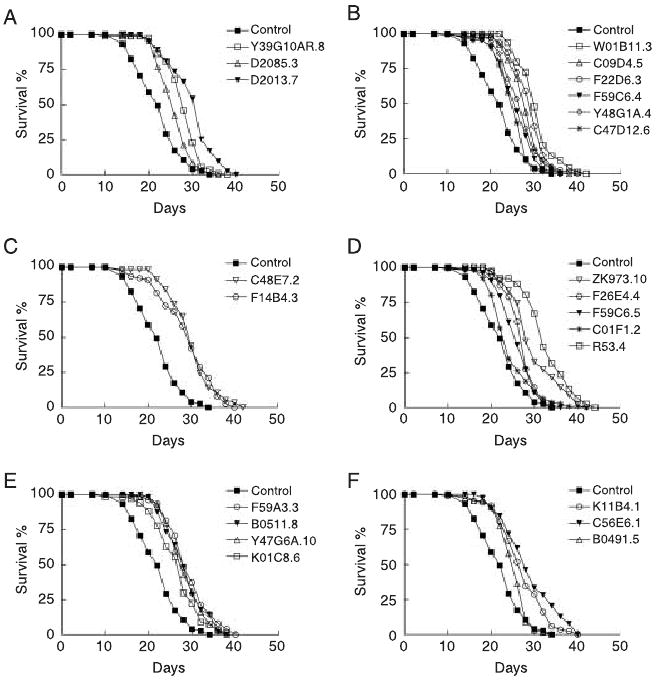
RNAi inactivation of 23 genes further extends eat-2(ad465) lifespan. Lifespan of animals treated with RNAi against genes involved in translation initiation (A); ribosome and tRNA biogenesis (B); transcription (C); mitochondrial respiration and adenosine triphosphate (ATP) synthesis (D); mitochondrial translation (E); and signaling and unknown functions (F).
The disposable soma theory suggests that aging takes place due to trade-offs between resources spent on somatic maintenance, which ensures prolonged survival, vs. resources for growth and reproduction, which enhance the fitness of the individual (Martin et al., 1996). To test whether the identified genes modulate lifespan by mediating such a trade-off, we examined survival under stressful conditions as well as fecundity. Animals at the fourth larval stage were treated with RNAi for 2 days. Adult animals were incubated at 35 °C for heat stress assays, or transferred to nematode growth medium (NGM) agar plates containing 360 mm juglone for oxidative stress assays. Various RNAi treatments increased tolerance to heat or oxidative stress (Table 2 and Supplementary Table S4) supporting the idea that common mechanisms may be shared between regulation of lifespan and stress tolerance (Johnson et al., 1996; Martin et al., 1996). We also measured the brood size of animals treated with different RNAi. Interestingly, all RNAi inactivation except C48E7.2, which encodes a subunit of RNA polymerase III, reduced fecundity (Table 2, Fig. 5 and Supplementary Table S4). Taken together these data support the idea that lifespan is determined by trade-offs between somatic maintenance vs. growth and reproduction.
Table 2.
Inactivation many of the 23 genes by RNAi during adulthood result in increased stress resistance and decreased fecundity
| Stress resistance | ||||
|---|---|---|---|---|
| Gene | Brief description | Heat* | Juglone† | Reduced fecundity‡ |
| Y39G10AR.8 | eIF-2γ | Yes | Yes | Yes |
| D2085.3 | eIF-2Bε | No | No | Yes |
| D2013.7 | eIF-3f | Yes | Yes | Yes |
| C09D4.5 | rpl-19 | Yes | Yes | Yes |
| W01B11.3 | Ribosome biogenesis protein Nop58p/Nop5p | Yes | No | Yes |
| F59C6.4 | 3′–5′ exoribonuclease subunit involved in rRNA processing | Yes | No | Yes |
| Y48G1A.4 | Nucleolar protein involved in 40S ribosome biogenesis | Yes | Yes | Yes |
| C47D12.6 | Threonyl-tRNA synthetase | Yes | No | Yes |
| F22D6.3 | Asparaginyl-tRNA synthetase | Yes | No | Yes |
| C48E7.2 | RNA polymerase III subunit | Yes | Yes | No |
| F14B4.3 | RNA polymerase I subunit | No | No | Yes |
| R53.4 | Mitochondrial F1F0-ATP synthase subunit f | Yes | Yes | Yes |
| F59C6.5 | NADH:ubiquinone oxidoreductase subunit NDUFB10/PDSW | Yes | Yes | Yes |
| ZK973.10 | NADH:ubiquinone oxidoreductase subunit NDUFS4 | Yes | No | Yes |
| C01F1.2 | Cytochrome C oxidase assembly protein | Yes | Yes | Yes |
| K01C8.6 | Mitochondrial rpl-10 | Yes | Yes | Yes |
| F59A3.3 | Mitochondrial rpl-24 | No | No | Yes |
| B0511.8 | Mitochondrial rps-30 | Yes | Yes | Yes |
| Y47G6A.10 | AAA+-type ATPase | Yes | Yes | Yes |
| F26E4.4 | Cell death regulator Aven | Yes | No | Yes |
| C56E6.1 | ABC superfamily transporter | No | No | Yes |
| K11B4.1 | Uncharacterized conserved protein | Yes | No | Yes |
| B0491.5 | Uncharacterized protein | Yes | Yes | Yes |
Two-day-old adults treated with various RNAi were incubated at 35 °C and their survival was measured. Significant heat stress resistance (P < 0.05, log-rank test) is highlighted.
Two-day-old adults treated with different RNAi were incubated on nematode growth medium (NGM) plates containing juglone (360 mm) and their survival were measured. Significant oxidative stress resistance (P < 0.05, log-rank test) was highlighted.
Brood sizes of animals treated with RNAi were measured at 20 °C. P values were calculated using the t-test. Significant brood size decreases (P < 0.05) were highlighted.
Fig. 5.
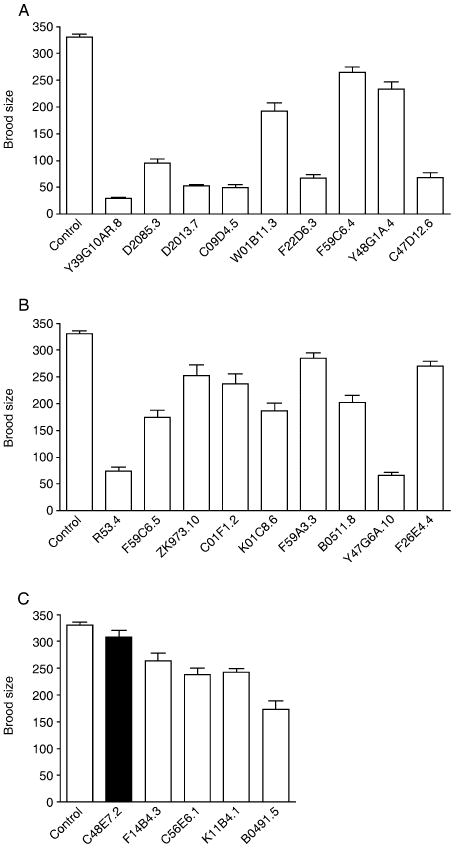
RNAi inactivation of most of the identified genes reduces fecundity. Brood sizes of animals treated with RNAi against genes involved in mRNA translation (A); mitochondrial functions (B); and transcription, signaling and unknown functions (C). RNAi inactivation of C48E7.2 does not affect brood size significantly.
Discussion
In his classic paper, Williams postulated the existence of pleiotropic genes that endow benefits early in life at the cost of deleterious effects later to explain the evolution of senescence (1957). Williams predicted ‘It would be expected that if development could be completely arrested there would be no senescence…’ Results of our study lend support to the idea that there is an intrinsic link between cellular processes that determine development and lifespan. Antagonistic pleiotropic genes have been proposed to function in two distinct ways (Partridge & Gems, 2006). The first type is when lifespan extension is observed by a gene's action during development. For example, inhibition of a gene involved in ATP synthesis during development is sufficient for adult lifespan extension (Dillin et al., 2002a). The second form of antagonistic pleiotropy is where the gene is essential for developmental processes (i.e. inhibition is harmful), but if its action is inhibited during adulthood it leads to lifespan extension. This study describes genes that belong to the latter category. Our data lend support to the evolutionary theories of aging that propose that the rate of aging is caused by the force of natural selection acting to optimize fitness early in life.
From a pool of 57 genes that showed developmental arrest when inactivated during development, we have identified 23 novel genes that extend lifespan in C. elegans when inhibited using RNAi only during adulthood. Remarkably, a number of these genes possess very close orthologs in flies and mammals including humans. These results uncover the identity of a number of genes that are consistent with the evolutionary theory of aging, and are likely to extend lifespan by conserved mechanisms across species.
Our results and previous studies indicate that down-regulation of key components of mRNA translation machinery extends adult lifespan in C. elegans (Curran & Ruvkun, 2007; Hansen et al., 2007; Henderson et al., 2006; Pan et al., 2007; Syntichaki et al., 2007), but the molecular mechanisms of lifespan extension remain unclear. It is possible that animals can sense reduced mRNA translation states and shift metabolism from growth to somatic maintenance. It is also possible that lifespan extension is caused by key factors that are regulated at the mRNA translation level. Previous studies showed that upon inhibition of translation initiation factors eIF4E and eIF4γ, the synthesis of most proteins is decreased. However, mRNAs encoding for some heat shock proteins are preferentially translated (Joshi-Barve et al., 1992). Therefore, identification of genes that are differentially translated upon inhibition of genes involved in protein synthesis may help explain the roles of mRNA translation in aging.
The disposable soma theory suggests that longevity is determined through the trade-offs between resources spent on somatic maintenance and reproduction (Martin et al., 1996). It has been estimated that the fraction of genes devoted to translation may be as high as 35–45% (Hershey & Merrick, 2000). We reason that inhibition of this costly process is likely to shift investment towards somatic maintenance and away from development, growth and reproduction. Our results are consistent with this notion, and we observed that inhibition of genes that regulate protein synthesis lead to increased stress resistance and a reduction in fecundity.
Previous studies showed that reduced activities of the electron transport chain and ATP synthase by RNAi during development but not adulthood lead to lifespan extension in C. elegans (Dillin et al., 2002b). However, we found that knocking-down genes involved in mitochondrial functions solely during adulthood can extend lifespan. One possible reason for this discrepancy could be the variable effectiveness of RNAi. Knocking down the genes identified in this study results in larval arrest, which was not the case in the previous study. Another possibility is that these genes may be involved in different aspects of mitochondrial functions. Further analysis on how mitochondria are involved in aging may help explain this discrepancy.
While our manuscript was under review, Curran & Ruvkun (2007) reported an RNAi screen for longevity genes from 2700 genes that play essential roles during development. They identified 64 genes that extend lifespan after inactivation by RNAi during adulthood. Four genes were common to both studies. Interestingly genes identified by Curran and Ruvkun are also enriched with those involved in protein synthesis and mitochondrial functions, suggesting these key processes link development of the organism and adult longevity.
Experimental procedures
Strains
Caenorhabditis elegans strains were cultured as described (Brenner, 1974). Genotypes of animals used were: daf-16(mu86) I, eat-2(ad465) II and daf-2(el370) III.
RNAi experiments
RNAi bacteria strains were cultured as previously described (Kamath et al., 2001). Worms were staged at old L4s and fed with Escherichia coli expressing different double-stranded RNAs. In all RNAi assays, E. coli carrying the empty RNAi vector L4440 was fed to animals as controls. Clone identity of all RNAi bacteria was verified by polymerase chain reaction (PCR) and DNA sequencing.
Lifespan assays
Lifespan assays were performed as previously described (Lithgow et al., 1995), except that 80 μL of 0.2 mm (+)-5-fluorodeoxyuridine (FUdR) was added onto plates during the reproductive phase to eliminate progeny. Worms were transferred onto fresh plates every 3–6 days. In all experiments, RNAi was introduced to old L4 larvae. All lifespan assays were performed at 20 °C. The first day of adulthood is Day 1 in the survival curves. Animals were scored as alive, dead or lost every other day. Animals that did not move in response to touching were scored as dead. Animals that died from causes other than aging, such as sticking to the plate walls, internal hatching or bursting in the vulval region, were scored as lost.
Statistical analysis
Statistical analyses were performed using the Prism 4 software (Graphpad Software, Inc., San Diego, CA, USA). Kaplan–Meier survival curves were plotted for each lifespan and compared using the log-rank test.
Fecundity analysis
Wild-type N2 L4 larvae growing at 20 °C were transferred to RNAi plates. Those animals were transferred every day to fresh RNAi plates and progeny produced during that 24-h period were counted.
Stress-resistance assays
Wild-type N2 L4 larvae were treated with RNAi and FUdR at 20 °C for 48 h. Adult animals were then collected for stress-resistance assays.
Thermotolerance
Thermotolerance assays were performed as previously described (Lithgow et al., 1995). Synchronized 2-day-old adults were shifted from 20 to 35 °C and survival scored by means of touch-provoked movement. Worms not responding to touch were scored as dead.
Oxidative stress
Juglone resistance was performed as previously described (de Castro et al., 2004). Ethanol-dissolved juglone solution was added to liquid NGM to a final concentration of 360 μm and poured immediately onto plates to dry. One hundred microliters of E. coli OP50 was spotted on the plates and dried. About 50 2-day-old adults were transferred to each plate 3 h after the plates were made. Worms were incubated at 20 °C and scored hourly until death.
Supplementary Material
Acknowledgments
We thank Drs. Gordon Lithgow, Julie Andersen, Matt Gill, Anders Olsen, Gawain McColl, Malene Hansen and Brian Zid for helpful discussions and comments on the manuscript. We also thank Drs. Gary Ruvkun and Sean Curran for sharing unpublished results. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources (NCRR). Di Chen was supported by a fellowship from the William Randolph Hearst Foundation. Pankaj Kapahi is supported by a grant from the Ellison Medical Foundation, Nathan Shock award, the American Federation for Aging Research foundation, and a gift from the Harold J. and Reta Haynes Family Foundation.
Footnotes
The following supplementary material is available for this article:
Fig. S1 Groups of genes.
Table S1 List of 57 genes that show larval arrest after RNAi inactivation.
Table S2 An RNAi screen identified 24 genes with functions in C. elegans development and aging.
Table S3 Lifespan extension in different genetic backgrounds.
Table S4 Heat stress, oxidative stress and fecundity assays.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1474-9726.2007.00305.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alone PV, Dever TE. Direct binding of translation initiation factor eIF2gamma-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2B epsilon. J Biol Chem. 2006;281:12636–12644. doi: 10.1074/jbc.M511700200. [DOI] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Berman JR, Kenyon C. Healthy animals with extreme longevity. Science. 2003;302:611. doi: 10.1126/science.1089169. [DOI] [PubMed] [Google Scholar]
- Aravind L, Ponting CP. Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci. 1998;7:1250–1254. doi: 10.1002/pro.5560070521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdichevsky A, Guarente L. A stress response pathway involving sirtuins, forkheads and 14-3-3 proteins. Cell Cycle. 2006;5:2588–2591. doi: 10.4161/cc.5.22.3513. [DOI] [PubMed] [Google Scholar]
- Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E, Hegi de Castro S, Johnson TE. Isolation of long-lived mutants in Caenorhabditis elegans using selection for resistance to juglone. Free Radic Biol Med. 2004;37:139–145. doi: 10.1016/j.freeradbiomed.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Crawford DK, Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002a;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002b;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88. [Google Scholar]
- Hinnebusch AG. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2000. pp. 185–244. [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Johnson TE, Lithgow GJ, Murakami S. Hypothesis: interventions that increase the response to stress offer the potential for effective life prolongation and increased health. J Gerontol A Biol Sci Med Sci. 1996;51:B392–B395. doi: 10.1093/gerona/51a.6.b392. [DOI] [PubMed] [Google Scholar]
- Joshi-Barve S, De Benedetti A, Rhoads RE. Preferential translation of heat shock mRNAs in HeLa cells deficient in protein synthesis initiation factors eIF-4E and eIF-4 gamma. J Biol Chem. 1992;267:21038–21043. [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003a;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003b;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci USA. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar R, Bandyopadhyay A, Maitra U. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40 S preinitiation complex. J Biol Chem. 2003;278:6580–6587. doi: 10.1074/jbc.M210357200. [DOI] [PubMed] [Google Scholar]
- Martin GM, Austad SN, Johnson TE. Genetic analysis of ageing: role of oxidative damage and environmental stresses. Nat Genet. 1996;13:25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- Medawar PB. An Unsolved Problem of Biology. London: H.K. Lewis; 1952. [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci USA. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Gems D. Beyond the evolutionary theory of ageing, from functional genomics to evo-gero. Trends Ecol Evol. 2006;21:334–340. doi: 10.1016/j.tree.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Shi J, Kahle A, Hershey JW, Honchak BM, Warneke JA, Leong SP, Nelson MA. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene. 2006;25:4923–4936. doi: 10.1038/sj.onc.1209495. [DOI] [PubMed] [Google Scholar]
- Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Williams GC. Pleiotropy, natural selection, and evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


