Abstract
Small bowel transplants provide an exceptional opportunity for long-term study of the microbial ecology of the human small bowel. The ileostomy created at time of transplant for ongoing monitoring of the allograft provides access to samples of ileal effluent and mucosal biopsies. In this study, we used qPCR to assay the bacterial population of the small bowel lumen of 17 small bowel transplant patients over time. Surprisingly, the posttransplant microbial community was found to be dominated by Lactobacilli and Enterobacteria, both typically facultative anaerobes. This represents an inversion of the normal community that is dominated instead by the strictly anaerobic Bacteroides and Clostridia. We found this inverted community also in patients with ileostomies who did not receive a transplant, suggesting that the ileostomy itself is the primary ecological determinant shaping the microbiota. After surgical closure of the ileostomy, the community reverted to the normal structure. Therefore, we hypothesized that the ileostomy allows oxygen into the otherwise anaerobic distal ileum, thus driving the transition from one microbial community structure to another. Supporting this hypothesis, metabolomic profiling of both communities demonstrated an enrichment for metabolites associated with aerobic respiration in samples from patients with open ileostomies. Viewed from an ecological perspective, the two communities constitute alternative stable states of the human ileum. That the small bowel appears to function normally despite these dramatic shifts suggests that its ecological resilience is greater than previously realized.
Keywords: 16S ribosomal RNA, alternative stable state, human microbiota, lactobacilli, intestinal allograft
Every human individual hosts an ecological experiment within. From the moment of birth, many of our organs are colonized by communities of microorganisms, especially our intestine. These microbes have diverse and sometimes profound effects on their human hosts. For example, they educate the immune system, aid in the digestion of foods, and provide a barrier to potentially invasive pathogenic counterparts (1). In a healthy human, the microbes digest complex plant polysaccharides and interact with gut epithelial cells in a neutral or even positive way. However, many factors can disturb the normal microbe-human balance.
A small bowel transplant (SBT) is a life-saving procedure for patients without sufficient bowel length to absorb enough nutrients to survive. Within the past decade, both the frequency and survival rates (now >70%) of SBT have increased due to greater awareness of this procedure and improved immunosuppression therapies (2). This patient population is at high risk for graft rejection, often followed by bacterial sepsis. However, the role of the microbiota in these severely immune-suppressed individuals is unknown.
Intestinal allograft rejection resembles Crohn's disease clinically and pathologically, and increased incidence of Crohn's disease has been found to be associated with mutations in the NOD2 gene. The NOD2 protein plays a key role in intestinal immune health, specifically in the sensing of microbial products and the production of antimicrobial peptides by the Paneth cells of the intestine. The likelihood of allograft rejection and sepsis in SBT patients with mutant NOD2 genes is ≈100 times higher than in patients with the wild-type gene (3). Therefore, we undertook this study (1) to characterize the microbiota colonizing this severely traumatized small bowel and (2) to test for correlations between shifts in that microbiota and clinical factors such as NOD2 genotype.
During SBT, the donor small bowel and its associated microbial community is traumatically disconnected, exposed to the environment, and surgically placed in another individual. In earlier SBTs, the donor's microbiota was removed by decontamination of the transplanted organ. Consequently, the small bowel would be completely recolonized with microbes from the upper GI tract and environment. In this case, the SBT can be viewed as the creation of a new ecosystem. In later SBT, decontamination was discontinued because of the growing understanding of the beneficial role of the microbiota, and the intestine was transplanted undisturbed from the donor to the recipient. In this case, SBT not only creates a new organ, but also inoculates the recipient with a putatively healthy microbiota.
At the end of the SBT surgery, an ileostomy (portal) through the abdominal wall is surgically created to allow access into the transplant site. Although its primary purpose is surveillance of the transplanted tissue for possible graft rejection, it also creates a rare opportunity to monitor the changes to the ileal microbial community through time during a clinically critical period. In no other medical situation is an ileostomy accessed so frequently or is such extensive clinical metadata available. This opportune combination is unparalleled.
Opportunities to sample the mid-GI tract have been rare; thus fecal samples have often been used as a surrogate. However, observations of the microbial ecosystem within the transplanted small intestine can provide insights of potential clinical and ecological significance. The roles of founder effects, environmental perturbations, host factors, and the resumption of bowel function, among others, can be observed. Correlations found can inform the protocol for future SBTs and suggest new research directions for studies of chronic gastrointestinal disorders, such as inflammatory bowel disease.
Many recent studies have begun to define the role of our human-associated microbiota, but several areas of research are relatively under-developed (4–11, 35). For example, we know little about this “ecological experiment” from the perspective of time (12), particularly time in our adult years (13) and time from the onset of disease to the recovery of health. In this study, we used a molecular approach to survey microbial diversity within the luminal contents of the transplanted small bowel, from several days after transplantation until graft rejection or establishment of normal function. We are unaware of another attempt to characterize the microbial community in this newly created ecosystem.
Results
SBTs and Sample Collection at GUMC.
All patients aiding in this research were enrolled through an on-going study at the Georgetown University Medical Center's Transplant Institute. The study was approved by the Institutional Review Board (IRB 2004–08) and informed consent was obtained for all patients. All SBT transplants were performed by a single surgeon (T.F.) during 2005–2007 using a method described in ref. 14. All intestinal allografts were from NOD2 wild type donors. The 17 individuals in this cohort were retrospectively selected based on two criteria: that they were adults and that they each had at least 10 posttransplant sampling points. This second criterion ensured that the subjects could be followed through an extended period, but in so doing it also might have biased the cohort toward individuals with better outcomes. The diverse clinical characteristics of the 17 subjects are presented in Table S1; sample collection time points, key clinical events, and raw qPCR bacterial data are included in Dataset S1. Of the 17 patients included, 13 survived >2 years after transplant.
Endoscopic surveillance of the allograft tissue via the ileostomy enabled us to collect a total of 251 samples of ileal effluent (contents of the lumen of the small intestine) from a location 5–10 cm into the ileum. Samples from the 17 patients were not time matched because the monitoring schedule, and thus the opportunity for effluent collection, was determined by the medical need to observe the allograft (transplanted small intestine) for healing and potential rejection.
Bacterial Diversity Post-Transplant.
Ileal bacterial diversity was assessed by sequencing the 16S rDNA in ileal effluent samples collected between 50 and 84 days posttransplant from nine randomly selected patients (SI Text). Four bacterial orders—Lactobacillales, Enterobacteriales, Bacteroidales, Clostridiales—represented 99.8% of the 1892 full-length bacterial 16S rDNA sequences obtained (Fig. S1B). The remaining 0.2% were four sequences from Fusobacteria and two from Actinobacteria.
Because only four orders of bacteria were found (with relatively low bacterial diversity, e.g., average Shannon index 1.3 for these libraries compared with 3.1 observed in ileal samples in ref. 15), we evaluated the use of qPCR in place of sequencing for these studies (Materials and Methods). qPCR methodology is less expensive, is more quantitative, and is more efficient in terms of time and operation. The relative proportions of those four taxonomic groups determined by qPCR (Fig. S1A) and by direct sequencing of full-length 16S rRNA sequences (Fig. S1B) were highly concordant, as determined by Lin's concordance correlation coefficient (CCC) (Fig. S1 C and D) (16). Therefore, qPCR was used for all subsequent analyses.
Facultative Anaerobes Dominate the Post-SBT Microbiota.
The bacterial populations present in the 229 posttransplant samples obtained before ileostomy closure were characterized using qPCR. The relative proportions of the four bacterial orders assayed were remarkably similar between individuals despite variability in the total bacterial count (Fig. 1).
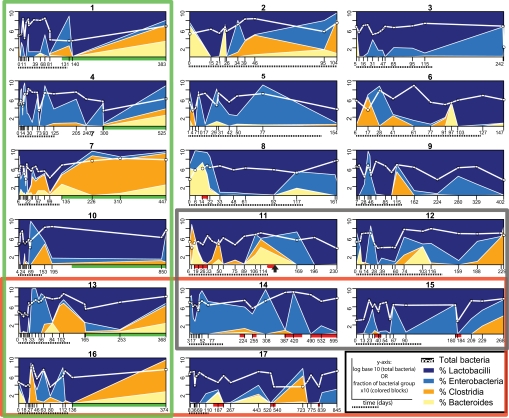
Fig. 1.
Bacterial populations as measured by qPCR for each of the 17 SBT patients at all available time points. The bacterial group fractions (colored blocks) were calculated by summing the four individual groups and representing that total as 100%. The different x axis scales reflect each patient's individual timetable. x axis: red bar, graft rejection; green bar, postsurgical closure of the ileostomy; dashed black line beneath, day 0–125, the period used for time-binning (Figs. S2 and S3). Multipatient groupings: green box, ileostomy closed; red box, NOD2 mutations (SI Text); black box, died during the study; black arrow (patient 14), a second SBT at day 128.
Unexpectedly, the Lactobacilli and Enterobacteria were generally much more abundant than the Bacteroides and Clostridia. This represents an inversion of the normal microbial profile reported for the human ileum (15, 17, 18, 23). In those studies, the strictly anaerobic Clostridia and Bacteroides dominated the community whereas the facultatively anaerobic Lactobacilli and Enterobacteria were but minor members. Although designation as a “facultative” or “strict” anaerobe cannot be assumed to apply to all members of a high-level bacterial taxon, these four particular bacterial orders are well characterized and are widely thought to fit these respiratory categorizations (19). Eight of the seventeen patients (1, 2, 3, 5, 8, 11, 13, 16) (Fig. 1) have higher levels of strict anaerobes at very early time points; 75% of these (1, 3, 5, 8, 13, 16) received a transplanted small bowel that had not been decontaminated and thus could have served as an inoculum.
Marked fluctuations in the relative proportions of the four groups and total bacteria occurred over the sampling period in every patient (Fig. 1). These are especially evident in the dominant groups, the Lactobacilli and Enterobacteria. Therefore, a battery of clustering algorithms and multivariate statistical tests (SI Text) were used to search for correlations between these fluctuations and specific clinical factors such as patient outcome (normal bowel function or death), ileostomy type (which determines whether mixing from the colon was possible or not), and NOD2 genotype. A normalized, time-binned dataset was prepared and used for these tests (Fig. S2). Attempts at clustering and statistical separation failed and bacterial population variability was variable between individuals, but was not associated with any clinical factor (Fig. S3).
Microbial Populations Revert to “Normal” After Ileostomy Closure.
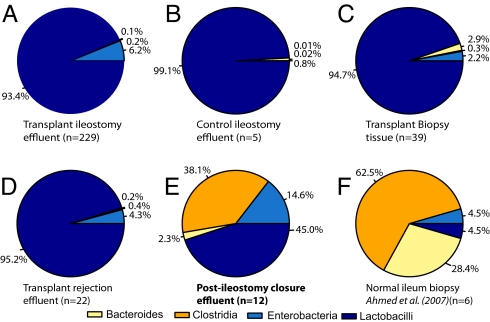
In 10 of the 17 subjects, the ileostomy was later surgically closed, thus ending the exposure of the transplanted tissue to the external environment. We were able to obtain only 12 postclosure ileal samples from six subjects due to the clinical difficulty of sampling postclosure (requiring a colonoscopy that traverses the entire colon from the rectum). In 10 of the 12 samples obtained, the microbial community contained a high proportion of strict anaerobes and fewer facultative anaerobes (Fig. 1, green bars; Fig. 2E)—the reverse of the distribution observed in the preclosure samples. The community composition of these postclosure samples is very similar to the microbial profiles published for mucosal biopsies of the normal human ileum (17) (data regraphed in Fig. 2F).
Fig. 2.
Summary of the relative bacterial populations found under various clinical conditions. In each chart, the population of each of the four bacterial orders is shown as the geometric mean of its percentage of the total population. (A) Ileal effluent from transplant patients, ileostomy present. (B) Ileal effluent from nontransplant patients, ileostomy present (Fig. S4). (C) Biopsy samples from transplant patients, ileostomy present (Fig. 3). (D) Ileal effluent from transplant patients during transplant rejection, ileostomy present. (E) Ileal effluent from transplant patients, ileostomy closed. (F) Regraphed selected data from table 3 in ref. 17 from biopsies of the terminal ileum of six healthy subjects.
Pre-Closure Populations Resemble Temporary Ileostomy Populations.
We noted that the high proportion of facultative anaerobes observed in the preclosure SBT samples might be due to the presence of the ileostomy rather than to the SBT itself. To test this hypothesis, ileal effluent samples from five other subjects with temporary ileostomies were analyzed using qPCR. The microbiota in these five nontransplant ileostomy control samples was strikingly similar to the patterns observed in the preclosure SBT transplant samples (Fig. 2A, B, and D and Fig. S4). Notably, the population of Bacteroides and Clostridia was higher in two patients (Fig. S4, patients 2 and 4). In these patients, the ileostomy had been created less than one week before sampling, whereas in the other three the ileostomies had been present for at least several months.
All five ileostomy control samples were collected externally at the ileostomy port, whereas the preclosure SBT samples had been obtained endoscopically from a location 5–10 cm into the ileum. Therefore, as a further control, effluent was collected externally from SBT patient 9 (with a permanent end ileostomy, i.e., no colon) at 2.5 years posttransplant. The same bacterial profile was observed in samples collected by both methods (Fig. S4). Furthermore, the data obtained for this patient demonstrated that this characteristic bacterial population was maintained for at least 2.5 years in the presence of an ileostomy.
Mucosal vs. Planktonic Bacterial Groups.
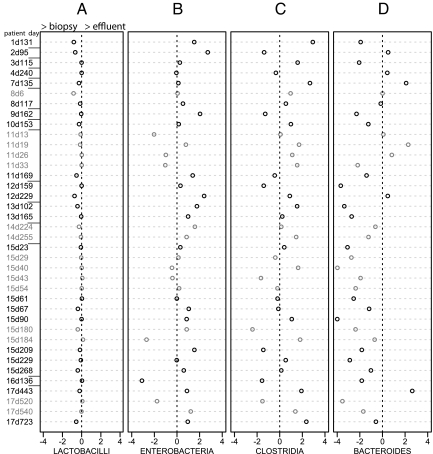
The microbiota adhering to the mucosal epithelium where host and microbe are directly interacting could be significantly different from the planktonic populations observed in the effluent samples. To investigate this possibility, we analyzed the bacteria adhering to 39 unwashed biopsy specimens collected from SBT patients during either graft health or rejection (Fig. 3). The data obtained indicate that the bacterial species composition of the effluent (Fig. 2A) and the mucosa (Fig. 2C) are quite similar. One bacterial order, the Bacteroides, showed higher levels in biopsy specimens (Fig. 2 C and Fig. 3D). This is consistent with previous observations that Bacteroides spp. preferentially occupy an adherent rather than planktonic niche in the human gut (20) and that they are more abundant in biopsy samples relative to fecal material (21). The exact ecological niche of Bacteroides is unclear as a recent study of fecal samples observed more planktonic Bacteroides than particulate-attached (22).
Fig. 3.
Comparison of effluent and biopsy-associated populations. The log10 of the ratio between biopsy samples (positive direction) and effluent samples (negative direction) [i.e., log10(biopsy percentage/effluent percentage)] for all four subgroups of bacteria measured with qPCR. Biopsy and effluent samples taken during a period of graft rejection are colored in gray, whereas pathologically normal tissue is colored in black. The naming of samples is patient number first, followed by “d” (day pos-transplant of collection). Log scale on the x axis means that 2 is 100× greater proportion of that bacterial group relative to the other sample type.
Metabolomics of Preclosure and Postclosure Effluent Samples.
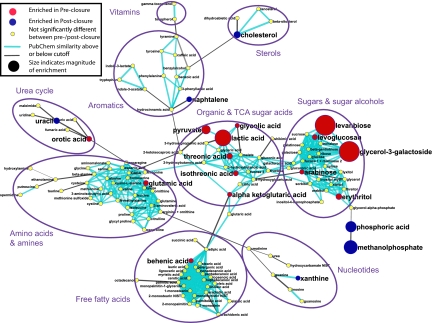
The different microbiota present in the preclosure and postclosure effluent suggested that the metabolism of these two microbial communities would also be distinctly different. To assess this, we used GC-TOF mass spectrometry-based metabolomic profiling (Materials and Methods), an approach that has been used in other studies (5) to assess global metabolite differences. The data are available at http://setupx.fiehnlab.ucdavis.edu/m1/main_public.jsp. Principal component analysis and partial least squared regression clearly demonstrated that the preclosure and postclosure samples were statistically separable and distinct (Fig. S5). Significant differences between the preclosure and postclosure samples (P < 0.1) were found for 18.3% of the metabolites (63 of 345) determined by ANOVA. 21 of these compounds were identifiable by their mass spectra (knowns), whereas 45 could not be identified (unknowns), but can be loosely categorized by known peaks in the mass spectra. A metabolic network diagram was constructed for all 139 identified compounds to highlight the 21 differentially regulated known metabolites, indicate their degree of enrichment in either preclosure or postclosure samples, and show their relationships in biochemical modules (Fig. 4). Notably, several key metabolites related to the tricarboxylic acid cycle (Krebs cycle) were enriched in the preclosure samples. These metabolites included pyruvate, alpha-ketoglutaric acid, and glutamic acid. Also enriched in these samples was lactic acid, a classic signature molecule produced by Lactobacilli. Extensive but unsuccessful literature searches were performed to attempt to understand a possible role for the highly enriched compounds in the preclosure samples, such as levanbiose and glycerol-3-galactoside. These metabolites represent bacterial processes that can not yet be assigned. Additionally, a range of unknown compounds with high mass spectral similarities to amino acids and sugars were found to be enriched in the postclosure samples (query of the public datasets via http://eros.fiehnlab.ucdavis.edu:8080/binbase-compound). These findings indicate that bacterial communities play a very active and not fully understood role in human metabolism.
Fig. 4.
Metabolomic network diagram of the 139 chemically identified metabolites. 62 metabolites (18.0%) showed significant enrichment in either preclosure or postclosure samples. Metabolites were mapped by structural similarity (see SI Text for details). Red, compounds enriched in preclosure samples; blue, compounds enriched in postclosure samples. Node size is proportional to the relative enrichment.
Discussion
Both 16S rDNA sequencing and qPCR were used to characterize the bacterial populations associated with SBTs. Four bacterial orders—Lactobacillales, Enterobacteriales, Bacteroidales, and Clostridiales—accounted for 99.8% of the bacteria detected. Our qPCR survey of their relative populations revealed that two distinct microbial communities are found after small bowel transplantation, one before the monitoring ileostomy has been surgically closed and the other after its closure. The strictly anaerobic Clostridia and Bacteroides dominate the postclosure community. This community is very similar to that reported for the human ileum and likely represents the normal ileal microbial population (15, 17, 18, 23). Before surgical closure of the ileostomy, the facultatively anaerobic Lactobacilli and Enterobacteria dominate—essentially an inversion of the normal population.
Surprisingly, the presence or absence of an ileostomy appeared to be the dominant variable determining this community shift, because the preclosure pattern is also found when an ileostomy is present independent of a SBT (Fig. S4). No coincident changes in diet or drug treatments occurred around ileostomy closure that could explain the dramatic microbial shift. Although a broad-spectrum antibiotic targeting Gram-negative bacteria and strict anaerobes was used prophylactically immediately before ileostomy closure, it would have been expected to decrease rather than increase the abundance of strict anaerobes. This observed preclosure pattern is also consistent with earlier studies that used ileostomy samples and culture-based techniques to characterize the intestinal microbiota (24, 25).
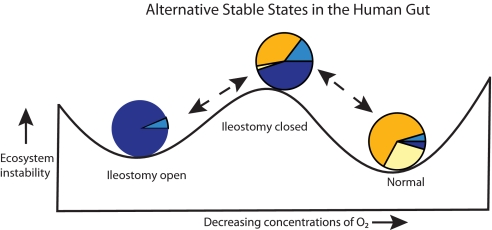
Viewing this shift in community structure from an ecological perspective, we hypothesize that the ileostomy is the primary ecological determinant of the unique preclosure SBT microbial community by virtue of its allowing oxygen into this normally anaerobic environment. Thus, the microbial community could revert to normal after ileostomy closure. The transition from the preclosure to the postclosure state can be modeled as an ecosystem system shift between two alternative stable states (Fig. 5) (26). A growing number of biological communities have been found to be able to exist in several possible stable configurations. A transition between states results when an environmental perturbation significantly alters a community variable or shifts a system parameter. Of particular interest to the field of conservation ecology is the question of how readily a degraded ecosystem can return to the native stable state.
Fig. 5.
Model of alternative stable states of the small bowel ecosystem. In this ball-in-cup diagram (26), the horizontal length represents all possible states of the ecosystem, the vertical length represents increasing ecosystem instability. The two valleys represent two alternative stable states of the small bowel microbiota: either dominated by strict anaerobes or by facultative anaerobes. The pie charts illustrating the microbial commmunity compositions are from Fig. 2A, D, and E.
For the specific case of the microbial community within the human ileum, we hypothesize that the introduction of oxygen via an ileostomy constitutes a parameter change that drives the transition from the normal community dominated by strict anaerobes to the preclosure state dominated by facultative anaerobes (Fig. 5). The reverse transition appears to be readily achievable. Five of the six patients observed after surgical ileostomy closure possessed a postclosure community that, like the normal ileal community, was dominated by strict anaerobes. The one patient that did not show this community transition differed from the other five by experiencing extended periods with high levels of inflammation both before and after ileostomy closure.
Although oxygen levels in the GI tract have been very difficult to quantify, it is a widely held that the distal GI tract is anaerobic. A few studies have documented extremely low oxygen tension levels throughout the distal GI tract (27, 28). Thus, the introduction of oxygen via the ileostomy represents a potentially significant alteration of the local environment, and one that can be subsequently reversed by surgical closure. The diffusion of oxygen inward from the ileostomy port could be sufficient to inhibit growth of the strict anaerobes, thus allowing the facultative anaerobes to predominate. An oxygen-dependent microbiota transition is well documented in studies from infants showing the succession from early colonization with facultative anaerobes to late colonization with strict anaerobes (12, 29).
Further support for this hypothesized role of oxygen is provided by metabolomic profiling of the preclosure and postclosure communities that demonstrated enrichment of metabolites of the aerobic tricarboxylic acid cycle (Krebs cycle) in preclosure samples. Lactate was also enriched preclosure, which could be the result of higher Lactobacilli levels and conceivably the absence or depletion of lactate-consuming Clostridia species (30). The majority of compounds that were significantly enriched in the postclosure samples could not be identified.
Historically, ileostomy samples have been used to study what was thought to be the normal composition of the human intestinal microbiota (24, 25). These studies, using culture-based methods, reported microbial profiles very similar to those in the preclosure and ileostomy control samples in this study, i.e., high populations of Lactobacilli and Enterobacteria, low populations of Clostridia and Bacteroides. Our results demonstrate that the microbiota assayed via ileostomy samples represent instead an alternative stable state, likely the result of the introduction of oxygen to a normally anaerobic environment. This suggests that data obtained with other model systems that may allow the introduction of oxygen, such as the fistulated cow (an anastomosis connecting the rumen to the abdominal wall) may reflect alternative states rather than the normal microbiota.
Our results indicate that an ileostomy, although serving a variety of clinical purposes, concomitantly alters the ileal microbial composition dramatically. The potential clinical complications resulting from this are presently unknown. In the preclosure SBT patients, the transplanted small bowel was functional despite the presence of the altered microbial community, suggesting that this inverted, Lactobacilli-dominated community is not detrimental, at least in the short term.
However, the microbial population shift after the ileostomy is closed is also drastic and potentially harmful. For example, patient 12 died 6 days after ileostomy closure from Enterobacterial sepsis. Several other instances of quick, lethal, postclosure sepsis have been observed at GUMC. These dramatic microbiota changes in immune-compromised patients suggest that the decision to create an ileostomy should be weighed against the potential complications from major microbial population changes or overgrowths. The development of noninvasive alternatives, such as fecal monitoring of calprotectin levels (31, 32) calls for reevaluation of the use of an ileostomy for monitoring these patients.
Conclusions
Our study of the microbiota associated with the transplanted human small bowel revealed that the small bowel can function with either of two alternative microbial populations. As part of the SBT surgical protocol, a temporary ileostomy is created to allow endoscopic monitoring of the transplant tissue. Until the ileostomy is surgically closed, the bacterial population present is dominated by facultative anaerobes, whereas after closure, and in the normal ileum, strict anaerobes prevail. These two population patterns can be viewed as alternative stable states of the human ileal ecosystem. Evidence presented indicates that the factor causing the shift to the preclosure state is the introduction of oxygen via the ileostomy into the normally anaerobic ileum. The small bowel transplant monitoring protocol generates samples of the ileal community that provide the unique opportunity to observe not only the preclosure state but also the transition back to the normal community after ileostomy closure. The alternative states described in this study suggest that the ecological possibilities within the human gut are broader than previously realized.
Materials and Methods
qPCR Reaction and Thermocycler Conditions.
Standard molecular methods such as DNA extraction and sequencing are described in detail in SI Text. Each qPCR contained 10 μL of 2x Takara Perfect Real Time master mix, 7.2 μL of water, 0.8 μL of a 10 μM F/R primer mix, and 2 μL of a 1:50 dilution of extracted DNA in DNase/RNase-free water. Cycling conditions: 95 °C for 20 sec; 40 repeats of the following steps: 95 °C for 4 sec, 30 sec annealing (Table S2). SYBR green fluorescence was detected with a BioRad Chromo4 Real Time PCR Detector on a Dyad Disciple Peltier Thermal Cycler. Melting curves were obtained from 55 °C to 90 °C, with fluorescence measurements taken at every 1 °C increase in temperature. Copies per μl were determined in the MJ Opticon Monitor Analysis Software, Version 3.1. All reactions were carried out in triplicate and a nontemplate control was performed in every analysis.
Metabolomics.
Metabolomic analysis was performed on two samples from each of the six patients for which both preclosure and postclosure samples were available. For each patient, the preclosure sample was the last sample before surgical ileostomy closure and the postclosure sample was the latest one available.
A 10-μL aliquot of effluent was suspended in 1 mL of a 5:2:2 solution of methanol:chloroform:water that had been de-gassed with N2 and chilled to −20 °C. The mixture was vortexed for 5 min and centrifuged at 16,000 × g for 5 min. The supernatant was discarded and the pellet was concentrated to dryness overnight at room temperature in a Centrivap cold trap vacuum concentrator. The samples were run according to methods described in ref. 33. The resulting metabolite profiles were analyzed using the semiautomated workflow in the UC Davis Genome Center metabolomics laboratory (34). Metabolite annotation was done using BinBase and the statistical analyses were performed using Statistica, version 6 (StatSoft). For metabolomic network graph methods see SI Text.
Supplementary Material
Acknowledgments.
We thank the 17 individuals that made this study possible; Charles Bevins and Scott Dawson for critical reading of the manuscript; Joseph Abdo, Cyril Chemali, and Gennadiy Novitsky at GUMC for technical assistance with sample and clinical data collection; Bertrand Perroud for assistance with the self-organizing-map software; Merry Youle for editing assistance; and Erin Esp, Senke Chen, Changjie Mathe, and Hans Mueller as part of the statistical consulting class (STA 401) at UC Davis for valuable feedback on the statistical methodology used here. This work was supported by Broad Foundation Grant IBD-0148 (to M.Z. and T.F.); start-up funds from University of California, Davis (to J.A.E.); and National Institutes of Health Grant R01 ES13932 (to O.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904847106/DCSupplemental.
References
- 1.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fishbein TM, Matsumoto CS. Intestinal replacement therapy: Timing and indications for referral of patients to an intestinal rehabilitation and transplant program. Gastroenterology. 2006;130:S147–151. doi: 10.1053/j.gastro.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Fishbein T, et al. NOD2-expressing bone marrow-derived cells appear to regulate epithelial innate immunity of the transplanted human small intestine. Gut. 2008;57:323–330. doi: 10.1136/gut.2007.133322. [DOI] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macfarlane S, Steed H, Macfarlane GT. Intestinal bacteria and inflammatory bowel disease. Crit Rev Clin Lab Sci. 2009;46:25–54. doi: 10.1080/10408360802485792. [DOI] [PubMed] [Google Scholar]
- 7.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 12.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fishbein TM, Florman S, Gondolesi G, et al. Intestinal transplantation before and after the introduction of sirolimus. Transplantation. 2002;73:1538–1542. doi: 10.1097/00007890-200205270-00004. [DOI] [PubMed] [Google Scholar]
- 15.Wang M, Ahrne S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54:219–231. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Lin LI. A concordance correlation-coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. [PubMed] [Google Scholar]
- 17.Ahmed S, et al. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. 2007;73:7435–7442. doi: 10.1128/AEM.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumgart M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 19.Madigan MT, Martinko JM, Brock TD. Brock biology of microorganisms. Upper Saddle River, NJ: Pearson Prentice Hall; 2006. pp. 379pp. 403pp. 702pp. 706–707. [Google Scholar]
- 20.Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: How do communities of bacterial symbionts become established in our intestine? Nat Immunol. 2004;5:569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- 21.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker AW, et al. The species composition of the human intestinal microbiota differs between particle-associated and liquid phase communities. Environ Microbiol. 2008;10:3275–3283. doi: 10.1111/j.1462-2920.2008.01717.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Heazlewood SP, Krause DO, Florin TH. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J Appl Microbiol. 2003;95:508–520. doi: 10.1046/j.1365-2672.2003.02005.x. [DOI] [PubMed] [Google Scholar]
- 24.Finegold SM, Sutter VL, Boyle JD, Shimada K. The normal flora of ileostomy and transverse colostomy effluents. J Infect Dis. 1970;122:376–381. doi: 10.1093/infdis/122.5.376. [DOI] [PubMed] [Google Scholar]
- 25.Gorbach SL, Nahas L, Weinstein L, Levitan R, Patterson JF. Studies of intestinal microflora. IV. The microflora of ileostomy effluent: A unique microbial ecology. Gastroenterology. 1967;53:874–880. [PubMed] [Google Scholar]
- 26.Beisner BE, Haydon DT, Cuddington K. Alternative stable states in ecology. Frontiers Ecol Environ. 2003;1:376–382. [Google Scholar]
- 27.He G, et al. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc Natl Acad Sci USA. 1999;96:4586–4591. doi: 10.1073/pnas.96.8.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lind Due V, Bonde J, Kann T, Perner A. Extremely low oxygen tension in the rectal lumen of human subjects. Acta Anaesthesiol Scand. 2003;47:372. doi: 10.1034/j.1399-6576.2003.00542.x. [DOI] [PubMed] [Google Scholar]
- 29.Adlerberth I. Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:13–29. doi: 10.1159/000146245. discussion 29–33. [DOI] [PubMed] [Google Scholar]
- 30.Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akpinar E, Vargas J, Kato T, et al. Fecal calprotectin level measurements in small bowel allograft monitoring: A pilot study. Transplantation. 2008;85:1281–1286. doi: 10.1097/TP.0b013e31816dcea2. [DOI] [PubMed] [Google Scholar]
- 32.Sudan D, et al. Calprotectin: A novel noninvasive marker for intestinal allograft monitoring. Ann Surg. 2007;246:311–315. doi: 10.1097/SLA.0b013e3180f61af4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiehn O, et al. Quality control for plant metabolomics: Reporting MSI-compliant studies. Plant J. 2008;53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 34.Fiehn O, Wohlgemuth G, Scholz M. Setup and annotation of metabolomic experiments by integrating biological and mass spectrometric metadata. Data Integrat Life Sci Proc. 2005;3615:224–239. [Google Scholar]
- 35.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.