Abstract
Temperature-sensitive (TS) mutations are a useful tool for elucidating gene function where a gene of interest is essential at multiple stages of development. However, the molecular mechanisms behind TS alleles vary. TS mutations of the myogenic regulator Myocyte enhancer factor-2 (MEF2) in Drosophila arise in the heteroallelic combination Mef230-5/Mef244-5. We show that the 30-5 mutation affects the N-terminal MADS domain, causing impaired DNA binding ability and failure of homozygous mutants to survive to adulthood. The 44-5 mutation deletes a downstream splice acceptor site and results in a truncated protein that is unable to activate MEF2 targets. 44-5 homozygotes consequently show severely impaired myogenesis and die as embryos. We propose that in heteroallelic mutants at the permissive temperature, 30-5/44-5 heterodimers form and have a sufficiently stable interaction with DNA to activate myogenic gene expression; at the restrictive temperature, 44-5 homodimers displace 30-5/44-5 heterodimers from target genes, thus acting in a dominant-negative manner. To test this, we show that 30-5/44-5 heterodimers can form, and we study additional Mef2 alleles for complementation with the 30-5 allele. An allele affecting the DNA binding domain fails to complement 30-5, whereas two alleles affecting downstream residues show temperature-dependent complementation. Thus, by combining one MEF2 isoform having weakened DNA binding ability with a second truncated MEF2 mutant that has lost its activation ability, a TS form of intragenic complementation can be generated. These findings will provide new insight and guidance into the functions of dimeric proteins and how they might be engineered to generate TS combinations.
TEMPERATURE-SENSITIVE (TS) mutants have a noted history in defining gene function. Using TS alleles, geneticists are able to attenuate gene function at a specific time and then observe the phenotypic effects of this genetic manipulation. TS mutants have been used for such diverse studies as identifying yeast genes critical in the various stages of the cell cycle (Hartwell et al. 1970) and in metazoans to dissect the requirements for genes that function at multiple developmental stages (see for example Cox and Baylies 2005). This powerful tool has proven especially useful in identifying and categorizing genes that function at multiple stages to support organism viability.
Defining the molecular mechanisms behind temperature sensitivity enables us to more thoroughly understand the synthesis, folding, collaboration, and function of a protein of interest. Sadler and Novick (1964) categorized bacteriophage TS mutants by the mechanism of their dysfunction (Sadler AND Novick 1964). The thermolabile (TL) class of mutants demonstrated a mutant phenotype when grown at a restrictive temperature or when shifted to a restrictive temperature during a later stage of development. The mutant phenotype in this instance was usually due to the destabilization of the encoded protein and the subsequent loss of protein function as a result of increased temperature. This could be due to decreased melting temperature from the loss of an hydrophobic amino acid or due to decreased ability to interact with DNA or other proteins as dimers or multimers.
Another category of TS mutants, temperature-sensitive folding (TSF) only showed a phenotype if incubated at the restrictive temperature during synthesis of the encoded protein. If the mutants were shifted to the restrictive temperature later, the protein did not lose conformation and no phenotype was apparent. These mutations were recessive and did not demonstrate intragenic complementation as did the thermolabile types. It was postulated that many mutants in this class affect the formation, folding, or initial integration of the encoded protein into larger complexes; once the complex is formed, the mutant protein is stabilized by its interactions with other subunits (Edgar and Lielausis 1963; Gordon and King 1993). Clearly, the mechanism of TS for a particular protein is strongly dependent upon its function as part of a multimeric or macromolecular complex and upon its half-life in the cell.
TS mutants have generally been isolated by screening through large numbers of organisms that have undergone random mutagenesis. More recently, researchers have looked for ways to engineer TS proteins. One approach was to introduce charged amino acids into the internal region of the CcdB toxin encoded by the F′ plasmid (Chakshusmathi et al. 2004). CcdB is a good candidate for TS screening because when transformed into Escherichia coli, it is lethal when functional. Several of the introduced mutations successfully destabilized the protein at the restrictive temperature; however, this approach was not always fully effective. Clearly, the engineering of new TS mutants will benefit from further characterization of the molecular basis of existing TS alleles.
We have recently described the isolation of a temperature-sensitive combination of alleles for the muscle transcription factor Myocyte enhancer factor 2 (MEF2) (Baker et al. 2005). Vertebrates possess four MEF2 genes, all of which contain a highly conserved, N-terminal MADS domain [MCM1, Agamous, Deficiens, serum response factor (SRF)] spanning amino acids 1–57, and a 29-amino-acid MEF2 domain immediately adjacent (reviewed in Black and Olson 1998; Black and Cripps 2009). Both mutagenesis studies and biophysical determination of the solution structure of MEF2A have clearly shown that the MADS domain is required for DNA binding; also, a portion of the MADS domain (residues 35–50) along with the adjacent MEF2 domain, is required for proper dimerization (Molkentin et al. 1996; Huang et al. 2000; Santelli and Richmond 2000).
Upon dimerization and DNA binding, MEF2 is a potent activator of target structural genes, both directly and in complex with other factors (Molkentin et al. 1995; Hamamori et al. 1997; Kelly Tanaka et al. 2008). Much of this transcriptional activation ability of MEF2 proteins arises from the function of residues located C terminal to the MEF2 domain, which are both required and sufficient for MEF2 transcriptional activity (Martin et al. 1994; Molkentin et al. 1995). Clearly, MEF2 is an highly modular protein with distinct functional domains.
Drosophila contain a single Mef2 gene that is expressed in muscles at all stages of development (Lilly et al. 1994; Nguyen et al. 1994; Taylor et al. 1995; Baker et al. 2005). Mef2 null mutants do not survive beyond embryogenesis and display a profound lack of differentiated muscle (Bour et al. 1995; Lilly et al. 1995), making this system useful for the study of MEF2 protein function. In this article, we define a molecular mechanism for the temperature sensitivity of a subset of Drosophila Mef2 mutant alleles. We show that a mutation affecting the MADS domain (allele 30-5) can complement a mutation affecting C-terminal residues (allele 44-5), but that this complementation is thermolabile. We provide evidence that the thermolability arises from competition between the attenuated DNA binding ability of the 30-5 isoform, and the strong DNA binding ability of the 44-5 isoform, which acts as a dominant-negative factor at the restrictive temperature. Our analyses of several additional Mef2 mutants confirm this model. These studies provide important new insight into critical regions of the MEF2 factor, and more broadly contribute to our understanding of the molecular basis of TS mutants.
MATERIALS AND METHODS
Drosophila stocks and crosses:
Drosophila stocks were grown on Carpenter's medium (Carpenter 1950) at the indicated temperatures. Mef2 mutants were obtained from Elliot Goldstein (Arizona State University; Goldstein et al. 2001) and balanced over a CyO, Cy Kr-GFP balancer chromosome (Casso et al. 1999). Viability studies were achieved by crossing CyO, Cy Kr-GFP/Mef2x with CyO, Cy Kr-GFP/Mef2y. We counted the number of heteroallelic escaper adults and the total number of progeny. Viability counts reflect the percentage of heteroallelic escapers detected, as a proportion of the number of such escapers expected if their viability were the same as their heterozygous siblings.
Immunohistochemistry:
Embryos were collected and stained as described by Patel (1994). Primary antibodies were rabbit anti-myosin heavy-chain (1:250; Kiehart and Feghali 1986), mouse anti-GFP (1:250; Invitrogen), and rat anti-tropomyosin (1:250; Peckham et al. 1992; Abcam Immunochemicals). Secondary detection was using the Vectastain Elite kit (Vector Laboratories) and diaminobenzidine (DAB) stain. Samples were cleared in glycerol and mounted for photography with an Olympus BX51 photomicroscope using DIC optics. Images were collated using Adobe Photoshop.
DNA and RNA methods:
To identify the lesions associated with each Mef2 allele, RNA was purified from larvae of the genotype CyO, Cy Kr-GFP/Mef2x. The RNA was purified using the QIAGEN RNeasy Mini kit and RNase-free DNase set and then subjected to reverse transcription followed by PCR, using Invitrogen Superscript III One-Step RT–PCR. Primers for the PCR were (5′-ATGGGCCGCAAAAAAATTCAAATATC-3′) and (5′-CTATGTGCCCCATCCGCC-3′), which were designed to amplify the entire Mef2 coding region. PCR products were cloned into the pGEM-T Easy vector (Promega), and 12 positive clones for each genotype were sequenced in their entirety, to ensure that a cDNA arising from the mutant Mef2 allele was sequenced several times. In all cases, we found several wild-type cDNA clones and several clones that consistently showed a departure from wild type.
To confirm the sequence alterations observed in the cDNA as arising from the Mef2 mutant allele, we isolated DNA from heterozygotes and amplified the appropriate genomic region by PCR. Primers used to characterize 44-5 splice variants were (5′-GCTGGAGATGTCGAACG-3′ and 5′-CTGCATATCCCACATCATCC-3′). For all mutants characterized, the lesion at the DNA level matched or accounted for the changes observed in the cDNAs.
Electrophoretic mobility shift assays:
Wild-type MEF2 protein was generated from pSK-DMef2 (Lilly et al. 1995) using T3 RNA polymerase and the TnT Coupled Transcription/Translation system (Promega). For mutant MEF2 proteins, the appropriate sequence change was introduced into pSK-DMef2 using the Invitrogen Gene-Tailor site-directed mutagenesis kit, and confirmed by sequencing. Protein was then generated as described for wild type. To use equivalent amounts of protein for wild-type and mutant shifts, parallel reactions were set up so that wild-type and mutant proteins were generated in the presence of either radioactive 35S-methionine, or nonradioactive methionine. Next, radioactive reactions were analyzed by SDS–PAGE and the radioactive bands corresponding to full-length MEF2 protein were quantified using the Cyclone Packard Phosphorimager. On the basis of the efficiency of MEF2 protein generation as determined in the radioactive reactions, the volume of nonradioactive MEF2 lysate was correspondingly adjusted in the gel-shift reactions to ensure that equal amounts of protein were used. Differences in the volumes used of programmed lysate were corrected by addition of unprogrammed lysate.
Electrophoretic mobility shift assays (EMSAs) were set up according to standard methods (Sambrook et al. 1989; Kelly Tanaka et al. 2008). For analysis of the individual DNA binding ability of MEF2 isoforms, reactions were incubated in water baths at either 18° or 29°, and the gels were run at a constant temperature of 18° or 29° by circulating water of the appropriate temperature through the electrophoresis rig. The dimerization assays were carried out at room temperature. After running, gels were dried and radioactive bands were quantified using the Packard Cyclone Phosphorimager. Data shown represent an average of at least two reactions for each mutant compared to wild type at each temperature.
Cell culture and transfections:
For a wild-type expression plasmid we used pPac-Pl-Mef2 (Kelly Tanaka et al. 2008). Mutant expression plasmids were generated by subcloning the mutant cDNAs into pPac-Pl. The reporter plasmid used was the −593/+2 Actin 57B-lacZ construct described by Kelly et al. (2002). Transfections and analyses were as described in Kelly Tanaka et al. (2008). All experiments were carried out in triplicate, and the average fold activation for each experiment is reported.
RESULTS
Temperature-dependent phenotypes of 30-5 and 44-5 alleles:
To define the basis of the 30-5/44-5 temperature sensitivity, we first studied the extent of embryonic muscle differentiation in homozygotes for each mutant Mef2 allele. These studies were performed at 18° and 29° and were compared to wild type and to the heteroallelic combination that we had previously described (Baker et al. 2005).
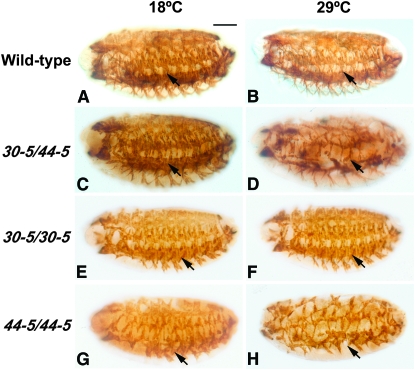
At stage 16, wild-type embryos stained for the accumulation of tropomyosin showed extensive skeletal muscle differentiation at both temperatures (Figure 1, A and B); and the heteroallelic combination showed relatively normal patterning of skeletal muscles at the permissive temperature, but a strong hypomorphic phenotype at the restrictive temperature (Figure 1, C and D).
Figure 1.—
Skeletal muscle development in wild-type and mutant Mef2 alleles. Stage 16 embryos were stained with anti-tropomyosin (A, B, E–H) or anti-myosin heavy-chain (C and D) to visualize skeletal muscle patterning and differentiation. Embryos were raised at the permissive temperature (18°) and the restrictive temperature (29°) to compare phenotypes at the two temperatures. All embryos are sagittal views with anterior to the left. Arrows indicate skeletal muscles. (A and B) Wild-type embryos develop normally at both the permissive and restrictive temperatures. In normal development, the three lateral transverse muscles 1–3 (LT 1–3) are present in each segment (arrows). (C) The heteroallelic combination 30-5/44-5 is phenotypically similar to wild type at 18°, yet is severely hypomorphic at 29° (D). Panels C and D are from Baker et al. 2005. (E and F) The 30-5 homozygous mutants closely resemble wild type at both temperatures, but are not viable to adulthood. (G and H) The 44-5 homozygous mutants are severely hypomorphic at both temperatures. Note that the 44-5 homozygotes most closely resemble the 30-5/44-5 combination raised at 29°. Bar, 100 μm.
For 30-5 homozygotes, muscle differentiation appeared relatively normal at both temperatures (Figure 1, E and F), suggesting that this allele on its own had significant wild-type function and did not show a strong temperature dependence in its activity. Given the relatively normal pattern of muscles in 30-5 animals, we determined whether they showed survival to adulthood at either 18° or 29°. 30-5 homozygotes never showed viability to the adult stage, although this could arise from second-site mutations that might have accumulated on the 30-5 chromosome. We therefore tested the viability of mutants heteroallelic for 30-5 and the Mef2 null allele P544 (Lilly et al. 1995). Since P544 was isolated in a different genetic screen to the Goldstein et al. (2001) alleles, the P544 chromosome is unlikely to share any second-site lethal mutations with 30-5. We found in this instance that 13% of 30-5/P544 mutants survived to adulthood at the permissive temperature (12 observed out of 89 expected) but none survived at the restrictive temperature (0 observed, 238 expected). We conclude that the 30-5 allele is a mild hypomorphic mutation, which shows a slight attenuation in its function with increasing temperature. This mild temperature-dependent effect does not account for the profound TS observed in 30-5/44-5 mutants.
For the 44-5 allele, homozygotes showed severe defects in muscle development at both temperatures (Figure 1, G and H). Again, there was not a strong effect of temperature upon the severity of the embryonic phenotype.
We conclude that these two alleles, while individually not strongly temperature dependent, somehow interacted to generate a temperature-dependent effect in heteroallelic combinations. We note that at the permissive temperature the 30-5/44-5 combination appears phenotypically similar to the 30-5 homozygotes, whereas at the restrictive temperature the heteroallelic combination appears similar to the 44-5 mutants. Thus we reasoned that the 44-5 allele has an antimorphic effect, but only at the restrictive temperature and only in combination with 30-5 mutants.
Sequence analysis of 30-5:
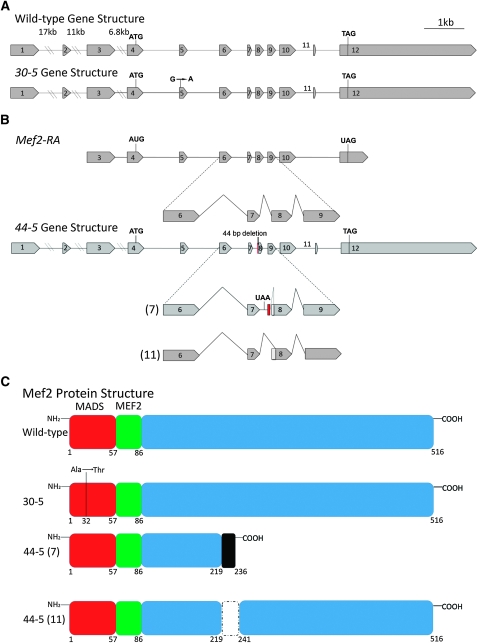
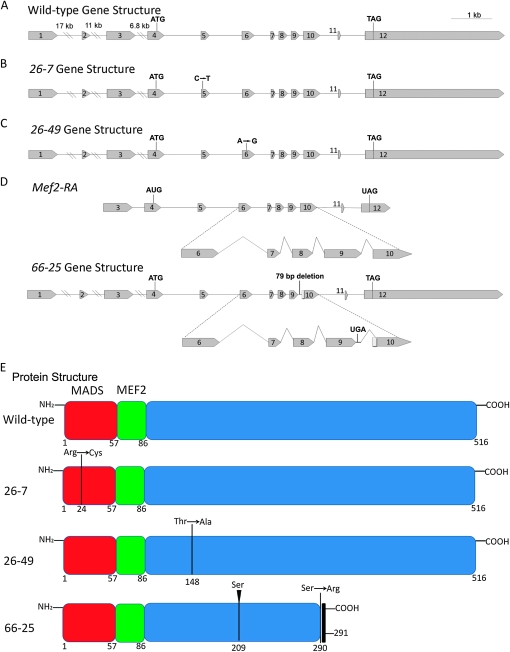
Our next goal was to understand the molecular mechanism underlying this temperature-sensitive effect. We isolated RNA from 30-5/+ heterozygotes and carried out an RT–PCR, amplifying Mef2 cDNA using primers flanking the coding region. We cloned and sequenced the resulting products and discovered a point mutation that was consistently observed in approximately half of the sequenced cDNAs. We found that the 30-5 cDNA contained a G-to-A point mutation in the MADS box (Figure 2A) resulting in an alanine-to-threonine conversion at amino acid 32 (Figure 2C). The same mutation was observed in clones of genomic DNA isolated from 30-5/+ heterozygotes. Given that 30-5 homozygotes develop a fairly normal pattern of skeletal muscles, we hypothesized that 30-5 mutants were capable of forming MEF2 dimers and activating downstream target genes, albeit with reduced efficiency compared to wild type.
Figure 2.—
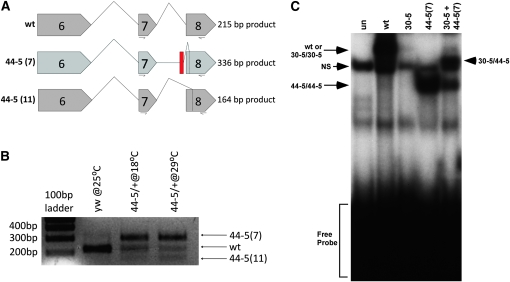
Gene structure, splicing patterns, and protein structure in Mef2 wild-type and mutant alleles. (A) Gene structure of wild type and 30-5. 30-5 contains a G-to-A point mutation in exon 5. (B, top) The wild-type Mef2-RA isoform represents one of the predominant Mef2 transcripts, the designation for which is from FlyBase (http://flybase.org). A detailed view of its splicing pattern between exons 6 and 9 is also shown, compared to the 44-5 gene structure and its two alternative splicing patterns. The 44-5 allele contains an additional 9 bp of nucleotides following intron 7 (shown in red) and 44 bp of nucleotides deleted from the 5′ end of exon 8, including the normal exon 8 splice acceptor. The 44-5 (7) cDNA retains intron 7, which includes a premature stop codon (UAA), and cryptically splices into the 45th bp of exon 8. The 44-5 (11) isoform utilizes the normal 5′ splice donor of exon 7, but splices cryptically into the 52nd bp of exon 8. (C) Wild-type MEF2 contains a 57-amino-acid MADS DNA-binding domain, and a 29-amino-acid MEF2 DNA-binding and dimerization domain. 30-5 contains an alanine-to-threonine substitution at amino acid 32. The black shaded box of 44-5 (7) is translated intron sequence and contains a premature stop codon at amino acid 236. 44-5 (11) contains an internal deletion of amino acids 219–241.
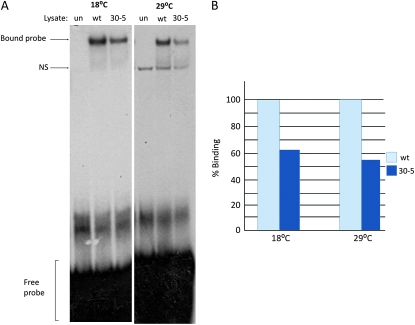
The 30-5 protein demonstrates decreased DNA binding ability: To test this hypothesis, we performed an EMSA using a radioactively labeled Actin 57B MEF2 site as a probe. The reaction was carried out at both 18° and 29°, and equal amounts of wild-type and 30-5 protein were used for binding reactions (see materials and methods for details). The results demonstrated that the 30-5 isoform consistently showed a reduced ability to bind to the probe relative to wild-type MEF2. However, this reduction in DNA binding was not affected by temperature (Figure 3, A and B). In cotransfection assays, 30-5 was able to activate Act57B in cell culture at both temperatures equally well (data not shown). These experiments, along with the phenotypic characterization of terminal differentiation in 30-5 mutants, support the idea that the 30-5 isoform in isolation is capable of activating the muscle differentiation pathway. However, due to its decreased DNA binding ability, the pathway is not activated robustly enough in vivo for the animal to survive to adulthood.
Figure 3.—
DNA binding ability of the 30-5 isoform at 18° and 29°. (A) Unprogrammed lysate (un), wild-type MEF2 (wt), and 30-5 MEF2 proteins were each mixed with a radioactively labeled MEF2 binding site from the enhancer region of Act57B. Reactions and electrophoresis were carried out at both 18° and 29°. These formed complexes that moved considerably slower than free probe through a polyacrylamide gel, indicating that both protein isoforms could bind to the MEF2 recognition site. However, the band intensity was significantly less intense when the 30-5 reaction was compared to wild-type, regardless of temperature. An additional nonspecific band (NS) is seen at 29° in the unprogrammed lysate as well as the experimental lanes. (B) Percentage of binding ability of 30-5 compared to wild type (100%) quantified by band intensity (see materials and methods). Note that the 30-5 isoform consistently bound to DNA less efficiently than wild type.
Sequence analysis and functional characterization of 44-5 isoforms:
We next isolated RNA from 44-5/+ heterozygotes and cloned and sequenced Mef2 cDNAs as performed previously for 30-5. We found two distinct mutant transcripts, 44-5 (7) and 44-5 (11) (Figure 2, B and C), both of which lacked the normal downstream splice acceptor site of exon 8. The 44-5 (7) transcript was missing the first 44 bases of exon 8 and contained extra nucleotides that did not match the Mef2 coding sequence, but did match sequence from intron 7. The 44-5 (11) transcript lacked the first 51 bases of exon 8. We then analyzed Mef2 genomic clones and discovered that in the 44-5 allele there was a 44-bp deletion, which removed the exon 8 splice acceptor site and a portion of exon 8 coding sequence; all but the last 3 bp of intron 7 were retained.
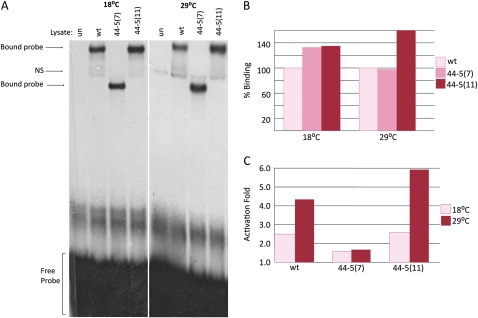
The effects of these altered transcripts upon the encoded MEF2 protein are potentially profound (Figure 2C). The 44-5 (11) variant predicts a 22-amino-acid internal deletion; and the 44-5 (7) splice variant encodes a polypeptide with a large C-terminal deletion. Both of these isoforms contain mutations in a region that has not been fully characterized, but which may encode the activation domain of MEF2. We predicted that 44-5 protein isoforms would be able to bind to DNA, but we were uncertain whether the binding ability of the truncated 44-5 (7) isoform would be reduced at the restrictive temperature. We found that each of the 44-5 (7) and 44-5 (11) isoforms bound to the Act57B MEF2 site at 18° and 29° extremely well (Figure 4, A and B). In fact, the 44-5 (11) isoform consistently bound to DNA even more effectively that wild-type MEF2.
Figure 4.—
DNA binding and activation ability of the 44-5 isoforms. (A) Unprogrammed lysate (un), wild-type MEF2 (wt), and the two 44-5 MEF2 isoforms were each mixed with a radioactively labeled MEF2 binding site from the enhancer region of Act57B and separated on a polyacrylamide gel. Reactions and electrophoresis were carried out at both 18° and 29°. All isoforms of MEF2 were able to bind strongly to the DNA probe; the 44-5 (7) complex is considerably smaller than wild type and 44-5 (11) since the protein is truncated. (B) Percentage of binding ability of 44-5 (7) and (11) compared to wild type (100%) quantified by band intensity. Both isoforms were able to bind to DNA as well as, if not better than, wild type at both temperatures. (C) Activation of a MEF2-dependent Act57B enhancer fused to a lac-Z reporter gene by either wild-type MEF2, 44-5 (7), or 44-5 (11) isoforms in cell culture. The ability of wild-type MEF2 to activate a downstream target gene increased at 29°. 44-5 (7) transactivation ability was not detectable above negative controls at either temperature. 44-5 (11) was able to activate reporter gene expression to a greater degree than wild type.
Since DNA binding ability of the 44-5 isoforms was not reduced, but terminal differentiation of 44-5 homozygotes was severely affected at both temperatures, we concluded that the 44-5 isoforms must not be able to activate downstream target genes. We tested this notion using cotransfection assays (Figure 4C), and found that the ability of the 44-5 (7) isoform to activate downstream targets was severely reduced at both temperatures. However, the 44-5 (11) isoform was capable of activating transcription as effectively as wild-type MEF2 at both temperatures.
That one of the 44-5 mutant isoforms was capable of activating a canonical MEF2 target gene was inconsistent with our demonstration of a severe muscle phenotype in 44-5 homozygotes. We therefore postulated that there was differential expression of the two mutant isoforms. To test this, we performed another RT–PCR on 44-5/+ heterozygotes, using a primer pair that would generate characteristic products for each of the mutant and wild-type isoforms (described in Figure 5A). This RT–PCR was carried out using mRNA isolated from animals raised at either 18° or 29°. We found that, at both temperatures, the 44-5 (7) isoform was the predominant mutant transcript (Figure 5B). While we have not confirmed at the protein level the prevalence of the truncated isoform in vivo, earlier studies have shown that truncated MEF2 isoforms are readily observed (Martin et al. 1994), although we note that the similar allele Mef2113 shows reduced stability of the protein (Ranganayakulu et al. 1995). In addition, the presence of longer isoforms might render a truncated MEF2 more stable. These results support the severity of the phenotypic data in 44-5 homozygotes at both temperatures, since the predominant isoform detected by RT–PCR can bind to target genes very well, but is incapable of transcriptional activation.
Figure 5.—
Quantification of the relative abundance of MEF2 44-5 transcripts at 18° and 29° and interaction of 44-5 protein with 30-5 protein. (A) Diagram of splicing patterns of wild type and the two 44-5 isoforms, showing the primer pairs used for RT–PCR. The 44-5 (7) retains intron sequence and its predicted product is therefore larger than wild type. 44-5 (11) is missing 51 bp of exon 8 and its predicted product is therefore smaller than wild type. (B) RT–PCR of wild type (y w) and 44-5/+ heterozygotes. 44-5 (7) is the dominant transcript in heterozygotes at both temperatures. (C) Electrophoretic mobility shift assay demonstrating dimerization of 30-5 protein with 44-5 protein. NS represents a nonspecific band arising from the lysate. Note that the shift corresponding to 30-5 homodimers is relatively weak in this instance. Arrowhead represents the 30-5/44-5 heterodimer interacting with DNA.
A model to explain temperature dependency of 30-5/44-5 mutants:
We have observed that the TS effect of 30-5/44-5 shows a phenotype that is closer to the homozygous 30-5 mutant at the permissive temperature, and closer to the 44-5 mutant at the restrictive temperature. Having established that the 30-5 isoform has a defect in DNA binding, and that the predominant 44-5 isoform can bind DNA but fails to activate transcription, we proposed the following model to explain the temperature sensitivity of this combination:
In 30-5/44-5 trans-heterozygotes, a 30-5/44-5 heterodimer can bind to DNA and activate downstream genes, due to the interaction of the 44-5 DNA binding ability and the 30-5 activation ability. This interaction occurs successfully at the permissive temperature and rescues the lethality of 30-5 homozygotes. At the restrictive temperature, molecular interactions are more restrictive to low-affinity binding, thus the 44-5 homodimers outcompete the 30-5/44-5 heterodimers for DNA binding. The superior binding of 44-5 to DNA at this temperature results in a more severely mutant phenotype, since the truncated isoform of 44-5 is incapable of activating target genes.
Central to this hypothesis is the notion that 44-5 and 30-5 isoforms heterodimerize. To determine if a physical interaction between 30-5 and 44-5 is possible, we carried out electrophoretic mobility shift assays, using either 30-5 protein, 44-5 (7) truncated protein, or a mixture of 30-5 and 44-5 (7) proteins that had been synthesized in the same in vitro reaction. When mixed with the MEF2 binding site from Act57B, the individual 30-5 and 44-5 (7) isoforms generated shifted bands that were consistent with their ability to interact with DNA, and consistent with their respective sizes—the truncated 44-5 (7) isoform bound to DNA had a greater mobility than the 30-5-DNA complex (Figure 5C). When the mixture of 30-5 and 44-5 (7) was incubated with the DNA probe, a band of intermediate mobility was also observed (Figure 5C). This band must represent an heterodimer of 30-5/44-5 (7) interacting with DNA. This result confirmed that the two isoforms can interact physically.
Analyses of additional Mef2 alleles support the model:
To further test our model for temperature sensitivity, we determined whether other Mef2 alleles with mutations affecting defined regions would show a temperature-dependent rescue of the 30-5 mutant phenotype. We predicted that an allele with a mutation affecting the DNA binding domain would not be strongly TS when combined with 30-5, whereas alleles encoding proteins with normal DNA binding domains and mutated C-terminal regions would demonstrate a TS effect similar to that observed with the 30-5/44-5 crosses.
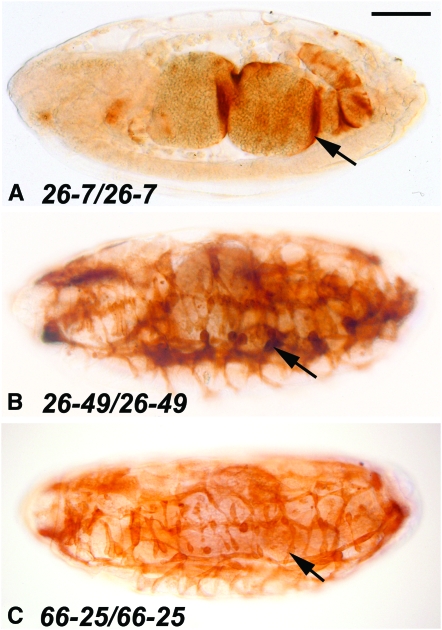
We chose three Mef2 alleles isolated by Goldstein et al. (2001) that demonstrated severely mutant phenotypes as homozygotes: 26-7 homozygotes were functionally null for muscle development (Figure 6A), whereas 26-49 and 66-5 alleles showed severely hypomorphic phenotypes (Figure 6, B and C). We performed sequence analyses of the three alleles, to determine whether the mutation affected either the DNA binding and dimerization region of the protein or a more C-terminal region (see materials and methods for details). For two alleles, the mutation was a point mutation in the coding region: when compared to wild type (Figure 7A) the 26-7 mutation affected the MADS box, encoding a missense mutation at amino acid 24 from Arg to Cys, CGC to UGC (Figure 7, B and E); this is the same point change as was observed for the 26-6 allele, isolated by Goldstein et al. (2001) and analyzed by Nguyen et al. (2002). The 26-49 mutation was outside of the MADS and MEF2 domains, encoding a missense mutation at amino acid 148 from Thr to Ala, ACG to GCG (Figure 7, C and E). For the 66-25 allele, our analysis of the cDNA revealed two alterations: there was an insertion of a serine codon at amino acid 209 of the coding sequence, and there was an insertion of 17 bp, introducing a UGA stop codon, after the ninth exon (Figure 7D). We analyzed genomic DNA from 66-25 and determined that the underlying mutation was a 79-bp deletion of the ninth intron, which removed the splice acceptor AG dinucleotide of exon 10. The result of this mutation is a truncation of the encoded protein (Figure 7E).
Figure 6.—
Skeletal muscle development in three new Mef2 mutant alleles. Stage 16 embryos raised at 29° were stained with anti-tropomyosin to visualize skeletal muscle patterning and differentiation. All embryos are sagittal views with anterior to the left. Embryos of the same genotypes raised at 18° showed similar phenotype to those shown here. (A) The 26-7 homozygous mutant showed a Mef2 null phenotype, since there was no skeletal muscle development detected, and only a small amount of stain was observed in the visceral muscle surrounding the gut (arrow). (B and C) The 26-49 and 66-25 homozygous mutants showed some level of skeletal muscle development (arrows); however, the phenotypes overall were indicative of these each being strongly hypomorphic alleles. Bar, 100 μm.
Figure 7.—
Analysis of gene and predicted protein structure for three new Mef2 mutant alleles. (A) Wild-type gene structure contains 12 exons. The ATG start codon is in the 4th exon and the TAG stop codon is in the 12th exon. (B) The 26-7 allele has a point mutation from C to T in exon 5, encoding an amino acid change from arginine to cysteine at amino acid 24. (C) The 26-49 allele has a point mutation from A to G in exon 6, encoding an amino acid change from threonine to alanine at amino acid 148. (D) The normal splicing pattern is shown for the Mef2-RA isoform. The 66-25 mutation is a 79-bp deletion after exon 9, which deletes the splice acceptor site and causes the gene to read into the intron and splice cryptically into the 10th exon. The result is to encode a stop codon 1 amino acid after the 9th exon. This allele also has an insertion of a serine codon after codon 209 (not shown on gene structure). (E) Predicted protein structure for wild-type and three new Mef2 mutant alleles as indicated: the 26-7 mutation encodes an amino acid change from arginine to cysteine at amino acid 24, which occurs in the DNA-binding MADS domain; the 26-49 mutation encodes an amino acid change from threonine to alanine at amino acid 148, which occurs in the downstream C-terminal region; the 66-25 mutation encodes an insertion of serine at amino acid 209 and a premature stop arising from readthrough of intron 9.
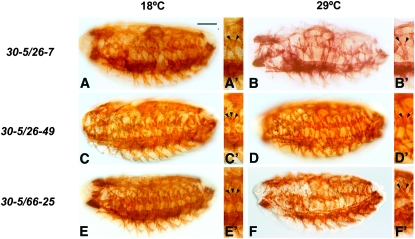
To determine whether a temperature-sensitive effect was observed when we crossed the three new alleles with 30-5, we used immunohistochemistry to assess tropomyosin accumulation in heteroallelic embryos raised at permissive or restrictive temperatures. For 30-5/26-7 there was a slight amelioration of the 26-7 homozygous phenotype, but the effect was not TS: at both temperatures the mutants still showed severely affected muscle development (Figure 8, A and B). By contrast there was a modest temperature-dependent rescue of both of the C-terminal region mutants, 26-49 and 66-25, when combined with 30-5 (Figure 8, C–F). In both cases, significant muscle fibers were observed in heteroallelic mutants, yet the phenotype was more severe at 29° (Figure 8, C–F).
Figure 8.—
Skeletal muscle phenotypes arising from heteroallelic crosses of new Mef2 alleles with the 30-5 allele. Stage 16 embryos were stained with anti-tropomyosin to visualize skeletal muscle patterning and differentiation. Embryos were raised at the permissive temperature (18°) and the restrictive temperature (29°) to compare phenotypes at the two temperatures. All embryos are sagittal views with anterior to the left. Arrows indicate skeletal muscles. Small figure panels show magnified images of single segments, highlighting the LT 1–3 muscles (arrowheads). (A and B) The 26-7/30-5 embryos were severely mutant at both temperatures. (A′ and B′) At both temperatures many segments contained only two LT 1–3 muscles (arrowheads), as opposed to three LT normally observed in wild type. (C and D) The 26-49/30-5 phenotype was temperature sensitive: at 18° the phenotype was closer to wild type (C), with predominantly three LT muscles per segment (C′). At 29° the phenotype was more severe (D), with some segments having only two LT muscles (D′). (E and F) The 66-25/30-5 crosses were also temperature sensitive, with similar phenotypes to those of the 26-49/30-5 crosses. At 18° the phenotype of the mutant (E) was closer to wild type, with three LT 1–3 muscles per segment (E′). At 29° the phenotype was more severe (F), with some segments having only two LT muscles (F′). Bar, 100 μm for main panels; 150 μm for insets.
To quantify this temperature sensitivity we studied the survival to adulthood of the heteroallelic mutant combinations. The original TS combination of 30-5/44-5 showed a dramatic difference in viability at the two temperatures, with 58% of embryos surviving to adulthood at the permissive temperature and 0% surviving at the restrictive temperature (Baker et al. 2005). For the 30-5/26-7 combination of two MADS domain mutant alleles, survival was 9% (31 observed out of 132 expected) at the permissive temperature and 0% (0 observed, 170 expected) at the restrictive temperature. When 30-5 was combined with the C-terminal region mutants the viability was more clearly TS: for the 30-5/26-49 cross, survival was 67% (134 observed, 200 expected) at the permissive temperature and 14% (33 observed, 239 expected) at the restrictive temperature; for the 30-5/66-25 cross, survival was 49% (93 observed, 189 expected) at the permissive temperature and 31% (61 observed, 196 expected) at the restrictive temperature.
These findings further support the model for temperature sensitivity: heteroallelic combination of one Mef2 mutant showing weakened DNA binding ability with a second Mef2 mutant showing weakened activation ability can generate a TS effect. Nevertheless, we note that the severity of the TS effect can still vary between different alleles in the same class.
DISCUSSION
MEF2 is a widely expressed, multifunctional dimeric protein that works in collaboration with other factors to influence a variety of developmental processes at multiple stages of development (reviewed in Potthoff and Olson 2007; Black and Cripps 2009). We have previously exploited the temperature sensitivity of the 30-5/44-5 combination to elucidate a role for MEF2 in adult myogenesis, where we showed that a strong reduction in MEF2 function could nevertheless support several aspects of adult muscle development (Baker et al. 2005). In this article we have characterized the molecular mechanism behind this TS combination, in an effort to acquire more information about the dynamics of temperature sensitivity and the mechanisms of MEF2 function.
Current models of temperature sensitivity take into account mutations that affect the proper formation or folding of a protein when synthesized at a restrictive temperature and mutations that destabilize a protein when shifted to a restrictive temperature (Sadler and Novick 1964). Mutations that are capable of destabilizing a protein can be found in DNA binding domains, protein–protein interaction domains or internal hydrophobic regions (Edgar and Lielausis 1963; Smith et al. 1980; Gordon and King 1993; Sundberg and Davis 1997). In addition, the temperature sensitivity of homodimeric proteins has received attention. In several instances, intragenic complementation has been observed for dimeric or multimeric proteins, and the classical literature has noted that in several cases such complementation is TS (Fincham 1966). This relationship was underlined by Sundberg and Davis (1997), who showed that mutations affecting different functional regions of the Saccharomyces cerevisiae spindle protein Spc110p could show effective TS intragenic complementation.
The 30-5/44-5 TS findings that we describe fit the latter model effectively. Combination of the 30-5 DNA-binding mutant with 44-5 or with either of two additional C-terminal mutants demonstrated a TS effect, whereas combination of 30-5 with an allele affecting the DNA binding domain did not show effective rescue and temperature sensitivity. We note that the severity of the various TS effects that we have observed is still the most striking with 30-5/44-5. This might be attributed to a less severe disruption of the activation domain in the C-terminal mutants 26-49 or 66-25. Indeed, the 26-49 isoform contains only a single-amino-acid substitution in the C terminus, but still produces a full-length protein. The 66-25 mutant protein isoform is truncated like that of 44-5 (7); however, it encodes approximately 71 more amino acids in the C-terminal region. As homozygotes, these alleles are severe, lethal hypomorphs, but in combination with 30-5 they complement well and demonstrate a TS effect.
Our findings for the temperature sensitivity of Mef2 allelic combinations could be applied to the analysis of other transcription factors. A combination of alleles for the Drosophila twist gene also shows TS in the heteroallelic arrangement (Thisse et al. 1987). Neither of these mutations affects the DNA binding domain (Baylies and Bate 1996); however they might nevertheless impact regions of the protein with distinct functions.
Regarding MEF2 functional domains, an important regulatory domain of MEF2 appears to lie between amino acids 219 and 241. This region has a potentially inhibitory function on the basis of the increased transcriptional activation activity of the 44-5 (11) isoform that lacks these amino acids. Previous work using murine MEF2s has identified a repression domain located in exon 9 (Gulick and Zhu 2004). Alternative splice variants lacking this domain have an increased ability to activate downstream MEF2 targets in cell culture as was seen in the cell culture assay for 44-5 (11). While the region identified by Gulick and Zhu (2004) probably does not correspond to amino acids 219–241, these finding nevertheless confirm that there is much still to be learned regarding the function of the MEF2 C-terminal region.
Acknowledgments
We are very grateful to the individuals who supplied reagents essential to this work: Elliot Goldstein, Daniel Kiehart, and Bruce Paterson. This work was supported by a grant from the National Institutes of Health (NIH) (GM61738 to R.M.C.). M.M.A. was supported by a MARC fellowship from the NIH (5T34GM008751). We acknowledge technical support from the Department of Biology's Molecular Biology Facility, supported by NIH grant no. 1P20RR18754 from the Institute Development Award (IDeA) Program of the National Center for Research Resources.
References
- Baker, P., K. K. Kelly Tanaka, N. Klitgord and R. M. Cripps, 2005. Adult myogenesis in Drosophila melanogaster can proceed independently of myocyte enhancer factor-2. Genetics 170: 1747–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylies, M. K., and M. Bate, 1996. twist: A myogenic switch in Drosophila. Science 272: 1481–1484. [DOI] [PubMed] [Google Scholar]
- Black, B. L., and E. N. Olson, 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell. Dev. Biol. 14: 167–196. [DOI] [PubMed] [Google Scholar]
- Black, B. L., and R. M. Cripps (Editors), 2009. Myocyte enhancer factor-2 transcription factors in heart development and disease in Heart Development and Disease. Academic Press, San Diego.
- Bour, B. A., M. A. O'Brien, W. L. Lockwood, E. S. Goldstein and R. Bodmer, 1995. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 9: 730–741. [DOI] [PubMed] [Google Scholar]
- Carpenter, J. M., 1950. A new semisynthetic food medium for Drosophila. Drosophila Inform. Serv. 24: 96–97. [Google Scholar]
- Casso, D., F.-A. Ramirez-Weber and T. B. Kornberg, 1999. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech. Dev. 88: 229–232. [DOI] [PubMed] [Google Scholar]
- Chakshusmathi, G., K. Mondal, G. S. Lakshmi, G. Singh, A. Roy et al., 2004. Design of temperature-sensitive mutants solely from amino acid sequence. Proc. Natl. Acad. Sci. USA 101(21): 7925–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, V. T., and M. K. Baylies, 2005. Specification of individual Slouch muscle progenitors in Drosophila requires sequential Wingless signaling. Development 132: 713–724. [DOI] [PubMed] [Google Scholar]
- Edgar, R. S., and I. Lielausis, 1963. Temperature-sensitive mutants of bacteriophage T4D: their isolation and genetic characterization. Genetics 49: 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham, J. R. S., 1966. Genetic Complementation. W. A. Benjamin, New York.
- Goldstein, E. S., S. L. Treadway, A. E. Stephenson, G. D. Gramstad, A. Keilty et al., 2001. A genetic analysis of the cytological region 46C-F containing the Drosophila melanogaster homolog of the jun proto-oncogene. Mol. Genet. Genomics 266: 695–700. [DOI] [PubMed] [Google Scholar]
- Gordon, C. L., and J. King, 1993. Genetic properties of temperature-sensitive folding mutants of the coat protein of phage P22. Genetics 136: 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick, T., and B. Zhu, 2004. Phosphorylation and alternative pre-mRNA splicing converge to regulate myocyte enhancer factor 2C activity. Mol. Cell. Biol. 24(18): 8264–8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L. H., J. Culotti and B. Reid, 1970. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA 66: 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamori, Y., W. U. Hung-Yi, V. Sartorelli and L. Kedes, 1997. The basic domain of myogenic basic helix-loop-helix (bHLH) proteins is the novel target for direct inhibition by another bHLH protein, Twist. Mol. Cell. Biol. 17(11): 6563–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K., J. M. Louis, L. Donaldson, F. L. Lim, A. D. Sharrocks et al., 2000. Solution structure of the MEF2A-DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J. 19: 2615–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, K. K., S. M. Meadows and R. M. Cripps, 2002. Drosophila MEF2 is a direct regulator of Actin 57B transcription in cardiac, skeletal, and visceral muscle lineages. Mech. Dev. 110(1-2): 39–50. [DOI] [PubMed] [Google Scholar]
- Kelly Tanaka, K. K., A. L. Bryantsev and R. M. Cripps, 2008. Myocyte enhancer factor 2 and chorion factor 2 collaborate activation of the myogenic program in Drosophila. Mol. Cell. Biol. 28(5): 1616–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart, D. P., and R. Feghali, 1986. Cytoplasmic myosin from Drosophila melanogaster. J. Cell Biol. 103: 1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly, B., S. Galewsky, A. B. Firulli, R. A. Schulz and E. N. Olson, 1994. D-MEF2: A MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc. Natl. Acad. Sci. USA 91: 5662–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly, B., B. Zhao, G. Ranganayakulu, B. M. Paterson, R. A. Schulz et al., 1995. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 267: 688–693. [DOI] [PubMed] [Google Scholar]
- Martin, J. F., J. M. Miano, C. M. Hustad, N. G. Copeland, N. A. Jenkins et al., 1994. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol. Cell. Biol. 14: 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin, J. D., B. L. Black, J. F. Martin and E. N. Olson, 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83: 1125–1136. [DOI] [PubMed] [Google Scholar]
- Molkentin, J. D., B. L. Black, J. F. Martin and E. N. Olson, 1996. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 16(6): 2627–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H. T., R. Bodmer, S. M. Abmayr, J. C. Mcdermott and N. A. Spoerel, 1994. D-mef2: a Drosophila mesoderm-specific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proc. Natl. Acad. Sci. USA 91: 7520–7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T., J. B. Wang and R. A. Schulz, 2002. Mutations within the conserved MADS box of the D-MEF2 muscle differentiation factor result in a loss of DNA binding ability and lethality in Drosophila. Differentiation 70: 438–446. [DOI] [PubMed] [Google Scholar]
- Patel, N., 1994. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes, in Methods in Cell Biology, edited by E. A. Fyrberg and L. S. B. Goldstein. Academic Press, Boston. [DOI] [PubMed]
- Potthoff, M. J., and E. N. Olson, 2007. MEF2: a central regulator of diverse developmental programs. Development 134(23): 4131–4140. [DOI] [PubMed] [Google Scholar]
- Peckham, M., R. M. Cripps, D. C. S. White and B. Bullard, 1992. Mechanics and protein content of insect flight muscles. J. Exp. Biol. 168: 57–76. [Google Scholar]
- Ranganayakulu, G., B. Zhao, A. Dokidis, J. D. Molkentin, E. N. Olson et al., 1995. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev. Biol. 171: 169–181. [DOI] [PubMed] [Google Scholar]
- Sadler, J. R., and A. Novick, 1964. The properties of repressor and the kinetics of its action. J. Mol. Biol. 12: 305–327. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Santelli, E., and T. J. Richmond, 2000. Crystal Structure of MEF2A Core Bound to DNA at 1.5 Å Resolution. J. Mol. Biol. 297: 437–449. [DOI] [PubMed] [Google Scholar]
- Smith, D. H., P. B. Berget and J. King, 1980. Temperature-sensitive mutants blocked in the folding or subunit assembly of the bacteriophage P22 tail-spike protein. I. Fine-structure mapping. Genetics. 2: 331–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg, H. A., and T. N. Davis, 1997. A mutational analysis identifies three functional regions of the spindle pole component Spc110p in Saccharomyces cerevisiae. Mol. Biol. Cell. 12: 2575–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, M. V., K. E. Beatty, H. K. Hunter and M. K. Baylies, 1995. Drosophila MEF2 is regulated by twist and is expressed in both the primordia and differentiated cells of the embryonic somatic, visceral and heart musculature. Mech. Dev. 50: 29–41. [DOI] [PubMed] [Google Scholar]
- Thisse, B., M. E. Messal and F. Perrin-Schmitt, 1987. The twist gene: Isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 15: 3439–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]