Abstract
Different genomic regions replicate at a distinct time during S-phase. The SuUR mutation alters replication timing and the polytenization level of intercalary and pericentric heterochromatin in Drosophila melanogaster salivary gland polytene chromosomes. We analyzed SuUR in different insects, identified conserved regions in the protein, substituted conserved amino acid residues, and studied effects of the mutations on SUUR function. SuUR orthologs were identified in all sequenced drosophilids, and a highly divergent ortholog was found in the mosquito genome. We demonstrated that SUUR evolves at very high rate comparable with that of Transformer. Remarkably, upstream ORF within 5′ UTR of the gene is more conserved than SUUR in drosophilids, but it is absent in the mosquito. The domain structure and charge of SUUR are maintained in drosophilids despite the high divergence of the proteins. The N-terminal part of SUUR with similarity to the SNF2/SWI2 proteins displays the highest level of conservation. Mutation of two conserved amino acid residues in this region impairs binding of SUUR to polytene chromosomes and reduces the ability of the protein to cause DNA underreplication. The least conserved middle part of SUUR interacting with HP1 retains positively and negatively charged clusters and nuclear localization signals. The C terminus contains interlacing conserved and variable motifs. Our results suggest that SUUR domains evolve with different rates and patterns but maintain their features.
IT is well established that replication timing in the S-phase generally correlates with the preceding transcriptional activity of the chromatin domain (Schubeler et al. 2002; MacAlpine et al. 2004; Donaldson 2005). As a rule, late replication is observed in transcriptionally silent and condensed chromosome regions, mostly composed of pericentric heterochromatin (PH). Late-replicating regions in euchromatin are represented by 100- to 200-kb chromatin domains (MacAlpine et al. 2004; White et al. 2004), which are often denoted as foci of late replication in the interphase nuclei (Berezney et al. 2000).
One of the peculiar features that advances Drosophila melanogaster as a model for studying late replication is its giant larval salivary gland polytene chromosomes that enable easy and precise identification of late-replicating regions. There are ∼240 regions showing late replication apart from the PH in D. melanogaster polytene chromosomes. These regions are scattered over the euchromatic chromosome arms and also display characteristic features of heterochromatin, such as dense packaging and low transcription level (Zhimulev et al. 2003a). Many late-replicating regions are underreplicated. Underreplication results from the early start of the G-phase before the S-phase is actually complete; hence many late-replicating chromosome sequences fail to complete replication by the end of each endocycle (Gall et al. 1971; Smith and Orr-Weaver 1991; Lilly and Spradling 1996). Morphologically, underreplication in these regions appears as “weak spots” or chromosome breaks on polytene chromosome squashes, which serve as a convenient cytological marker of late replication and underreplication. These regions are collectively referred to as intercalary heterochromatin (IH). Many of them are known to be bound by repressive Pc-G protein complexes and are mainly composed of deeply silenced genes (Zhimulev and Belyaeva 2003; Zhimulev et al. 2003a; Belyakin et al. 2005).
Underreplication is also known to be significantly affected by a product of the SuUR gene. This gene encodes a protein that is specifically associated with PH and IH (Makunin et al. 2002; Zhimulev et al. 2003b; Pindyurin et al. 2007). The only known mutation of the gene, SuURES, is caused by an ∼6-kb insertion into the last exon (Makunin et al. 2002). SuURES larvae show altered replication timing in late-replicating regions. Namely, replication in these regions completes sooner than in the wild-type strain, so the polytenization level in IH is restored to that of the euchromatin. This is also accompanied by an increase in the degree of polytenization of many sequences in PH and by the concomitant structuring of the chromocenter (Belyaeva et al. 1998; Moshkin et al. 2001; Zhimulev et al. 2003a). Conversely, an increase in the SuUR gene copy number enhances the underreplication in IH regions (Zhimulev et al. 2003a; Belyakin et al. 2005). Ectopic expression of SUUR in follicular cells suppresses the amplification of chorion gene clusters (Volkova et al. 2003). Finally, strong SUUR overexpression in third instar larval salivary glands leads to structural changes (“swellings”) in chromosome morphology of PH and IH regions (Zhimulev et al. 2003c).
SuUR gene has four exons and a very short promoter region devoid of recognizable regulatory elements in addition to two presumptive E2F-binding sites (Makunin et al. 2002). Recently, an upstream open reading frame (uORF) was identified in the 5′ UTR of SuUR (Hayden and Bosco 2008). The gene encodes a 962-aa protein without any homologs reported in protein databases (Makunin et al. 2002). Nevertheless, the N terminus of the protein shows moderate similarity to the ATPase/helicase domain of chromatin-remodeling proteins from the SWI2/SNF2 group. ATP-dependent chromatin-remodeling factors are known to serve as molecular motors that alter the accessibility of DNA in chromatin, thereby regulating many aspects of transcription and replication (Havas et al. 2001). While the strongest similarity between SUUR and SNF2/SWI2 proteins is observed within Walker A and Walker B motifs involved in ATP binding and hydrolysis (Walker et al. 1982), the SUUR sequence differs significantly from the canonical motifs (Makunin et al. 2002). It is unknown whether SUUR could bind and hydrolyze ATP, but the fragment containing the first 360 amino acid residues shows a dominant-negative effect and displaces endogenous SUUR from polytene chromosomes (Kolesnikova et al. 2005).
Previously, we demonstrated that the C-terminal fragment SUUR495–962 controls underreplication, although it is unable to induce structural changes in chromatin when overexpressed. On the contrary, the N-terminal fragments SUUR1–599 and SUUR1–779 had no effect on endoreplication, but were able to bind PH and IH regions and to induce formation of chromosome swellings such as the full-length SUUR (Kolesnikova et al. 2005). Here we demonstrate that SUUR is present in other Drosophila species and that it affects the break formation in salivary gland polytene chromosomes in D. simulans. Comparative analysis of SUUR in 11 Drosophila species showed that the protein belongs to a group of fast-evolving genes although its domain organization is conserved in Drosophila. We introduced targeted point mutations in two conserved regions within N- and C-terminal parts of SUUR and analyzed how these substitutions affect the protein function. We showed that point mutations in the N-terminal region of SUUR abolish its specific binding to the late-replicating regions of polytene chromosomes and decrease the ability of the protein to suppress polytenization in these regions. We also performed a more precise functional mapping of the C-terminal region of SUUR, which was known to cause underreplication.
MATERIALS AND METHODS
Drosophila stocks and genetics:
Fly stocks were kept on standard Drosophila cornmeal medium at 25°. The following stocks carrying GAL4 drivers were used: da-GAL4 for ubiquitous expression (Wodarz et al. 1995), Sgs3-GAL4 for expression in salivary glands starting from the mid-third instar (PS1–PS11) (Cherbas et al. 2003), AB1-GAL4 for expression in salivary glands from early embryogenesis (Drysdale et al. 2005), arm-GAL4 for weak variegated expression in salivary glands (Kolesnikova et al. 2005), and C323-GAL4 for expression in follicle cells (Manseau et al. 1997). The w; SuURES stock was described in Belyaeva et al. (1998). Oregon-R was used as a wild-type stock. We used D. erecta and D. virilis from the laboratory stock collection and D. ananassae (strain 14024-0371.13) from the Tucson Drosophila Stock Center.
Molecular procedures:
All molecular procedures were performed as described in Sambrook and Russell (2001). DNA-modifying enzymes were purchased from New England Biolabs. Genomic DNA from D. erecta was amplified by PCR using SuUR-specific primers, and sequencing of PCR products was done on the ABI PRISM 310 Genetic Analyzer (Applied Biosystems) at the DNA Sequencing Center of the Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia (http://sequest.niboch.nsc.ru). RNA from ovaries was isolated using Trizol (Gibco BRL). A RT–PCR kit (Promega) was used according to the manufacturer's recommendations. Primer sequences used for the RT–PCR and amplification of SuUR genomic sequences in different Drosophila species are available upon request.
Constructs for transformations:
Transgenic constructs are described in the supporting information (File S1). Plasmids were co-injected with pUChsΔ2-3wc (pTURBO) transposase helper plasmid into y1 w67 embryos, and several independent transgenic lines were obtained for each construct (Rubin and Spradling 1982).
Immunostaining of polytene chromosomes:
Indirect immunofluorescent analysis of polytene chromosomes was performed as described in Poux et al. (2001). We used E45 antibodies raised against the middle part of SUUR (Makunin et al. 2002) and antibodies against hemagglutinin tag (HA) provided by V. Pirrotta. The E45 antibodies and HA antiserum were used at a 1:50 and 1:10 dilutions, respectively. For immunostaining of D. simulans polytene chromsomes and analysis of chromosome binding of SUURNmut protein, the double-squash approach was used when a positive control was present on the same slide with the investigated polytenes.
Data analysis:
We used the BLAT program (Kent 2002; http://genome.ucsc.edu) to map the SuUR orthologs in the genomic sequences available. Multiple protein alignments were constructed using ClustalW (Thompson et al. 1994; http://www.ebi.ac.uk/clustalw). K-Estimator 6.1v software (Comeron 1999; http://en.bio-soft.net/format/KEstimator.html) was used to calculate the number of synonymous (Ks) and nonsynonymous (Ka) substitutions. The SAPS program was used for the statistical analysis of protein sequences (Brendel et al. 1992; http://www.isrec.isb-sib.ch/software/SAPS_form.html). The phylogenetic tree was built in MEGA4 (Kumar et al. 2008). Identification of protein motifs and structure predictions were performed using MotifScan (http://myhits.isb-sib.ch/cgi-bin/motif_scan) and Predict Protein (Rost et al. 2004; http://www.predictprotein.org).
RESULTS
Identification of SUUR protein in insects:
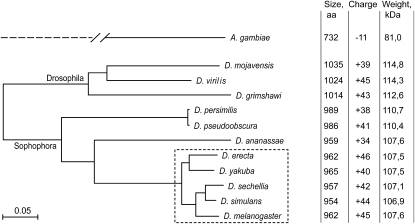
Southern blot hybridization of SuUR cDNA with genomic DNAs from various Drosophila species produced signals in species from the melanogaster subgroup only (data not shown). Among these, D. erecta was one of the most distant species from D. melanogaster (Figure 1). We amplified and sequenced the genomic DNA from the SuUR locus in D. erecta (GenBank accession no. AJ539550). The exon–intron structure of the SuUR gene in D. erecta was confirmed by comparison of the genomic sequence and the sequence of SuUR cDNA fragment obtained from D. erecta total ovarian RNA by RT–PCR. Splice sites are conserved between D. melanogaster and D. erecta.
Figure 1.—
Phylogenetic tree based on SUUR proteins and the main properties of each protein. The tree was built in MEGA4 software using protein alignment with the following settings: neighbor-joining method, complete deletion of gaps, Poisson correction, and uniform rates among sites. Species from the melanogaster subgroup are boxed with a dashed line.
We also used predicted SUUR sequences from nine recently sequenced Drosophila species for which genomic sequences are available at the UCSC Genome Browser website (Figure 1). We noted that the annotations of the SuUR gene produced by some annotation projects differ significantly from the exon–intron structure of the gene in D. melanogaster in five of nine species: D. simulans, D. yakuba, D. ananassae, D. persimilis, and D. virilis. The differences include the prediction of an additional exon in the 5′-end of the gene, which merged the uORF with the main ORF, and the prediction of additional introns and lack thereof, notably by the Genescan annotations (Burge and Karlin 1997). We determined the exon–intron structure of SuUR in D. yakuba, D. ananassae, and D. virilis by sequencing RT–PCR products obtained from total fly RNA. Sequences of PCR products confirmed discrepancies in Genescan annotation; therefore, we used our version of SuUR annotation. For D. simulans and D. persimilis, we transferred annotation of SuUR from the closely related species D. melanogaster and D. pseudoobscura, respectively. Sequences of SuUR ORFs used in this study are given in Figure S1. These data confirmed the integrity of the conserved uORF predicted in SuUR 5′ UTR for all analyzed species (Hayden and Bosco 2008). Our review of SuUR annotation in Drosophila species demonstrates that the computer gene annotations should be used with great care.
The SuUR gene is not annotated outside of Drosophila. However, the BLAST search identified a weak similarity (∼25% identities, E-value 7e−6) with ENSANGP00000027713.1 protein from Anopheles gambiae (contemporary gene name AGAP005819; coordinates: chr2L:21,832,968–21,835,239; AgamP3 genome assembly). The similarity was limited to the N-terminal region of SUUR (aa 51–276). The rest of the protein sequence in A. gambiae is highly diverged, making comparison of the full-size proteins impossible. In contrast to the Drosophila SUUR, the ENSANGP00000027713.1 protein has no negatively or positively charged regions in the middle part of the protein. There is no apparent uORF upstream of the ENSANGP00000027713.1 main ORF. In D. melanogaster, the CG6310 gene is located downstream of SuUR. Similarly, the mosquito CG6310 homolog ENSANGT00000010378.2 (contemporary name AGAP005820; chr2L:21,835,318–21,836,802; AgamP3 genome assembly) is located downstream of ENSANGP00000027713.1, indicating that the latter indeed represents a highly diverged version of Drosophila SuUR. As Drosophila SUUR, the mosquito protein contains noncanonical Walker A and Walker B motif sites: it has substitution in GKT sequence from the putative nucleotide-binding loop and in DExH box. The predicted mosquito protein is smaller and has a negative charge while Drosophila SUUR has a positive total charge (Figure 1). We were unable to identify SUUR orthologs in other sequenced nondipteran insect species.
SUUR contributes to the formation of chromosome breaks in D. simulans:
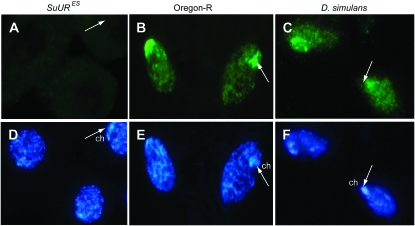
In many Drosophila species, the salivary gland polytene chromosomes display specific chromosome breaks and constrictions due to underreplication (Zhimulev 1998), suggesting that SUUR contributes to underreplication in these species. Immunostaining of D. simulans salivary gland polytene chromosomes with anti-SUUR antibodies does not produce any pronounced pattern. A chromocenter-specific signal could be detected only in rare nuclei. Immunofluorescent analysis of SUUR localization on polytene chromosomes of D. melanogaster × D. simulans hybrid larvae shows absence of staining in ∼90% of nuclei, with ∼10% of nuclei demonstrating a staining pattern characteristic of D. melanogaster. Notwithstanding, we did observe strong SUUR staining in follicle-cell nuclei preparations of whole-mount ovaries of D. simulans (Figure 2C). In both wild-type D. melanogaster and D. simulans the antibodies produce staining throughout the nucleus with a strong signal immediately adjacent to the chromocenter while virtually no staining is observed in the SuURES mutant (Figure 2).
Figure 2.—
Immunodetection of SUUR in follicular cells with E45 antibodies. (A) Negative control: no staining is observed in follicle cells of SuURES. (B) Positive control: strong staining is observed in the nucleus and especially the chromocenter (ch, indicated by arrows) in follicle cells of D. melanogaster (Oregon-R). (C) In D. simulans, the antibodies produce staining similar to that observed in Oregon-R. (D–F) Hoechst staining.
To prove that SUUR does contribute to underreplication in D. simulans, we crossed AB1-GAL4>UAS-SuUR1–458 D. melanogaster females with D. simulans males. The AB1-GAL4>UAS-SuUR1–458 transgenic combination provides expression of the N-terminal half of SUUR in salivary glands from an early developmental stage. Expression of this fragment (SUUR1–458) under early AB1-GAL4 driver has a dominant-negative effect, and results in the complete disappearance of weak spots from the polytene chromosomes, similar to the SuUR mutant phenotype (Kolesnikova et al. 2005). Consistently, overexpression of SUUR1–458 leads to the disappearance of weak spots on both homologs in D. melanogaster × D. simulans hybrid progeny (Figure S2). It needs to be pointed out that the chromosomes of both D. simulans and D. melanogaster (Oregon-R) × D. simulans hybrids demonstrate weak spots in the same regions as D. melanogaster. This result argues in favor of a common mechanism of weak spot formation in both species and that the SUUR protein has a key role in this process.
SUUR orthologs display high levels of substitutions in different Drosophila species:
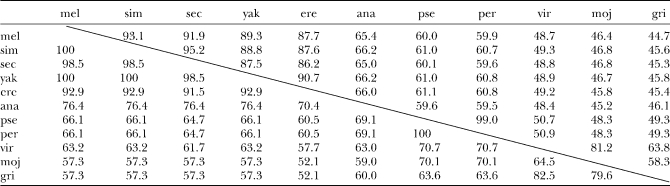
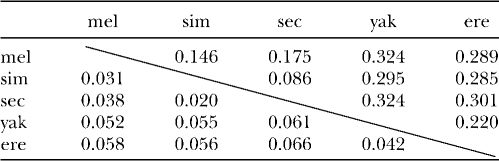
Comparison of SUUR orthologs from Drosophila species revealed high numbers of amino acid substitutions, insertions, and deletions, even in closely related species (Table 1, Figure S3). Strikingly, the level of amino acid conservation is much higher within uORF, which encompasses 68 residues in D. melanogaster, than is observed for SUUR main ORF (Table 1). We calculated the numbers of synonymous (Ks) and nonsynonymous (Ka) substitutions per site for the species from the melanogaster subgroup, using K-Estimator software (Comeron 1999) (Table 2). We excluded distantly related species from this analysis because of the ambiguity in alignment, especially in the middle part of the protein (see below). The number of nonsynonymous substitutions per site in the SuUR gene between D. melanogaster and D. yakuba is 0.052. This is very similar to the Ka value characteristic for the fast-evolving genes in Drosophila (Schmid and Tautz 1997). The size and charge of the SUUR protein are retained in the course of evolution despite the high substitution rate (Figure 1). The secondary structure predictions even in very distant species, such as D. melanogaster and D. grimshawi, turned out to be mostly identical. Numerous helices and extended sheets were predicted in the N-terminal part while the rest of the protein was less structured (Figure S4).
TABLE 1.
Pairwise comparison of amino acid identities for SUUR protein and uORF
Identity for SUUR protein is shown above the diagonal, and identity for uORF from SuUR transcript is shown below the diagonal. mel, D. melanogaster; sim, D. simulans; sec, D. sechellia; yak, D. yakuba; ere, D. erecta; ana, D. ananassae; pse, D. pseudoobscura; per, D. persimilis; vir, D. virilis; moj, D. mojavensis; gri, D. grimshawi.
TABLE 2.
Synonymous (above diagonal) and nonsynonymous (below diagonal) substitutions per site in SuUR coding region
mel, D. melanogaster; sim, D. simulans; sec, D. sechellia; yak, D. yakuba; ere, D. erecta.
The phylogenetic tree created for available SUUR proteins is fairly consistent with the tree obtained in genomewide analysis (Stark et al. 2007); e.g., D. yakuba and D. erecta are grouped together, and the D. pseudoobscura branch is shorter (Figure 1). While the Drosophila subgenus branch appeared somewhat longer in the SUUR tree in comparison to the whole-genome tree, it could be just a consequence of a rooting problem: unfortunately, the sequence of the D. willistoni genome was not available at the UCSC Genome Browser website at the time of our analysis, and use of SUUR sequence from this species could affect the position of the tree root, and hence could affect the length of the Drosophila subgenus branch.
Distribution of substitutions across the protein is nonuniform (Figure 3A, Figure S3). The N-terminal region of SUUR is the most conserved part of the protein. It has a relatively low level of substitutions, no insertions, and no deletions even in distantly related Drosophila species. The middle part of SUUR (D. melanogaster residues 280–581) shows the lowest level of amino acid identity across species (Table 3). Two distantly related species from the subgenus Drosophila, D. mojavensis and D. virilis, have long insertions in this region of SUUR (Figure S3). Despite an extremely high level of primary sequence divergence, the negatively and positively charged regions located in the middle part (Figure 3B) consistently maintained their properties in other species (Table 3). For example, the SAPS program (http://www.isrec.isb-sib.ch/software/SAPS_form.html) predicts statistically significant spacing between positively charged residues on the sides of a negatively charged region of SUUR in D. melanogaster, D. simulans, D. sechellia, D. yakuba, D. erecta, D. pseudoobscura, and D. persimilis. In D. mojavensis, D. virilis, and D. grimshawi, this negatively charged region is interrupted by a single positively charged residue (Figure S3). In D. ananassae, both negatively and positively charged clusters are smaller (Table 3). Intriguingly, these regions display a very similar total charge in different SUUR orthologs, although the vast majority of the charged residues per se are not conserved (Figure S3).
Figure 3.—
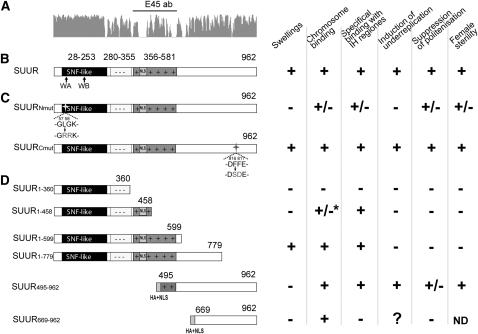
Organization and features of different SUUR isoforms. (A) Conservation plot, based on the SUUR sequences from 11 Drosophila species, and the protein region used to generate E45 antibodies (E45 ab). (B) Domain organization of SUUR protein. The SNF-like domain is solid, the negatively charged region is marked with - -, and the positively charged amino acid region is darkly shaded with + +. NLS, nuclear localization signal. Arrows show the position of regions with similarity to Walker A (WA) and Walker B (WB) motifs of SNF2/SWI2 proteins. (C) Point mutations introduced within the N- and C-terminal part of SUUR are indicated by arrows. (D) Truncated fragments of SUUR are described by Kolesnikova et al. (2005) except for SUUR669–962. HA+NLS, hemagglutinin tag and nuclear localization signal sequence. An asterisk indicates that granules in intercalary and pericentric heterochromatin were observed.
TABLE 3.
Conservation of charge for middle part
| Negative charge region
|
Positive charge region
|
|||||
|---|---|---|---|---|---|---|
| Species | Position | Identity | Charge | Position | Identity | Charge |
| mel | 280–355 | −17 | 356–581 | +39 | ||
| sim | 280–350 | 86.8 | −16 | 351–573 | 85.3 | +37 |
| sec | 280–350 | 85.5 | −16 | 351–576 | 84.0 | +35 |
| yak | 280–356 | 80.5 | −18 | 357–583 | 78.0 | +35 |
| ere | 280–356 | 79.2 | −18 | 357–580 | 75.8 | +41 |
| ana | 280–351 | 56.5 | −13 | 352–580 | 39.6 | +29 |
| pse | 280–359 | 46.9 | −18 | 360–609 | 40.9 | +37 |
| per | 280–359 | 46.3 | −18 | 360–612 | 40.0 | +36 |
| vir | 280–371 | 30.4 | −16 | 372–659 | 24.3 | +43 |
| moj | 280–369 | 26.9 | −17 | 370–676 | 24.8 | +43 |
| gri | 280–371 | 31.5 | −16 | 372–664 | 21.4 | +44 |
Amino acid identity was calculated between D. melanogaster and other species. mel, D. melanogaster; sim, D. simulans; sec, D. sechellia; yak, D. yakuba; ere, D. erecta; ana, D. ananassae; pse, D. pseudoobscura; per, D. persimilis; vir, D. virilis; moj, D. mojavensis; gri, D. grimshawi.
The middle part of SUUR largely coincides with the region (aa 339–671) known to interact with another heterochromatic protein, HP1, in the yeast two-hybrid assay (Pindyurin et al. 2008). Surprisingly, this part undergoes very rapid evolution. Even the sequence that displays similarity to the HP1-interacting motif (LRVSL, aa 429–433; Pindyurin et al. 2008) diverged significantly in Drosophila species (Figure S3), with only two species, D. yakuba and D. erecta, containing this motif unaltered.
A search for the known protein motifs in SUUR using PredictProtein and MotifScan identified type I or type II nuclear localization signals (NLS) in the middle part of the protein in all species, except for D. mojavensis in which no bipartite NLS was found. In addition to NLS, a motif homologous to the AT hook was present in the middle of the protein in five species, from D. melanogaster to D. ananassae. No known motifs were identified within the C-terminal part of SUUR, although this region encompassed alternating stretches of conserved and nonconserved sequences (Figure S3).
Targeted mutagenesis of SUUR and the effect of mutations on endoreplication:
On the basis of the protein alignment, we substituted conserved amino acid residues in two regions of SUUR protein. In the N-terminal part, we introduced L57R/G58R substitutions (SUURNmut) within a conserved region with similarity to the Walker A motif of ATPase/helicase domain (Figure 3, B and C). It has previously been established that ectopic expression of truncated SUUR protein containing amino acids 1–779 (SUUR1–779) does not suppress endoreplication in salivary glands (Figure 3D), indicating that the protein domain (or its crucial part) involved in the suppression of endoreplication is located downstream of the nonsense mutation in SUUR1–779. Therefore, we substituted two invariably conserved amino acid residues within this region, F816S and F817D, to obtain SUURCmut. Mutated ORFs were cloned into the pUAST vector (Brand and Perrimon 1993), and several independent transformants were generated for each construct.
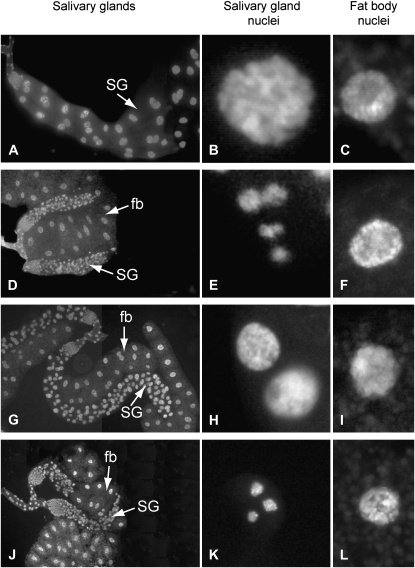
We examined the effects of ectopically expressed mutated proteins in the UAS-GAL4 system (Brand and Perrimon 1993). Permanent strong expression of SUUR in salivary glands under the AB1-GAL4 driver suppresses endoreplication and results in miniature salivary glands (Volkova et al. 2003). Expression of SUURNmut under the control of AB1-GAL4 causes only partial suppression of endoreplication. The nuclei of salivary glands from AB1-GAL4>UAS-SuURNmut larvae are larger than those with ectopic expression of the full-length SUUR from AB1-GAL4>UAS-SuUR larvae, although they are smaller than Oregon-R salivary gland nuclei (Figure 4).
Figure 4.—
Effects of point mutations in SUUR on its ability to suppress endoreplication in salivary glands upon overexpression from early development. Left column, salivary gland; middle column, salivary gland nuclei; right column, fat body nuclei photographed under the same magnification. (A–C) Oregon-R. (D) Ectopic expression of SUUR protein in salivary glands under the AB1-GAL4 driver suppresses polytenization, which leads to the formation of miniature salivary gland. The size of the salivary gland nuclei in AB1-GAL4>UAS-SuUR larvae is dramatically reduced (E) as compared to the fat body nuclei (F) and wild-type salivary gland nuclei (B). (G) Ectopic expression of SUURNmut with the AB1-GAL4 driver results in partial suppression of polytenization. Salivary gland nuclei in AB1-GAL4>UAS-SuURNmut larvae (H) are similar in size to those of the fat body (I), only slightly smaller than in Oregon-R (B), but significantly larger than in AB1-GAL4>UAS-SuUR larvae (E). (J–L) Effects of ectopic expression of SUURCmut on polytenization in the salivary gland are indistinguishable from those of the full-length SUUR. Salivary gland (SG) nuclei and fat body (fb) nuclei were stained with Hoechst.
Ectopic expression of UAS-SuUR in follicular cells under the control of the C323-GAL4 driver suppresses amplification of chorion genes and results in complete female sterility (Volkova et al. 2003). When SUURNmut was ectopically expressed under the C323-GAL4 driver, we observed weak suppression of the female sterile phenotype (20 crosses were set for 10 independent transgenic stocks, and in 2 crosses from different stocks single escapers were observed).
Contrary to our expectations, substitutions in the C-terminal part of the SUUR had no detectable effects on the protein. Overexpression of SUURCmut under the AB1-GAL4 driver results in miniature salivary glands similar in size to those observed upon ectopic overexpression of full-length SUUR in AB1-GAL4>UAS-SuUR larvae (Figure 4). Ectopic expression of SUURCmut under C323-GAL4 resulted in complete female sterility similar to the ectopic expression of full-length SUUR.
Mutation in N terminus impairs the protein's ability to associate with chromosomes and alters the chromatin structure:
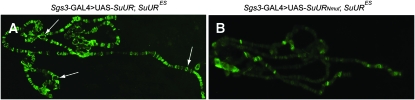
In wild-type polytene chromosomes, SUUR is detected in late-replicating regions. When UAS-SuUR is expressed under the control of the weak mosaic arm-GAL4 driver, 20% of salivary gland nuclei demonstrate weak spots in the IH regions and an immunostaining pattern similar to those of wild-type SUUR (Kolesnikova et al. 2005). It is a convenient system for the expression of the protein at the level similar to that of the wild type. When SUURNmut is expressed under the control of the arm-GAL4 driver, no protein is detected in PH or IH or elsewhere on the arm-GAL4>UAS-SuURNmut; SuURES chromosomes except for a weak signal in nucleolus (Figure S5), and no weak spots were observed (data not shown). To test whether SUURNmut is capable of any chromosome binding, we employed a strong salivary-gland-specific Sgs3-GAL4 driver, which is active in mid-third instars when most of the replication in the salivary gland has ceased. Sgs3-GAL4>UAS-SuUR; SuURES larvae chromosomes display distinct binding signals in all bands and the chromocenter (Figure 5A). In contrast, in Sgs3-GAL4>UAS-SuURNmut; SuURES larvae, the immunolocalization signal for SUURNmut is weak and dim (Figure 5B). These results suggest that the introduced substitutions within the N-terminal regions of SUUR dramatically decrease the binding of the protein to chromosomes.
Figure 5.—
SUURNmut displays reduced binding to chromosomes when overexpressed under the control of the Sgs3-GAL4 driver. Chromosomes from larvae expressing full-length SUUR and SUURNmut were photographed using identical exposure times. (A) On Sgs3-GAL4>UAS-SuUR; SuURES polytene chromosomes, SUUR is found in almost all bands. Swellings are indicated by arrows. (B) On Sgs3-GAL4>UAS-SuURNmut; SuURES polytene chromosomes, binding of mutant protein is drastically reduced.
Ectopic expression of UAS-SuUR under Sgs3-GAL4 induces swellings in IH regions (Zhimulev et al. 2003c). In contrast, no changes in chromosome morphology are observed when SUURNmut is expressed with the same Sgs3-GAL4 driver (data not shown).
The C-terminal part of SUUR binds to polytene chromosomes:
Mutation of conserved amino acid residues F816S and F817D in the C-terminal region has no pronounced effect on the ability of the SUUR protein to cause underreplication. Earlier we showed that the C-terminal SUUR fragment SUUR495–962 suppresses endoreplication while ectopic expression of the SUUR fragment SUUR1–779 lacking residues 780–962 does not (Kolesnikova et al. 2005). We decided to test the overexpression effects of the smaller conserved C-terminal region of SUUR (aa 669–962). We cloned the fragment of the SuUR ORF that contained the last 293 codons fused to the HA tag and NLS into the pUAST vector (see File S1, Figure 3D) to obtain the UAS-SuUR669–962 construct (hereafter, SUUR669–962).
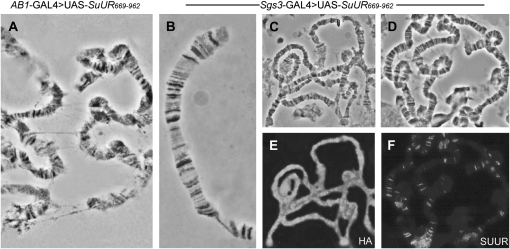
When SUUR669–962 was expressed from the onset of development under control of the AB1-GAL4 driver, the size of the salivary glands remained unaffected. However, the analysis of polytene chromosomes from AB1-GAL4>UAS-SuUR669–962 larvae revealed general disorganization of polytene chromosomes (Figure 6A). In contrast to the wild-type chromosomes where ectopic fibers typically link IH regions, in AB1-GAL4>UAS-SuUR669–962 chromosomes we observed numerous ectopic fibers that were formed along the chromosome arms.
Figure 6.—
Chromosome binding of SUUR669–962 upon ectopic expression and its effects on chromatin structure. (A) Ectopic expression from early development in AB1-GAL4>UAS-SuUR669–962 larvae results in the formation of ectopic fibers. (B) Polytene chromosomes from Sgs3-GAL4>UAS-SuUR669-962 larvae resemble wild type in that they show no foam or swellings. (C and E) In Sgs3-GAL4>UAS-SuUR669-962 larvae, the chromosomes are nonspecifically covered with the ectopically expressed truncated protein, as detected with anti-HA antibodies. (D and F) Endogenous SUUR in Sgs3-GAL4>UAS-SuUR669–962 larvae can be detected at its typical chromosomal sites with E45 antibodies. (A–D) Phase contrast. (E and F) Immunostaining.
Ectopic expression of SUUR669-962 under arm-GAL4 resulted in a range of uniformly staining chromosomes of varying intensities (data not shown), as detected with anti-HA antibodies. The antibodies do not stain polytene chromosomes of the wild-type strain (data not shown). Also, it has been shown elsewhere that neither HA tag nor NLS bind polytene chromosomes on their own (Jaquet et al. 2002), suggesting that the observed localization pattern reflects a property of the SUUR669–962 fragment. Overexpression of SUUR669–962 with Sgs3-GAL4 driver results in extremely strong nonspecific binding of SUUR to the chromosomes, regardless of the banding pattern (Figure 6E). Notably, under these conditions polytene chromosome morphology remained unchanged (Figure 6B), even though the chromosomes appeared totally covered by the SUUR669–962 (Figure 6E). Endogenous SUUR was found to be specifically associated with its typical chromosomal sites (Figure 6F), indicating that the C-terminal part of the protein does not have a dominant-negative effect. Our results indicate that, while the C-terminal part of the protein is essential for underreplication, it does not significantly suppress endoreplication on its own.
DISCUSSION
SUUR orthologs are present in the genomes of 11 Drosophila species. Notably, all these species display chromosome breaks and constrictions marking local DNA underreplication in salivary gland polytene chromosomes (Zhimulev 1998). We observed a dominant-negative effect of SUUR1–458 overexpression in the salivary glands of hybrid D. melanogaster × D. simulans larvae, which manifested as a disappearance of chromosome breaks. This indirectly supports the idea that D. simulans SUUR protein is functional and affects late replication in heterochromatic regions in a way similar to that of D. melanogaster SUUR.
The potential SUUR ortholog in the mosquito lacks functionally important protein domains such as positively and negatively charged clusters, so its function in the mosquito remains in question. A BLAST search of the A. gambiae protein at NCBI resulted in high-confidence hits to predicted proteins in the yellow fever mosquito Aedes aegypti (E-value e−25 and e−22) and in the southern house mosquito Culex quinquefasciatus (E-value e−22). Interestingly, a BLAST search of SUUR from some Drosophila and Anopheles species detected mammalian ERCC6 protein, which is important in transcription-coupled excision repair (Troelstra et al. 1992) as the best hit. Mutations in ERCC6 lead to Cockayne syndrome (Mallery et al. 1998; Laugel et al. 2008). The similarity is restricted to the N-terminal part of the protein, and E-values range from e−11 for the search with the D. mojavensis protein to e−6 with A. gambiae.
The Ka/Ks ratio for SUUR orthologs varies from 0.16 to 0.23 within species from the melanogaster subgroup. The Ka/Ks ratio between D. melanogaster and D. yakuba orthologs is 0.16, suggesting that about one-half of the 1850 Drosophila-specific proteins evolve under stronger selection pressure than SUUR, judging from recent genomewide analysis of the Drosophila proteome (Zhang et al. 2007). The same study demonstrated that proteins with orthologs in distant species tend to evolve under stronger selection pressure than Drosophila-specific proteins. SUUR protein has a high substitution rate similar to the fast-evolving genes in Drosophila (Schmid and Tautz 1997). The evolution rate of the SuUR gene is comparable to that of transformer (tra), a gene involved in the primary somatic sex-determination pathway (O'Neil and Belote 1992). Specifically, the amino acid identity level in TRA and SUUR in D. melanogaster and D. simulans is, respectively, 92.4% and 93.1% and that in D. melanogaster and D. erecta is, respectively, 87.0% and 87.7% (O'Neil and Belote 1992). In other sequenced insect species, such as Bombyx mori and Apis mellifera, no SuUR orthologs were identified. On one hand, this is very typical for the fast-evolving genes: there is no true ortholog of tra in A. mellifera; however, a distant tra homolog, csd, is present (Beye et al. 2003; Cho et al. 2006). On the other hand, A. mellifera was not reported to have polyploid tissues. Possibly, the SUUR ortholog is absent in honeybees not because of the rapid evolution of the gene, but because of the fact that a mechanism involving SUUR does not exist in this species.
Conserved uORF was identified within 5′ UTR of SuUR (Hayden and Bosco 2008). Both the uORF and the main ORF in SuUR are maintained in drosophilids, and high divergence of SUUR in mosquito coincides with the lack of uORF in this species. This observation further supports the possibility that SuUR expression could be controlled via uORF. Although no examples of uORFs affecting downstream ORF protein production in Drosophila or other insect species have been demonstrated to date, 44 conserved uORFs were recently identified in D. melanogaster (Hayden and Bosco 2008).
Domain organization of SUUR is conserved in all Drosophila species analyzed, and different domains display different rates of amino acid substitutions. The most conserved region of the protein is found at its N terminus where it coincides with the region possessing similarity to the SNF2/SWI2 domain. When this region is absent in SUUR, the protein can no longer display some of its prominent effects in the overexpression system: SUUR495–962 (Figure 3D) fails to induce swellings, weakly induces underreplication, and only mildly suppresses polytenization (Kolesnikova et al. 2005). Even though the Walker A and Walker B motifs within this region are noncanonical, they are invariably conserved in all species analyzed (Figure S3). Substitution of two amino acid residues in the ATPase-like region, L57R/G58R, attenuates SUUR ectopic expression phenotypes due to its decreased binding to IH regions (this is best seen when we expressed SUURNmut under the control of arm-GAL4). To some extent SUURNmut functionally resembles SUUR495–962 much more than SUUR, because it fails to form chromosome swellings and only partially suppresses polytenization. Interestingly, the SUUR1–360 fragment does not bind to polytene chromosomes (Kolesnikova et al. 2005), but the substitutions in this region decrease the binding of the full-length protein.
The least conserved middle region of the protein encompasses negatively and positively charged clusters, and it is important for the specific binding of SUUR with chromosomes (Kolesnikova et al. 2005). The middle part of SUUR maintains charged regions as well as their net charge across all Drosophila species, despite the deletions, insertions, and high rate of charged amino acid substitutions. Ratios of Ka/Ks for this part of the protein are ∼0.5 for species from the melanogaster subgroup (Table S1), indicating that even this part of the protein apparently is under mild negative selection. A possible HP1-interacting motif has been described within the middle part of SUUR (Pindyurin et al. 2008), but it shows no conservation beyond the melanogaster subgroup.
The C terminus of SUUR is moderately conserved, but no characterized protein domains were identified in this region. Comparison of two C-terminal fragments, SUUR495–962 and SUUR669–962, further supports our conclusion that the middle part of SUUR (specifically, the positively charged region) is indispensable for the specific chromosome binding and suppression of underreplication (Figure 3). Expression of SUUR669–962 does not induce underreplication, although we speculate that ectopic fibers formed along the chromosome arms upon early overexpression of SUUR669–962 might result from very weak nonspecific underreplication. However, substitutions of the conserved aromatic residues 816 and 817 within its C terminus do not disrupt the ability of SUUR to induce underreplication.
Thus, even though SUUR falls into a class of fast-evolving genes, the protein has some highly conserved regions and maintains its domain structure in drosophilids. Substitution of two conserved amino acids or a truncation of an N-terminal half of the protein could modify the overexpression phenotypes equally well. Presence of an uORF in the SuUR gene that is far more conserved than the SUUR protein opens exciting possibilities for studying gene regulation.
Acknowledgments
The authors thank Andrey Gorchakov for critical reading of the manuscript and stimulating discussion and Vincenzo Pirrotta for providing pHA plasmid and the antibodies. This work was supported by the following Russian Foundation for Basic Research (RFBR) grants: RFBR 08-04-00521-a and RFBR 08-04-01105-a.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.104844/DC1.
References
- Belyaeva, E. S., I. F. Zhimulev, E. I. Volkova, A. A. Alekseyenko, Y. M. Moshkin et al., 1998. Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc. Natl. Acad. Sci. USA 95: 7532–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyakin, S. N., G. K. Christophides, A. A. Alekseyenko, E. V. Kriventseva, E. S. Belyaeva et al., 2005. Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc. Natl. Acad. Sci. USA 102: 8269–8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney, R., D. D. Dubey and J. A. Huberman, 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484. [DOI] [PubMed] [Google Scholar]
- Beye, M., M. Hasselmann, M. K. Fondrk, R. E. Page and S. W. Omholt, 2003. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114: 419–429. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brendel, V., P. Bucher, I. R. Nourbakhsh, B. E. Blaisdell and S. Karlin, 1992. Methods and algorithms for statistical analysis of protein sequences. Proc. Natl. Acad. Sci.USA 89: 2002–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C., and S. Karlin, 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Cherbas, L., X. Hu, I. Zhimulev, E. Belyaeva and P. Cherbas, 2003. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development 130: 271–284. [DOI] [PubMed] [Google Scholar]
- Cho, S., Z. Y. Huang, D. R. Green, D. R. Smith and J. Zhang, 2006. Evolution of the complementary sex-determination gene of honey bees: balancing selection and trans-species polymorphisms. Genome Res. 16: 1366–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J. M., 1999. K-estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15: 763–764. [DOI] [PubMed] [Google Scholar]
- Donaldson, A. D., 2005. Shaping time: chromatin structure and the DNA replication programme. Trends Genet. 21: 444–449. [DOI] [PubMed] [Google Scholar]
- Drysdale, R. A., M. A. Crosby, W. Gelbart, K. Campbell, D. Emmert et al., 2005. FlyBase: genes and gene models. Nucleic Acids Res. 33: D390–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall, J. G., E. H. Cohen and M. L. Polan, 1971. Reptitive DNA sequences in Drosophila. Chromosoma 33: 319–344. [DOI] [PubMed] [Google Scholar]
- Havas, K., I. Whitehouse and T. Owen-Hughes, 2001. ATP-dependent chromatin remodeling activities. Cell. Mol. Life Sci. 58: 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden, C. A., and G. Bosco, 2008. Comparative genomic analysis of novel conserved peptide upstream open reading frames in Drosophila melanogaster and other dipteran species. BMC Genomics 9: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquet, Y., M. Delattre, A. Spierer and P. Spierer, 2002. Functional dissection of the Drosophila modifier of variegation Su(var)3–7. Development 129: 3975–3982. [DOI] [PubMed] [Google Scholar]
- Kent, W. J., 2002. BLAT: the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova, T. D., I. V. Makunin, E. I. Volkova, V. Pirrotta, E. S. Belyaeva et al., 2005. Functional dissection of the Suppressor of UnderReplication protein of Drosophila melanogaster: identification of domains influencing chromosome binding and DNA replication. Genetica 124: 187–200. [DOI] [PubMed] [Google Scholar]
- Kumar, S., J. Dudley, M. Nei and K. Tamura, 2008. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinformatics 9: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugel, V., C. Dalloz, A. Stary, V. Cormier-Daire, I. Desguerre et al., 2008. Deletion of 5′ sequences of the CSB gene provides insight into the pathophysiology of Cockayne syndrome. Eur. J. Hum. Genet. 16: 320–327. [DOI] [PubMed] [Google Scholar]
- Lilly, M. A., and A. C. Spradling, 1996. The Drosophila endocycle is controlled by cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10: 2514–2526. [DOI] [PubMed] [Google Scholar]
- MacAlpine, D. M., H. K. Rodriguez and S. P. Bell, 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 18: 3094–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makunin, I. V., E. I. Volkova, E. S. Belyaeva, E. N. Nabirochkina, V. Pirrotta et al., 2002. The Drosophila Suppressor of Underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics 160: 1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallery, D. L., B. Tanganelli, S. Colella, H. Steingrimsdottir, A. J. van Gool et al., 1998. Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am. J. Hum. Genet. 62: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manseau, L., A. Baradaran, D. Brower, A. Budhu, F. Elefant et al., 1997. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev. Dyn. 209: 310–322. [DOI] [PubMed] [Google Scholar]
- Moshkin, Y. M., A. A. Alekseyenko, V. F. Semeshin, A. Spierer, P. Spierer et al., 2001. The bithorax complex of Drosophila melanogaster: underreplication and morphology in polytene chromosomes. Proc. Natl. Acad. Sci. USA 98: 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil, M. T., and J. M. Belote, 1992. Interspecific comparison of the transformer gene of Drosophila reveals an unusually high degree of evolutionary divergence. Genetics 131: 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pindyurin, A. V., C. Moorman, E. de Wit, S. N. Belyakin, E. S. Belyaeva et al., 2007. SUUR joins separate subsets of PcG, HP1 and B-type lamin targets in Drosophila. J. Cell Sci. 120: 2344–2351. [DOI] [PubMed] [Google Scholar]
- Pindyurin, A. V., L. V. Boldyreva, V. V. Shloma, T. D. Kolesnikova, G. V. Pokholkova et al., 2008. Interaction between the Drosophila heterochromatin proteins SUUR and HP1. J. Cell Sci. 121: 1693–1703. [DOI] [PubMed] [Google Scholar]
- Poux, S., D. McCabe and V. Pirrotta, 2001. Recruitment of components of Polycomb group chromatin complexes in Drosophila. Development 128: 75–85. [DOI] [PubMed] [Google Scholar]
- Rost, B., G. Yachdav and J. Liu, 2004. The PredictProtein server. Nucleic Acids Res. 32: W321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. V. Russell, 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schmid, K. J., and D. Tautz, 1997. A screen for fast evolving genes from Drosophila. Proc. Natl. Acad. Sci. USA 94: 9746–9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler, D., D. Scalzo, C. Kooperberg, B. van Steensel, J. Delrow et al., 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet. 32: 438–442. [DOI] [PubMed] [Google Scholar]
- Smith, A. V., and T. L. Orr-Weaver, 1991. The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development 112: 997–1008. [DOI] [PubMed] [Google Scholar]
- Stark, A., M.F. Lin, P. Kheradpour, J.S. Pedersen, L. Parts et al., 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelstra, C., A. van Gool, J. de Wit, W. Vermeulen, D. Bootsma et al., 1992. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell 71: 939–953. [DOI] [PubMed] [Google Scholar]
- Volkova, E. I., A. A. Yurlova, T. D. Kolesnikova, I. V. Makunin and I. F. Zhimulev, 2003. Ectopic expression of the Suppressor of Underreplication gene inhibits endocycles but not the mitotic cell cycle in Drosophila melanogaster. Mol. Genet. Genomics 270: 387–393. [DOI] [PubMed] [Google Scholar]
- Walker, J. E., M. Saraste, M. J. Runswick and N. J. Gay, 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1: 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, E. J., O. Emanuelsson, D. Scalzo, T. Royce, S. Kosak et al., 2004. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc. Natl. Acad. Sci. USA 101: 17771–17776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz, A., U. Hinz, M. Engelbert and E. Knust, 1995. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell 82: 67–76. [DOI] [PubMed] [Google Scholar]
- Zhang, G., H. Wang, J. Shi, X. Wang, H. Zheng et al., 2007. Identification and characterization of insect-specific proteins by genome data analysis. BMC Genomics 8: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhimulev, I. F., 1998. Polytene chromosomes, heterochromatin, and position effect variegation. Adv. Genet. 37: 1–566. [DOI] [PubMed] [Google Scholar]
- Zhimulev, I. F., and E. S. Belyaeva, 2003. Intercalary heterochromatin and genetic silencing. BioEssays 25: 1040–1051. [DOI] [PubMed] [Google Scholar]
- Zhimulev, I. F., E. S. Belyaeva, I. V. Makunin, V. Pirrotta, E. I. Volkova et al., 2003. a Influence of the SuUR gene on intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma 111: 377–398. [DOI] [PubMed] [Google Scholar]
- Zhimulev, I. F., E. S. Belyaeva, I. V. Makunin, V. Pirrotta, V. F. Semeshin et al., 2003. b Intercalary heterochromatin in Drosophila melanogaster polytene chromosomes and the problem of genetic silencing. Genetica 117: 259–270. [DOI] [PubMed] [Google Scholar]
- Zhimulev, I. F., E. S. Belyaeva, V. F. Semeshin, V. V. Shloma, I. V. Makunin et al., 2003. c Overexpression of the SuUR gene induces reversible modifications at pericentric, telomeric and intercalary heterochromatin of Drosophila melanogaster polytene chromosomes. J. Cell Sci. 116: 169–176. [DOI] [PubMed] [Google Scholar]